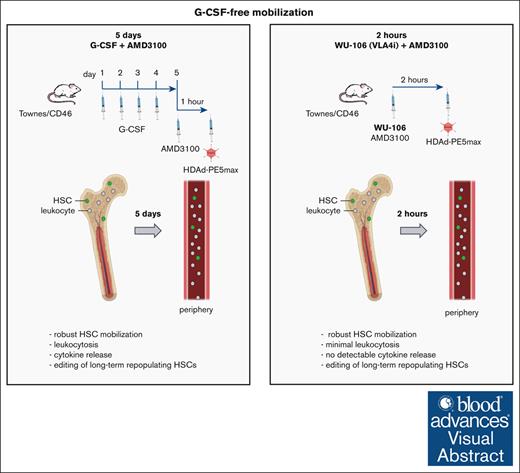
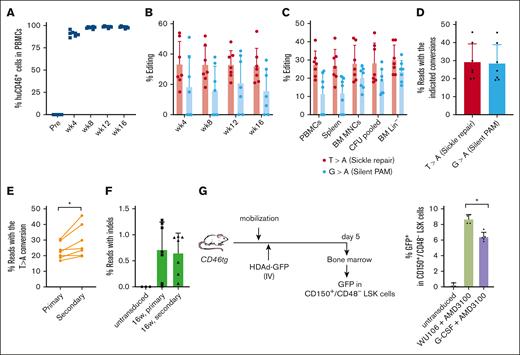
Key Points
Using WU-106/AMD3100 mobilization, in vivo transduction of long-term persisting HSCs in SCD mice can be done within 2 hours.
Compared with G-CSF/AMD3100 mobilization, the WU-106/AMD3100-based method is safer and as efficient for phenotypic correction of the disease in mice.
Visual Abstract
We have reported the direct repair of the sickle cell mutation in vivo in a disease model using vectorized prime editors after hematopoietic stem cell (HSC) mobilization with granulocyte colony-stimulating factor (G-CSF)/AMD3100. The use of G-CSF for HSC mobilization is a hurdle for the clinical translation of this approach. Here, we tested a G-CSF-free mobilization regimen using WU-106, an inhibitor of integrin α4β1, plus AMD3100 for in vivo HSC prime editing in sickle cell disease (SCD) mice. Mobilization with WU-106 + AMD3100 in SCD mice was rapid and efficient. In contrast to the G-CSF/AMD3100 approach, mobilization of activated granulocytes and elevation of the key proinflammatory cytokine interleukin-6 in the serum were minimal. The combination of WU-106 + AMD3100 mobilization and IV injection of the prime editing vector together with in vivo selection resulted in ∼23% correction of the SCD mutation in the bone marrow and peripheral blood cells of SCD mice. The treated mice demonstrated phenotypic correction, as reflected by normalized blood parameters and spleen size. Editing frequencies were significantly increased (29%) in secondary recipients, indicating the preferential mobilization/transduction of long-term repopulating HSCs. Using this approach, we found <1% undesired insertions/deletions and no detectable off-target editing at the top-scored potential sites. Our study shows that in vivo transduction to treat SCD can now be done within 2 hours involving only simple IV injections with a good safety profile. The same-day mobilization regimen makes in vivo HSC gene therapy more attractive for resource-poor settings, where SCD does the most damage.
Introduction
We have developed an in vivo approach for hematopoietic stem cell (HSC) transduction with helper-dependent adenovirus (HDAd) vectors that use natural receptors highly expressed on primitive HSCs, such as CD46 and desmoglein 2.1-3 In vivo HSC transduction with HDAd vectors requires mobilization of HSCs from the bone marrow (BM). These vectors are then injected IV while mobilized HSCs circulate in the peripheral blood. We have demonstrated in mice and rhesus macaques that a large fraction of HSCs transduced in the periphery returns to the BM and persists there long-term.2,4 Without mobilization, HSC transduction in the BM is only marginal.4 It is likely that extracellular matrix proteins and a variety of other cell types limit the access of systemically applied HDAd vectors to HSCs through physical barriers for vector diffusion, target receptor masking, vector phagocytosis, and other factors. Drugs that are used for HSC mobilization change the BM microenvironment and alter HSC adherence to the BM stroma, resulting in the egress of primitive HSCs into the bloodstream at high numbers.
We recently reported efficient correction of the sickle cell disease (SCD) mutation in the CD46/Townes mouse model of SCD after in vivo HSC prime editing with a CD46-targeted and chimeric helper-dependent adenovirus vector referred to as HDAd5/35++.5 In this study, we used a mobilization regimen consisting of 4 daily doses of subcutaneous (SC) granulocyte colony-stimulating factor (G-CSF) injection, followed by SC injection of AMD3100/plerixafor on day 5. This regimen robustly mobilizes HSCs from the BM to the periphery and is therefore, widely used for harvesting HSCs for transplantation purposes.6,7 However, the use of G-CSF as a mobilizing agent has a number of disadvantages. It requires multiple doses, is known to alter the function of the HSC niche as well as bone formation and mobilizes variable numbers of HSCs. A critical problem associated with G-CSF is the unselective mobilization of committed cells, leading to the sequestration of vectors and high peripheral cytokine levels released from granulocytes that interact with HDAd. Leukocytosis and the release of proinflammatory cytokines from granulocytes would be particularly critical in patients with SCD.8 G-CSF can trigger vaso-occlusive crises and even death in both adults and adolescents with SCD. The search for a G-CSF–free mobilization regimen is therefore crucial for in vivo HSC gene therapy for SCD. A number of HSC cell surface molecules could be targeted to modify interactions between HSCs and BM stroma including C-X-C chemokine receptor type 4 (CXCR4), CXCR2, very late antigen 4 (VLA-4), cluster of differentiation 44 (CD44), L-selectin, lymphocyte function-associated antigen-1, CD117 (c-kit), and Robo4.9
Recently, we tested truncated GRO-β (growth-regulated oncogene-β), a CXCR2 agonist, in combination with AMD3100, a CXCR4 inhibitor. We demonstrated that it mediates efficient mobilization and in vivo transduction of HSCs with β-thalassemia correction in mice using an integrated HDAd vector system.10
Here, we focused on a G-CSF–free mobilization approach that targets VLA-4 in combination with AMD3100. VLA-4 (CD49d/CD29, integrin α4β1) and its ligand vascular cell adhesion molecule 1 (VCAM-1) mediate cell-cell adhesion. The observation that HSCs are rapidly released (within an hour) into the peripheral circulation after targeted disruption of the VCAM-1/VLA-4 axis indicates that this interaction provides an alternative pathway for HSC mobilization that is independent of the CXCR4/CXCL12 axis.11 The J.F.D. Laboratory developed and optimized novel polyethylene glycol–conjugated inhibitors of VLA-4 for HSC mobilization. A lead VLA-4 inhibitor among them is WU-106.12,13 By simultaneous administration with plerixafor, WU-106/plerixafor resulted in rapid and reversible mobilization of HSCs into the peripheral circulation.11
We performed an in vivo HSC prime editing study in SCD mice using WU-106 + AMD3100, which allows HSC mobilization/in vivo transduction within 2 hours. We demonstrated the therapeutic efficacy and better safety of the rapid mobilization regimen compared with the multiday G-SCF/AMD3100 approach.
