Through transcriptomic analysis, we identified 7 kinases whose expressions were strongly predictive of compromised survival in MDS.
The KISS could improve risk stratification and imply novel therapeutic opportunities in MDS.
Visual Abstract
The human kinome, which comprises >500 kinases, plays a critical role in regulating numerous essential cellular functions. Although the dysregulation of kinases has been observed in various human cancers, the characterization and clinical implications of kinase expressions in myelodysplastic syndromes (MDS) have not been systematically investigated. In this study, we evaluated the kinome expression profiles of 341 adult patients with primary MDS and identified 7 kinases (PTK7, KIT, MAST4, NTRK1, PAK6, CAMK1D, and PRKCZ) whose expression levels were highly predictive of compromised patient survival. We then constructed the kinase stratification score (KISS) by combining the weighted expressions of the 7 kinases and validated its prognostic significance in 2 external MDS cohorts. A higher KISS was associated with older age, higher peripheral blood and marrow blast percentages, higher Revised International Prognostic Scoring System (IPSS-R) risks, complex karyotype, and mutations in several adverse-risk genes in MDS, such as ASXL1, EZH2, NPM1, RUNX1, STAG2, and TP53. Multivariate analysis confirmed that a higher KISS was an independent unfavorable risk factor in MDS. Mechanistically, the KISS-high patients were enriched for gene sets associated with hematopoietic and leukemic stem cell signatures. By investigating the Genomics of Drug Sensitivity in Cancer database, we identified axitinib and taselisib as candidate compounds that could potentially target the KISS-high myeloblasts. Altogether, our findings suggest that KISS holds the potential to improve the current prognostic scheme of MDS and inform novel therapeutic opportunities.
Introduction
Myelodysplastic syndromes (MDS) are a heterogeneous constellation of myeloid neoplasms, originating from the clonal proliferation of malignant hematopoietic stem cells (HSCs).1 Although the initial clinical manifestations of MDS are usually characterized by ineffective hematopoiesis and peripheral blood cytopenias, the disease can eventually evolve into acute myeloid leukemia (AML) in ∼30% of patients and frequently becomes fatal.1-3
Currently, the prognosis of patients with newly diagnosed MDS is most commonly evaluated with the Revised International Prognostic Scoring System (IPSS-R)4; however, it is observed that patients may still have variable clinical outcomes even if they are categorized within the same risk category.5 As the genomic landscape of MDS becomes more elucidated with the advances in the sequencing technology, the IPSS-Molecular has recently been proposed to further fine tune the risk stratification of MDS.6,7 Nevertheless, it is both financially and computationally demanding to obtain the genomic information required by this more complex genetically inspired risk model. Endeavors to identify standardized molecular markers that could improve the outcome prediction for patients with myeloid neoplasms had been undertaken before as well. Most notably, Pellagatti et al performed an integrative transcriptomic analysis of 125 patients with MDS and devised the gene expression profiling–based Coxnet signature comprising 20 genes to refine the risk classification in MDS.8 Ng et al first analyzed the global gene expression of leukemic stem cells (LSCs) derived from patients with AML in murine xenotransplantation models to identify prognostic biomarkers closely related to stemness and then applied a statistical regression algorithm to generate the 17-gene LSC score (LSC17) that could improve risk stratification in patients of diverse AML subtypes.9 Nevertheless, currently, no clinico-genomic or transcriptomic risk model could inform readily applicable treatment implications.
The treatment options for MDS have evolved substantially in recent years, in parallel with our deeper understanding of the pathophysiology of MDS. Although hypomethylating agents and allogeneic hematopoietic stem cell transplantation (allo-HSCT) remain the standard of care for patients with high-risk MDS, venetoclax, the selective B-cell lymphoma 2 (BCL2) inhibitor, has also been demonstrated to improve the response rates further.10 However, relapses or progression to AML are still common, especially in those who are unfit for allo-HSCT. Therefore, there exists an unmet need for