Materials and methods
Reagents for in vivo transduction and selection
HDAd vector
The HDAd-PE5max vector has been described previously.5 New stocks were produced by amplifying from the initial stock using a short-shafted Ad5/35S++ helper virus. HDAd-GFP contains a GFP (green fluorescent protein) gene under the control of a human EF1α (elongation factor-1 alpha) promoter.4 Helper virus contamination levels were found to be <0.05%. Titers were 2 × 1012 to 4 × 1012 viral particles (vp) per mL.
Animal studies
All experiments involving animals were conducted in accordance with the institutional guidelines set forth by the University of Washington. The University of Washington receives accreditation from the Association for the Assessment and Accreditation of Laboratory Animal Care International and all live animal work conducted at the university is in accordance with the Office of Laboratory Animal Welfare Public Health Assurance policy, United States Department of Agriculture (USDA) Animal Welfare Act and Regulations, the Guide for the Care and Use of Laboratory Animals and the University of Washington Institutional Animal Care and Use Committee policies. The study was approved by the University of Washington, Institutional Animal Care and Use Committee (protocol number 3108-01). C57BL/6-based transgenic mice that contained the human CD46 genomic locus and provided CD46 expression at a level and in a pattern similar to that in humans (hCD46+/+ mice) have been described earlier.14
SCD mouse model
A Townes male mouse (HBBtm2(HBG1,HBB∗)Tow or hα/hα::βS/βS) was purchased from Jackson Laboratory (JAX stock no. 013071) and bred with human CD46 transgenic female mice. After 3 rounds of breeding, mice homozygous for CD46, HBBS, and HBA were obtained and used in the experiments. The following primers were used for genotyping: (1) Hemoglobin subunit beta (HBB) primers: 5’-ATGTCAGAAGCAAATGTGAGGAGCA-3’, 5’-AATTCTGGCTTATCGGAGGCAAG-3’, and 5’-TTGAGCAATGTGGACAGAGAAGG-3’; (2) Hemoglobin subunit alpha (HBA) primers: 5’-TCCTGCAGGGTGAGGAAGGAAGG-3’, 5’- TCTATGCACATCAATTAGCAGAGGC-3’, and 5’-CCCCAAGGCACTCCAGGGACATAG-3’; and (3) CD46 primers: 5’-GCCAGTTCATCTTTTGACTCTATTAA-3’, and 5’-AATCACAGCAATGACCCAAA-3’.
HSC mobilization and in vivo transduction
G-CSF/AMD3100
HSCs were mobilized in mice by SC injections of human recombinant G-CSF (250 μg/kg per mouse per day, 4 days), followed by an SC injection of AMD3100 (5 mg/kg) on day 5. In addition, animals received dexamethasone (10 mg/kg, intraperitoneal [IP]) 16 and 2 hours before virus injection to blunt the innate toxicity associated with IV HDAd injection. Forty-five minutes after AMD3100, animals were IV injected with virus vectors through the retro-orbital plexus (4 × 1010 vp per mouse).
WU-106/AMD3100
WU-106 (10 mg/kg, SC, in saline) was given together with AMD3100 (5 mg/kg, SC), 2 hours before the first HDAd injection. As described above, dexamethasone (10 mg/kg) was injected IP 16 hours and 2 hours before virus injection.
In vivo selection
Selection was started on day 6 after transduction. Mice were injected with O6BG (15 mg/kg, IP) 2 times, 30 minutes apart. One hour after the second injection of O6BG, the mice were injected (IP) with 5 mg/kg BCNU. On days 19 and 33, 2 more rounds of selection were performed using BCNU doses of 9 and 10 mg/kg, respectively.
Secondary BM transplantation
BM cells from ex vivo- or in vivo-transduced CD46tg mice were isolated aseptically. Lin– cells were isolated and transplanted as described above. Secondary recipients were kept for 16 weeks after transplantation for terminal point analyses.
Statistical analyses
The Student's t test was used for comparisons between the 2 groups. For comparisons of multiple groups, 1-way and 2-way analysis of variance with Bonferroni posttest for multiple comparisons was used. Statistical analysis was performed using GraphPad Prism version 10.0.3 (GraphPad Software Inc, La Jolla, CA). The statistical significance was indicated as ∗∗∗P ≤ .001, ∗∗P ≤ .01, and ∗P ≤ .05.
The following methods are described in the Supplementary Information: colony-forming unit (CFU) assay, measurement of gene editing by Sanger sequencing and Next-generation sequencing (NGS), off-target analysis, flow cytometry, hemoglobin (Hb) electrophoresis by isoelectric focusing (IEF), CD62p enzyme-linked immunosorbent assay, tissue analysis, blood analyses, and data availability.
Results
The goal of this study was to evaluate the safety and efficacy of a new mobilization regimen (WU-106 + AMD3100) in the context of in vivo HSC gene therapy for SCD in an adequate mouse model (Townes mice). In this mouse model, murine α-globin genes were replaced with human α-globin and murine adult β-globin genes were replaced with human sickle βS and fetal γ-globin genes linked together. To make the Townes model suitable for HDAd5/35++ transduction, which requires human CD46 as a receptor, we bred Townes mice with human CD46 transgenic mice. After 3 rounds of backcrossing, mice homozygous for human CD46 and 2 human (α, βS/γ) globin genes were used for the experiments. Triple homozygous CD46/Townes mice displayed sickle-like erythrocytes, severe anemia, ∼40% reticulocytes in peripheral blood, leukocytosis, and thrombocytosis.15 CD46/Townes mice were fragile and to make them stronger we fed them a protein/fat/vitamin-rich diet (Sickle diet, 59M3) (supplemental Figure 1). The effect of the sickle diet was visible as ∼20% body weight gain within 1 month of initiation. Long-term body weights of animals (aged 10-12 weeks) on diet were comparable with those of control CD46-transgenic mice. Although the mortality rate of triple homozygous mice before initiation of the diet was >50%, only ∼25% of mice on the 59M3-diet died over an 8-month period (N > 30).
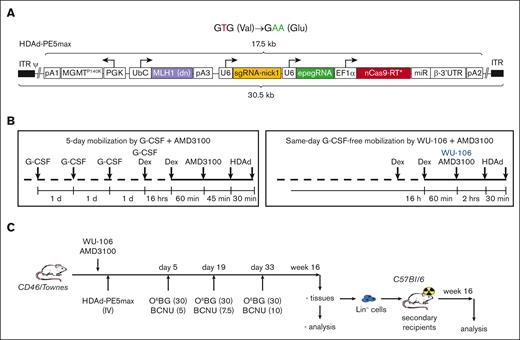
The HDAd5/35++–based gene therapy vector (HDAd-PE5max) contains prime editing elements that allow correction of the SCD mutation.5 In addition to fixing the sickle mutation (T > A conversion), a G > A silent mutation was introduced to prevent the prime editor from continuous editing by destroying the protospacer-adjacent motif (PAM) (Figure 1A, top panel). HDAd-PE5max also carries an expression cassette for the O⁶-alkylguanine DNA alkyltransferase mutant MGMTP140K, which allows for in vivo selection of transduced/edited HSCs by low-dose chemotherapy drugs (O6BG/BCNU) (Figure 1A, bottom panel). The length of all expression cassettes adds up to 17.5 kilobase, a cargo size that can be readily accommodated in HDAd5/35++ vectors with an insert capacity of 35 kilobase.
HDAd-PE5max vector and schematic of the experiment. (A) Top panel: expected prime editing of SCD mutation. In addition to fixing the sickle mutation (T > A conversion), a G > A silent mutation is introduced to prevent the prime editor from continuous editing by destroying the PAM. New nucleotides are shown as green letters. Bottom panel: the HDAd-PE5max vector contains engineered prime editor guide RNA (epegRNA), codon–optimized editing enzyme, dominant negative MLH1 gene, and MGMTP140K cassette for in vivo selection. EF1α, elongation factor 1α promoter; miR RNA/β-3’UTR, miR-183-5p and miR-218-5p target sites embedded into β-globin to suppress nCas9-RT expression in HDAd producer cells; pA, polyadenylation signals (pA1, BGH pA; pA2, SV40 pA; pA3, rabbit β-globin pA); U6, U6 RNA polymerase III promoter; UbC, human ubiquitin C promoter. (B) Mobilization regimens. Left panel: G-CSF + AMD3100 regimen. G-CSF (250 μg/kg) was given SC for 4 days, followed by 1 SC injection of AMD3100 (5 mg/kg) on the 5th day. Dexamethasone (10 mg/kg) was injected IP 16 hours and 1.75 hours before the first HDAd dosing to blunt cytokine responses. HDAd-PE5max was injected IV in 2 doses (each 4 × 1010 vp per animal) at 45 and 75 minutes after AMD3100. Right panel: WU-106 + AMD3100 regimen. WU-106 (10 mg/kg, SC) and AMD3100 (5 mg/kg, SC) were injected together. HDAd was administered IV 2 and 2.5 hours after WU-106 + AMD3100 (in 2 doses each 4 × 1010 vp per animal). Cytokine prophylaxis with dexamethasone was done 16 hours and 2.5 hours before the first HDAd injection. (C) Mobilized CD46/Townes mice were transduced with HDAd-PE5max via IV injection. Three rounds of selection were conducted on days 5, 19, and 33 after transduction by IP injection with the indicated doses of O6BG/BCNU. The primary mice were euthanized at week 16 after transduction. BM Lin– cells were isolated from primary mice and infused into lethally irradiated C57BL/6 mice. The secondary transplanted mice were followed for 16 weeks for terminal analyses.
HDAd-PE5max vector and schematic of the experiment. (A) Top panel: expected prime editing of SCD mutation. In addition to fixing the sickle mutation (T > A conversion), a G > A silent mutation is introduced to prevent the prime editor from continuous editing by destroying the PAM. New nucleotides are shown as green letters. Bottom panel: the HDAd-PE5max vector contains engineered prime editor guide RNA (epegRNA), codon–optimized editing enzyme, dominant negative MLH1 gene, and MGMTP140K cassette for in vivo selection. EF1α, elongation factor 1α promoter; miR RNA/β-3’UTR, miR-183-5p and miR-218-5p target sites embedded into β-globin to suppress nCas9-RT expression in HDAd producer cells; pA, polyadenylation signals (pA1, BGH pA; pA2, SV40 pA; pA3, rabbit β-globin pA); U6, U6 RNA polymerase III promoter; UbC, human ubiquitin C promoter. (B) Mobilization regimens. Left panel: G-CSF + AMD3100 regimen. G-CSF (250 μg/kg) was given SC for 4 days, followed by 1 SC injection of AMD3100 (5 mg/kg) on the 5th day. Dexamethasone (10 mg/kg) was injected IP 16 hours and 1.75 hours before the first HDAd dosing to blunt cytokine responses. HDAd-PE5max was injected IV in 2 doses (each 4 × 1010 vp per animal) at 45 and 75 minutes after AMD3100. Right panel: WU-106 + AMD3100 regimen. WU-106 (10 mg/kg, SC) and AMD3100 (5 mg/kg, SC) were injected together. HDAd was administered IV 2 and 2.5 hours after WU-106 + AMD3100 (in 2 doses each 4 × 1010 vp per animal). Cytokine prophylaxis with dexamethasone was done 16 hours and 2.5 hours before the first HDAd injection. (C) Mobilized CD46/Townes mice were transduced with HDAd-PE5max via IV injection. Three rounds of selection were conducted on days 5, 19, and 33 after transduction by IP injection with the indicated doses of O6BG/BCNU. The primary mice were euthanized at week 16 after transduction. BM Lin– cells were isolated from primary mice and infused into lethally irradiated C57BL/6 mice. The secondary transplanted mice were followed for 16 weeks for terminal analyses.
We used 2 regimens for HSC mobilization in the CD46/Townes mice. The standard regimen that we used before5 consisted of 4 days of SC G-CSF injection, followed by an SC injection of AMD3100 on day 5 (Figure 1B, left panel). HDAd-PE5max was injected IV 45 and 75 minutes after AMD3100 (in 2 doses of each 4 × 1010 vp per mouse). Dexamethasone was given IP to blunt HDAd-induced cytokine release. In the second regimen, WU-106 and AMD3100 were given together SC followed by IV HDAd-PE5max injection 2 hours and 2.5 hours later (also in 2 doses of each 4 × 1010 vp per mouse) (Figure 1B, right panel). These time points were selected based on previous mobilization kinetics studies in DBA/2J mice, which showed the highest number of mobilized CFUs at 2 hours drug after injection.13 On days 5, 19, and 33, mice received IP O6BG/BCNU injections at the indicated doses (Figure 1C). Notably, selection during this time was based on the transient presence of the episomal HDAd-PE5max vector in HSCs. At week 16, in vivo transduced mice were euthanized for BM/spleen analyses, and BM lineage-negative (Lin–) cells were transplanted into secondary recipients, which were followed for another 16 weeks to assess whether editing occurred in long-term repopulating HSCs (LT-HSCs).
Mobilization efficacy and safety
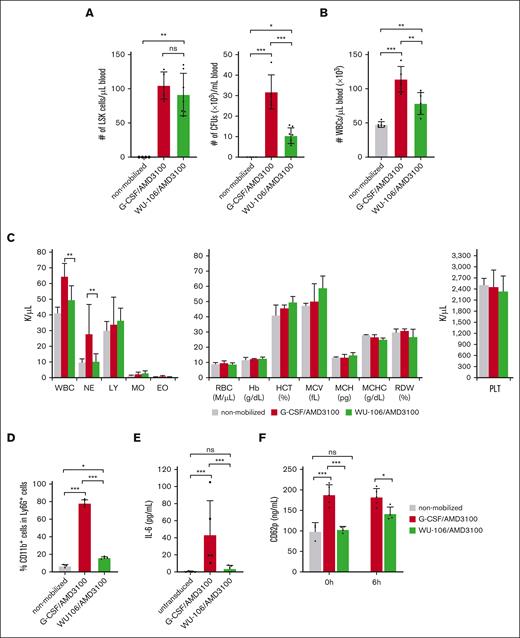
HSC mobilization was analyzed at the time of the first HDAd injection (ie, 60 minutes after AMD3100 in the G-CSF + AMD3100 regimen and 2 hours after WU-106 + AMD3100 injection) based on the number of Lin–/Sca1+/cKit+ (LSK) cells and HSPC-derived CFUs in the peripheral blood (Figure 2A, left and right panels, respectively). Although the number of LSK cells was comparable for both mobilization regimes, the number of CFUs was significantly less in WU-106/AMD3100–mobilized CD46/Townes mice. Notably, both LSK cells and CFUs contained subfractions of cells with different levels of stemness/pluripotency.
Mobilization efficacy and safety. (A) Left panel: number of LSK cells perμl blood at 2 hours after WU-106 + AMD3100 injection or 45 minutes after AMD3100 injection in the G-CSF + AMD3100 regimen (before the HDAd injection). LSK cells were measured by flow cytometry. Right panel: CFU were measured by methylcellulose-based colony assay with blood samples drawn at the above-listed time points. PBMCs from 5 μL mobilized peripheral blood were plated for the CFU assay. Total colonies were counted 11 days later. Each dot represents 1 mouse. (B) White blood cells (WBCs) in blood measured by hemocytometer after lysing RBCs. (C) Blood cell analysis. Hematological parameters measured by Hemavet. The platelet (PLT) counts are shown in the right panel. (D) Percentage of activated (CD11b+/Ly6G+) neutrophils at the peak of mobilization. (E) Serum cytokine levels 6 hours after HDAd injection were measured using a cytometric bead array. IL-6 levels are shown. (F) Serum CD62p (P-selectin) concentrations measured by enzyme-linked immunosorbent assay at the pak of mobilization (right before HDAd injection) “0 hours” and 6 hours after HDAd injection (“6 hours”). Studies in panels A-D were carried out in mice without HDAd injection at the peak of mobilization. Serum cytokine levels and platelets (E-F) were measured 6 hours after HDAd injection. For comparisons of multiple groups, a 2-way analysis of variance (ANOVA) with Bonferroni posttest for multiple comparisons was used. Statistical analysis was performed using GraphPad Prism version 10.0.3. ∗∗∗P ≤ .001, ∗∗P ≤ .01, ∗P ≤ .05. EO, eosinophils; Hb, hemoglobin; HCT, hematocrit; LY, lymphocytes; MCH, mean corpuscular Hb; MCHC, MCH concentration; MCV, mean corpuscular volume; MO, monocytes; NE, neutrophils.
Mobilization efficacy and safety. (A) Left panel: number of LSK cells perμl blood at 2 hours after WU-106 + AMD3100 injection or 45 minutes after AMD3100 injection in the G-CSF + AMD3100 regimen (before the HDAd injection). LSK cells were measured by flow cytometry. Right panel: CFU were measured by methylcellulose-based colony assay with blood samples drawn at the above-listed time points. PBMCs from 5 μL mobilized peripheral blood were plated for the CFU assay. Total colonies were counted 11 days later. Each dot represents 1 mouse. (B) White blood cells (WBCs) in blood measured by hemocytometer after lysing RBCs. (C) Blood cell analysis. Hematological parameters measured by Hemavet. The platelet (PLT) counts are shown in the right panel. (D) Percentage of activated (CD11b+/Ly6G+) neutrophils at the peak of mobilization. (E) Serum cytokine levels 6 hours after HDAd injection were measured using a cytometric bead array. IL-6 levels are shown. (F) Serum CD62p (P-selectin) concentrations measured by enzyme-linked immunosorbent assay at the pak of mobilization (right before HDAd injection) “0 hours” and 6 hours after HDAd injection (“6 hours”). Studies in panels A-D were carried out in mice without HDAd injection at the peak of mobilization. Serum cytokine levels and platelets (E-F) were measured 6 hours after HDAd injection. For comparisons of multiple groups, a 2-way analysis of variance (ANOVA) with Bonferroni posttest for multiple comparisons was used. Statistical analysis was performed using GraphPad Prism version 10.0.3. ∗∗∗P ≤ .001, ∗∗P ≤ .01, ∗P ≤ .05. EO, eosinophils; Hb, hemoglobin; HCT, hematocrit; LY, lymphocytes; MCH, mean corpuscular Hb; MCHC, MCH concentration; MCV, mean corpuscular volume; MO, monocytes; NE, neutrophils.
Mobilization with WU-106 + AMD3100 resulted in significantly less leukocytosis, as reflected by the number of white blood cells (counted on a hemocytometer) (Figure 2B). Hematological analyses using Hemavet showed that lower white blood cells were mostly due to less mobilization of neutrophils by WU-106/AMD3100 (Figure 2C, left panel). Differences in erythroid parameters and platelet numbers were not significant between the nonmobilized, G-CSF + AMD3100- and WU-106 + AMD3100-mobilized animals (Figure 2C, right panel). Activation of neutrophils was measured by flow cytometry of CD11b on Ly6G+ cells at the corresponding peak of mobilization (1 hour after AMD3100 for G-CSF + AMD3100 and 2 hours after AMD3100 + WU-106) (Figure 2D). Although ∼75% of neutrophils were CD11b+ after G-CSF + AMD3100, the average percentage of activated neutrophils after WU-106 + AMD3100 was 16%. The percentage of CD11b+/Ly6G+ cells in nonmobilized mice was ∼6.5%. Increased numbers of activated granulocytes/neutrophils and/or their activation can trigger the release of proinflammatory cytokines after contact/uptake of HDAd particles.16 Therefore, we measured the serum concentrations of proinflammatory cytokines 6 hours of injection of HDAd vectors using a cytometric bead array. We found increased levels of the key cytokine IL-6 in the peripheral blood of G-CSF + AMD3100 mobilized animals, but not in WU-106 + AMD3100-mobilized animals (Figure 2E). Other cytokines, including tumor necrosis factor, IL-10, IL-12p70, interferon-γ, and membrane cofactor protein-1, were not detectable by the assay. Another read-out for HDAd–associated innate toxicity is platelet activation measured by serum CD62p (P-selectin) enzyme-linked immunosorbent assay. At the peak of mobilization (before HDAd injection), CD62p levels were significantly higher for the G-CSF + AMD3100 group, whereas the level in mice mobilization with WU-106 was comparable to nonmobilized mice (Figure 2F, “0 hour”). This indicates that G-CSF administration leads to platelet activation, a phenomenon reported also by others.17 As expected, HDAd injection resulted in the interaction with and activation of platelets. However, serum CD62p levels 6 hours after HDAd were significantly lower in the WU-106 + AMD3100 group (Figure 2F, “6 hour”).
In summary, WU-106/AMD3100 treatment resulted in efficient HSC mobilization. There was less undesired mobilization of activated neutrophils, which in turn reduced the levels of the proinflammatory cytokine IL-6 after HDAd injection. Furthermore, platelet activation by IV-injected HDAd was less pronounced in the WU-106/AMD3100 mobilization regimen.
Editing and hemoglobin profiles in primary mice
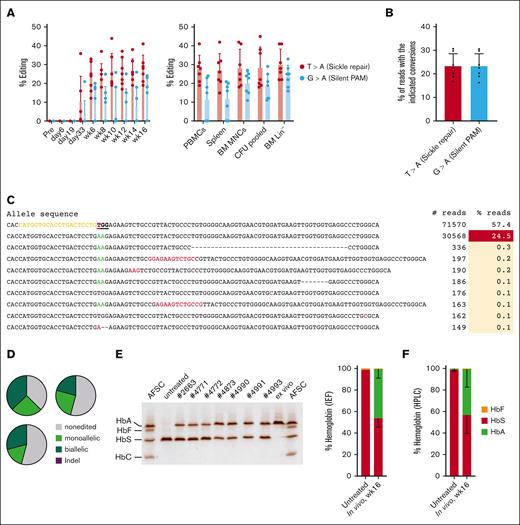
Next, we analyzed target site editing in peripheral blood mononuclear cells (PBMCs) of WU-106/AMD3100 mobilized HDAd-PE5max–injected CD46/Townes mice (“primary mice”) (Figure 3A, left panel). Editing data after G-CSF + AMD3100 mobilization have been published previously.5 The percentages of T > A (ie, the repair of the SCD mutation) and G > A editing (to introduce a silent PAM mutation) were measured. SCD mutation editing became detectable by Sanger sequencing before the last round of O6BG/BCNU treatment (day 33) and, after selection, reached levels of ∼25% to 30% that remained stable until the end of the observation period. At week 16, target site editing was found in ∼30% of alleles in DNA from PBMCs, spleen, BM mononuclear cells (BM MNCs), BM Lin– cells, and progenitor colonies (formed from Lin– cells) (Figure 3A, right panel). Editing levels were confirmed by NGS of amplicons derived from BM MNCs. NGS data showed editing of ∼23% for both the sickle repair and silent PAM sites (Figure 3B). This is comparable with frequencies measured previously in G-CSF + AMD3100 mobilized animals that were treated in the same way.5
Target-site correction and sickle cell hemoglobin (HbS) to normal adult hemoglobin (HbA) conversion in HDAd-PE5max transduced mice after WU-106 + AMD3100 mobilization. (A) Target-site editing measured using Sanger sequencing. Editing at the sickle mutation site (T > A) and silent PAM site (G > A) are shown. Left panel: editing in PBMCs. Right panel: editing measured at week 16 after in vivo transduction in PBMCs, spleen, BM MNCs, pooled CFUs, and BM Lin–. Each dot represents an individual animal. (B) Target base conversions in BM MNCs (week 16), as measured by NGS. Each dot represents an individual animal. (C) Top alleles with frequencies >0.1% in mouse #4985, measured by NGS. (D) Allelic analysis of single Lin– cell–derived progenitor colonies. Editing was measured at day 11 after plating transduced Lin– cells. Shown are data from 3 mice with the indicated ear tag number. Twenty-four colonies from each mouse were analyzed. (E-F) Analyses of Hb composition. Whole blood samples at week 16 after in vivo transduction were analyzed. Samples from untreated CD46/Townes mice were used as a control. (E) Separation of Hb variants by IEF electrophoresis. Each lane represents 1 mouse (ear tag number labeled) or Hb AFSC controls. Bands in the AFSC controls, indicating 4 different Hb variants, are labeled. “ex vivo” indicates a control sample with near complete HbS to HbA conversion published previously.5 The sample was collected from a mouse transplanted with CD46/Townes Lin– cells ex vivo and transduced with HDAd-PE5max. The intensity of the bands was analyzed using ImageJ software and is summarized by the bar graph on the right. (F) Percentages of Hb variants measured by high-pressure liquid chromatography. For comparisons of multiple groups, 1-way and 2-way ANOVA with Bonferroni posttest for multiple comparisons was used. Statistical analysis was performed using GraphPad Prism version 10.0.3. ∗∗∗P ≤ .001, ∗∗P ≤ .01, ∗P ≤ .05.
Target-site correction and sickle cell hemoglobin (HbS) to normal adult hemoglobin (HbA) conversion in HDAd-PE5max transduced mice after WU-106 + AMD3100 mobilization. (A) Target-site editing measured using Sanger sequencing. Editing at the sickle mutation site (T > A) and silent PAM site (G > A) are shown. Left panel: editing in PBMCs. Right panel: editing measured at week 16 after in vivo transduction in PBMCs, spleen, BM MNCs, pooled CFUs, and BM Lin–. Each dot represents an individual animal. (B) Target base conversions in BM MNCs (week 16), as measured by NGS. Each dot represents an individual animal. (C) Top alleles with frequencies >0.1% in mouse #4985, measured by NGS. (D) Allelic analysis of single Lin– cell–derived progenitor colonies. Editing was measured at day 11 after plating transduced Lin– cells. Shown are data from 3 mice with the indicated ear tag number. Twenty-four colonies from each mouse were analyzed. (E-F) Analyses of Hb composition. Whole blood samples at week 16 after in vivo transduction were analyzed. Samples from untreated CD46/Townes mice were used as a control. (E) Separation of Hb variants by IEF electrophoresis. Each lane represents 1 mouse (ear tag number labeled) or Hb AFSC controls. Bands in the AFSC controls, indicating 4 different Hb variants, are labeled. “ex vivo” indicates a control sample with near complete HbS to HbA conversion published previously.5 The sample was collected from a mouse transplanted with CD46/Townes Lin– cells ex vivo and transduced with HDAd-PE5max. The intensity of the bands was analyzed using ImageJ software and is summarized by the bar graph on the right. (F) Percentages of Hb variants measured by high-pressure liquid chromatography. For comparisons of multiple groups, 1-way and 2-way ANOVA with Bonferroni posttest for multiple comparisons was used. Statistical analysis was performed using GraphPad Prism version 10.0.3. ∗∗∗P ≤ .001, ∗∗P ≤ .01, ∗P ≤ .05.
The expected GTG > GAA conversion was observed in the overwhelming majority of edited alleles (Figure 3C). We analyzed the editing in colonies derived from single BM Lin– cells from 3 individual mice (Figure 3D). An average of 29% and 22% of colonies showed biallelic and monoallelic T > A corrections, respectively.
We measured human Hb variants in erythrocyte lysates of CD46/Townes mice using IEF and high-pressure liquid chromatography. IEF was developed by Linus Pauling in 1949 and showed that forms of sickle cell Hb (HbS) had higher isoelectric points than Hb from normal adult erythrocytes (HbA), suggesting that HbS is more positively charged than HbA and therefore, runs faster in IEF gels. Based on the IEF, 46.6% of HbS was converted to normal HbA by prime editing (Figure 3E). Similar results were obtained using high-pressure liquid chromatography (Figure 3F). Higher levels of correction at the protein level compared with genomic editing frequencies are likely resulted from the longer lifespans of corrected red blood cells (RBCs) over sickling erythrocytes.
Phenotypic correction of SCD in a mouse model
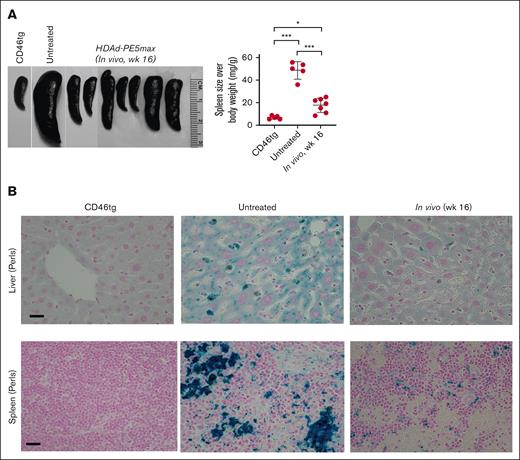
At week 16 after in vivo HSC transduction with the HDAd-PE5max vector, the phenotypic features of SCD were analyzed in CD46/Townes mice. In hematological analyses, untreated CD46/Townes mice showed high leukocytosis, abnormal platelet counts, low RBC count, and low hematocrit and Hb production compared with CD46tg healthy mice (Figure 4A). Importantly, CD46/Townes mice that were mobilized with WU-106/AMD3100 and injected with HDAd-PE5max exhibited normal hematological parameters at levels comparable to those of healthy controls. In an in vitro sickling assay with sodium metabisulfite, a reagent that reduces oxygen tension, ∼80% of RBCs showed a typical sickle shape in the blood smears of untreated CD46/Townes mice (Figure 4B, upper panels). In contrast, in treated animals <40% displayed sickle cell morphology. Improved erythropoiesis upon treatment was also visible after staining the blood smears for reticulocytes (Figure 4B-C, lower panels). Notably, WU-106/AMD3100 mobilization followed by in vivo transduction/selection did not alter body weight, number of BM MNCs and Lin– cells compared to untreated animals (Figure 4D-F). Splenomegaly resulting from extramedullary hemopoiesis, a characteristic SCD symptom, was seen in untreated CD46/Townes mice (Figure 5A). In contrast, a marked reduction in spleen size was seen in prime-edited animals. Histological analyses of the liver and spleen from treated CD46/Townes mice showed largely regressed parenchymal iron deposition (Figure 5B). In summary, in vivo prime editing after in vivo transduction of WU-106/AMD3100-mobilized HSCs resulted in the therapeutically relevant conversion of HbS to normal HbA and a greatly ameliorated phenotype in CD46/Townes mice.
Phenotypic correction after WU-106/AMD3100 mobilization and in vivo HSC prime–editing blood analyses. (A) Complete blood cell count using week 16 samples after in vivo transduction (n = 7). Blood samples from healthy CD46.tg mice (n = 3) and untreated CD46/Townes mice (n = 4) were used as the controls. For comparisons of multiple groups, 1-way and 2-way ANOVA with Bonferroni posttest for multiple comparisons was used. Statistical analysis was performed using GraphPad Prism version 10.0.3. ∗∗∗P ≤ .001, ∗∗P ≤ .01, ∗P ≤ .05. (B) Representative microphotographs of the blood cell smears from week 16 samples. First panel: smears of total blood cells subjected to a sickling assay; second panel: blood cell smears stained with Giemsa; third panel: staining of blood smears for reticulocytes with brilliant cresyl blue, which stains nuclear remnants of basophilic ribonucleoproteins in reticulocytes (black arrow). The scale bars are 20 μm. (C) Upper panel: percentage of sickle cells in the blood smears. Each dot represents the percentage in an individual mouse. Lower panel: percentage of reticulocytes in blood smears. (D-F) Comparisons of body weight (D), number of BM MNCs (E), and number of Lin– cells in untreated and in vivo transduced animals. ns, not significant.
Phenotypic correction after WU-106/AMD3100 mobilization and in vivo HSC prime–editing blood analyses. (A) Complete blood cell count using week 16 samples after in vivo transduction (n = 7). Blood samples from healthy CD46.tg mice (n = 3) and untreated CD46/Townes mice (n = 4) were used as the controls. For comparisons of multiple groups, 1-way and 2-way ANOVA with Bonferroni posttest for multiple comparisons was used. Statistical analysis was performed using GraphPad Prism version 10.0.3. ∗∗∗P ≤ .001, ∗∗P ≤ .01, ∗P ≤ .05. (B) Representative microphotographs of the blood cell smears from week 16 samples. First panel: smears of total blood cells subjected to a sickling assay; second panel: blood cell smears stained with Giemsa; third panel: staining of blood smears for reticulocytes with brilliant cresyl blue, which stains nuclear remnants of basophilic ribonucleoproteins in reticulocytes (black arrow). The scale bars are 20 μm. (C) Upper panel: percentage of sickle cells in the blood smears. Each dot represents the percentage in an individual mouse. Lower panel: percentage of reticulocytes in blood smears. (D-F) Comparisons of body weight (D), number of BM MNCs (E), and number of Lin– cells in untreated and in vivo transduced animals. ns, not significant.
Phenotypic correction after WU-106/AMD3100 mobilization and in vivo HSC prime editing: spleen and liver analyses. (A) Spleen size (left panel) and spleen weight relative to the body weight (right panel). Each symbol represents an individual mouse. Two-way ANOVA with Bonferroni posttest for multiple comparisons was used. Statistical analysis was performed using GraphPad Prism version 10.0.3. ∗∗∗P ≤ .001, ∗∗P ≤ .01, ∗P ≤ .05. (B) Hemosiderosis: spleen and liver sections stained with Perl Prussian blue. Iron deposition is shown as the cytoplasmic blue pigments of hemosiderin in the spleen tissue sections. The scale bars are 20μm.
Phenotypic correction after WU-106/AMD3100 mobilization and in vivo HSC prime editing: spleen and liver analyses. (A) Spleen size (left panel) and spleen weight relative to the body weight (right panel). Each symbol represents an individual mouse. Two-way ANOVA with Bonferroni posttest for multiple comparisons was used. Statistical analysis was performed using GraphPad Prism version 10.0.3. ∗∗∗P ≤ .001, ∗∗P ≤ .01, ∗P ≤ .05. (B) Hemosiderosis: spleen and liver sections stained with Perl Prussian blue. Iron deposition is shown as the cytoplasmic blue pigments of hemosiderin in the spleen tissue sections. The scale bars are 20μm.
Analyses of secondary recipients
To demonstrate mobilization of long-term repopulating HSCs by WU-106/AMD3100 and subsequent in vivo editing of these cells, we transplanted BM Lin– cells harvested at week 16 after in vivo HSC transduction of CD46/Townes mice into lethally irradiated C57Bl/6 mice. The ability of the transplanted cells to drive multilineage reconstitution in secondary recipients was assessed over a period of 16 weeks. Engraftment levels based on human CD46 expression in PBMCs remained stable at ∼95% (Figure 6A). Target site editing measured by Sanger sequencing was stable for 16 weeks at a level of ∼30% in PBMCs, the spleen, BM MNCs, Lin– cells, and pooled CFU cells (Figure 6B-C). Editing levels and purity were analyzed by NGS of week 16 BM MNCs from secondary recipients (Figure 6D-F). On average, 29.1% of the alleles were edited with the expected T > A conversion (Figure 6D). The frequency of insertions/deletions (indels) was <1% in samples from primary and secondary recipients (Figure 6F). Notably, the target-site editing was significantly higher than that in primary mice (P = .028), suggesting preferential mobilization/editing of long-term repopulating stem cells (Figure 6E).
Analyses of secondary recipients. Lin− cells from in vivo transduced mice were transplanted into lethally irradiated C57Bl/6 mice (N = 7). The cells from 1 donor were injected into 1 recipient. (A) Engraftment based on human CD46 expression in PBMCs measured by flow cytometry. (B) Editing of target sites in PBMCs at the indicated weeks after transplantation measured by Sanger sequencing. (C) Editing measured by Sanger sequencing at week 16 after transplantation of PBMCs, splenocytes, BM MNCs, pooled CFU cells, and BM Lin− cells. (D) Target base conversions T > A (sickle repair) and G > A (silent PAM) measured by NGS in week 16 BM MNCs. (E) Comparison of correction levels between the primary mice and secondary recipients. Individual primary (donor) and secondary (recipient) mice were matched using a paired Student's t test. ∗P ≤ .05. (F) Indel frequencies measured by NGS in week 16 BM MNCs of primary mice and secondary mice. (G) GFP expression in LT-HSCs was characterized by the following phenotype: CD150+/CD48–/LSK. Left panel: schematic of the experiment. CD46-transgenic mice were either mobilized with G-CCSF + AMD3100 or WU-106/AMD3100 as described in Figure 1. HDAd-GFP was injected IV 45 and 75 minutes after AMD3100 (in 2 doses of each 4 × 1010 vp per mouse) for G-CSF/AMD3100 mobilized mice, or 2 hours and 2.5 hours after WU-106 AMD310 (also in 2 doses of each 4 × 1010 vp per mouse). Five days later, the mice were euthanized and BM MNCs were analyzed by flow cytometry for GFP expression in LT-HSCs. Right panel: percentage of GFP+ LT-HSC cells. One-way ANOVA with Bonferroni posttest for multiple comparisons was used. ∗P ≤ .05.
Analyses of secondary recipients. Lin− cells from in vivo transduced mice were transplanted into lethally irradiated C57Bl/6 mice (N = 7). The cells from 1 donor were injected into 1 recipient. (A) Engraftment based on human CD46 expression in PBMCs measured by flow cytometry. (B) Editing of target sites in PBMCs at the indicated weeks after transplantation measured by Sanger sequencing. (C) Editing measured by Sanger sequencing at week 16 after transplantation of PBMCs, splenocytes, BM MNCs, pooled CFU cells, and BM Lin− cells. (D) Target base conversions T > A (sickle repair) and G > A (silent PAM) measured by NGS in week 16 BM MNCs. (E) Comparison of correction levels between the primary mice and secondary recipients. Individual primary (donor) and secondary (recipient) mice were matched using a paired Student's t test. ∗P ≤ .05. (F) Indel frequencies measured by NGS in week 16 BM MNCs of primary mice and secondary mice. (G) GFP expression in LT-HSCs was characterized by the following phenotype: CD150+/CD48–/LSK. Left panel: schematic of the experiment. CD46-transgenic mice were either mobilized with G-CCSF + AMD3100 or WU-106/AMD3100 as described in Figure 1. HDAd-GFP was injected IV 45 and 75 minutes after AMD3100 (in 2 doses of each 4 × 1010 vp per mouse) for G-CSF/AMD3100 mobilized mice, or 2 hours and 2.5 hours after WU-106 AMD310 (also in 2 doses of each 4 × 1010 vp per mouse). Five days later, the mice were euthanized and BM MNCs were analyzed by flow cytometry for GFP expression in LT-HSCs. Right panel: percentage of GFP+ LT-HSC cells. One-way ANOVA with Bonferroni posttest for multiple comparisons was used. ∗P ≤ .05.
To assess in vivo transduction of long-term repopulating HSCs, we mobilized CD46-transgenic mice with G-CSF + AMD3100 or WU-106/AMD3100, injected a GFP-expressing HDAd5/35++ vector IV at the peak of mobilization, and measured GFP expression in BM CD150+/CD48- LSK cells, a cell fraction enriched for LT-HSCs 18 (Figure 6G; supplemental Figure 2). We found significantly more GFP+ cells in this fraction in WU-106 + AMD3100 mobilized mice.
Off-target editing analyses
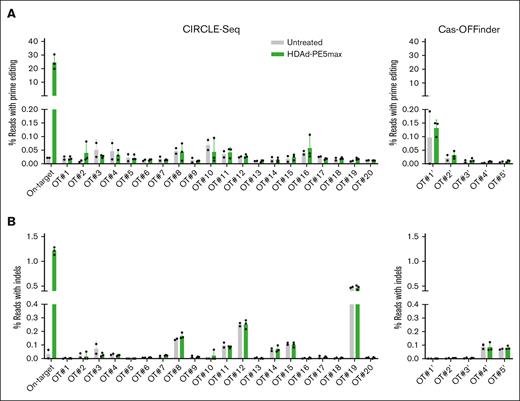
We used amplicon deep sequencing to investigate the fidelity of HDAd-PE5max in the context of rapid mobilization and in vivo HSC prime editing. Potential off-target sites were experimentally identified by CIRCLE-seq (circularization for in vitro reporting of cleavage effects by sequencing), a highly sensitive in vitro method for genome-wide off-target screening,19 or computationally nominated by Cas-OFFinder20 (Figure 7).20 Twenty-five candidates described previously5 were amplified from the BM MNC genomic DNA of 3 in vivo transduced mice with the highest on-target editing and 2 untreated control mice (a total of 125 amplicons). The amplicons were subjected to amplicon NGS. The results showed that the percentages of reads containing a PAM single nucleotide variant were <0.2% and no marked differences were observed between untreated and treated animals, demonstrating no significant off-target prime editing at these sites (Figure 7A). Because prime editing requires more stringent conditions than CRISPR/Cas9-mediated binding/nicking, we also analyzed the frequencies of indels at these sites. We found that indel levels in HDAd-PE5max-transduced mice were close to the background shown in the control mouse (Figure 7B). Together, these data show no significant off-target editing after WU-106/AMD3100 mobilization and in vivo HSC prime editing.
Analyses of potential off-target editing by targeted amplicon deep sequencing. Genomic DNA from 3 mice with the highest on-target editing and 2 untreated control mice was used. (A) Off-target prime editing at the top 20 potential sites experimentally identified by CIRCLE-seq (left panels), and the top 5 sites predicted in silico by Cas-OFFinder (right panels). After sequencing, off-target prime editing was calculated based on the percentage of reads with G/T/C > A conversion at position +4 (corresponding to the sickle mutation), counting the predicted nicking site as position +1. If position +4 is already an A in the wild-type allele, a calculation was performed based on the percentage of G/T/C > A conversion at position +5 (corresponding to the silent PAM mutation). Each symbol represents an individual animal. (B) Percentage of reads with indels. The indel levels were analyzed using a Cas-Analyzer. Genomic DNA samples were the same as those in panel A.
Analyses of potential off-target editing by targeted amplicon deep sequencing. Genomic DNA from 3 mice with the highest on-target editing and 2 untreated control mice was used. (A) Off-target prime editing at the top 20 potential sites experimentally identified by CIRCLE-seq (left panels), and the top 5 sites predicted in silico by Cas-OFFinder (right panels). After sequencing, off-target prime editing was calculated based on the percentage of reads with G/T/C > A conversion at position +4 (corresponding to the sickle mutation), counting the predicted nicking site as position +1. If position +4 is already an A in the wild-type allele, a calculation was performed based on the percentage of G/T/C > A conversion at position +5 (corresponding to the silent PAM mutation). Each symbol represents an individual animal. (B) Percentage of reads with indels. The indel levels were analyzed using a Cas-Analyzer. Genomic DNA samples were the same as those in panel A.
Discussion
We report that the combination of WU-106 + AMD3100 mobilization and IV injection of an HDAd-PE5max vector together with in vivo selection resulted in efficient prime editing in long-term repopulating HSCs. Another key aspect of our approach is that it is G-CSF–free. Thus, it does not result in massive mobilization of neutrophils/progenitors, which in turn minimizes IL-6 release and IL-6-related toxicity. Mobilization/in vivo HSC transduction approach can be done within several hours.
Using the standard G-CSF/AMD3100 regimen, ∼100 000 LSK cells or 30 000 CFUs per mL peripheral blood were mobilized 40 minutes after AMD3100 injection in mice. Similarly, in rhesus macaques, G-CSF/AMD3100 resulted in 30 000 to 100 000 mobilized CD34+/CD45RA–/CD90+ cells per mL peripheral blood 8 hours after AMD3100 injection.21 We have shown in mice and non-human primates (NHPs) that at least 20% of mobilized HSCs are transduced after IV HDAd injection and that a large fraction of these cells return to the BM.2 However, G-CSF is contraindicated in patients with SCD. Data from the J.F.D. group demonstrated that the combination of small-molecule VLA-4 and CXCR4 inhibitors (plerixafor or motixafortide) achieves safe, rapid, and reliable mobilization of sufficient quantities of high-quality stem cells in mice and NHPs. Here, we used 1 of the PEGylated VLA-4 inhibitors, WU-106, plus AMD3100, for in vivo HSC transduction in SCD CD46/Townes mice. This G-CSF–free mobilization regimen is superior to G-CSF/AMD3100 in several aspects. First, mobilization and HDAd injection can be done within 2 hours instead of 5 days. If adding cytokine prophylaxis before HDAd injection, the whole procedure can be performed in 1 day and can therefore be considered as outpatient treatment. However, at this stage of development, the approach still requires 3 cycles of O6BG/BCNU selection within the first month after HDAd injection. Second, the favorable pharmacokinetics of WU-106 mediates rapid and transient (∼6 hours) mobilization. This allows for efficient transduction with HDAd vectors (half-life in blood: ∼2 hours) and efficient return to the BM without continuous mobilization. Notably, when we used mobilization drugs such as the anti-α4 integrin monoclonal antibody natalizumab with a half-life in human of ∼11 days, we saw continuous mobilization with very low transgene markings of BM HSCs (data not shown). Third, WU-106/AMD3100 was safer in the context of our in vivo approach. G-CSF/AMD3100 nonspecifically mobilizes HSCs, myeloid progenitors, and maturing myeloid cells, such as granulocytes that carry granules filled with proteases and cytokines, which can be released upon contact with HDAd particles in the periphery.22 WU-106/AMD3100 mobilization resulted in lower numbers of activated granulocytes (CD11b+/Ly6G+) than mobilization with G-CSF/AMD3100. Serum levels of the proinflammatory cytokine IL-6 were therefore, on average, 10-fold lower than those in GCSF/AMD3100 + HDAd mice. Furthermore, serum levels of CD62p/P-selectin, a marker for platelet activation, were significantly lower in HDAd-injected mice that were mobilized by WU-106/AMD3100.
Additionally, selective mobilization of HSCs would reduce vector sequestration by committed cells. For upcoming NHP studies, we hope that we can largely address the cytokine release problem by pursuing a combination of (1) cytokine prophylaxis before and shortly after IV HDAd injection (involving the IL-1 and IL-6 antagonists anakinra and tocilizumab), (2) minimizing the release of cytokines by macrophages by modifying capsid proteins, and (3) more selective HSC mobilization approaches that reduce the number of granulocytes in blood circulation (eg, WU-106/AMD3100). We directly compared the mobilization and hematological parameters for WU-106/AMD3100 and G-CSF/AMD3100. Based on the number of LSK cells present in the periphery at 45 minutes after AMD3100 (for G-CSF/AMD3100) or 2 hours after WU-106/AMD3100, mobilization efficiency was comparable. However, there were significantly fewer CFUs mobilized at this time with the WU-106/AMD3100 regimen. It is possible that the kinetics of CFU mobilization are different between the 2 approaches, or that the CFU assay does not adequately reflect the number of long-term repopulating cells. The latter hypothesis is supported by the fact that the target site editing in secondary recipients at week 16 were, on average 30%, in the present study and significantly higher than those in the previous study with G-CSF/AMD3100,5 indicating that WU-106/AMD31000 preferentially mobilizes long-term persisting HSCs. This is consistent with our recently published data using a derivative of WU-106, CWHM-823.13 In addition when compared with the G-CSF/AMD3100 approach that involved the same strain of mice (CD46/Townes), the same vector (HDAd-PE5max), and the regimen of O6BG/BCNU selection, the degree of phenotypic correction in primary mice was similar. This is, therefore, the first demonstration of the therapeutic use of WU-106/AMD3100 to treat SCD by in vivo HSC gene therapy.
The requirement for O6BG/BCNU in vivo selection of transduced HSPCs is currently a limitation of the approach, specifically in patients with SCD being at increased risk of developing leukemia and clonal hematopoiesis,23,24 even though we did not observe clonal hematopoiesis in vivo after Sleeping Beauty transposase-mediated gene addition in mice that underwent O6BG/BCNU selection.4,10,25 In addition to strategies that increase the number of edited HSCs in the BM, our current efforts are also focused on alternative in vivo selection approaches, including the in vivo creation of truncation in the erythropoietin receptor to confer a selective advantage to edited erythroid progenitor cells by mediating hypersensitivity to erythropoietin.26 Another strategy that potentially can be used to expand edited HSCs is based on epitope engineering of HSC membrane proteins using base or prime editors in combination with monoclonal antibodies that target only unedited cells.27 We also recently described the expansion of in vivo HDAd-transduced HSPCs by constitutive expression of a truncated high mobility group A2 (tHMGA2) without additional treatment (unpublished data).
To be considered for clinical trials in humans, the new protocol involving WU-106/AMD3100 should undergo rigorous safety testing in mice, humanized mice, and NHPs (including preimmune animals). These studies should assess acute toxicity (eg, based on serum cytokines, complete blood count, etc.), toxicity from transduction of nontarget tissues, an adaptive immune response against the vector and the payload as well as against edited proteins, longitudinal, long-term on- and off-target editing (especially after multiplex editing), genomic rearrangements between off-target sites, monitoring of hematopoietic clonality (longitudinal, long-term), editing in nontarget tissues, specifically germ line, after isolation of parenchymal cells from major organs, long-term persistence of edited HSCs and PBMCs, and others.
Although the WU-106/Plerixafor mobilization will be used for harvesting HSCs from patients with SCD and possibly for mobilization-based BM conditioning in the context of ex vivo HSC gene therapy,9,28 our data indicate that this same-day mobilization regimen will further simplify our in vivo HSC transduction approach and improve its safety profile for the treatment of SCD and other genetic diseases.
Acknowledgments
The authors acknowledge Sucheol Gil and Theo Koob for mouse genotyping and assisting with animal experiments. The study was supported by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (R01HL128288 and R01HL141781) (A.L.), Ensoma Bio (A.L. and H.-P.K.), and Bill & Melinda Gates Foundation (INV-038139) (A.L.). The study was also supported by grants from the National Cancer Institute (R35 CA210084 (J.F.D.) and R50 CA211466-02 (M.R.).
Authorship
Contribution: C.L. and A.L. provided the conceptual framework for the study; C.L. designed and performed the experiments; A.K.A. participated in experiments; P.R., M.R., D.K., and J.F.D. produced WU-106; J.F.D. and H.-P.K. provided critical comments on the manuscript and directions regarding the mobilization experiments; and C.L. and A.L. wrote the manuscript.
Conflict-of-interest disclosure: A.L. and H.-P.K. are the scientific cofounders of Ensoma Bio. H.-P.K. is a paid adviser for Ensoma Bio. J.F.D. receives research funding from NeoImmuneTech, Macrogenics, Incyte, Bioline Rx; has equity ownership in Magenta Therapeutics, Wugen; and is a board member for hC Bioscience Inc and RiverVest Venture Partners. The remaining authors declare no competing financial interests.
Correspondence: Chang Li, University of Washington, 1705 NE Pacific St, HSB K263, Box 357720, Seattle, WA 98195; email: cli1239@uw.edu.
References
Author notes
Next-generation sequencing data have been deposited to the NCBI SRA with accession code PRJNA1101957. This SRA submission will be released upon publication.
The full-text version of this article contains a data supplement.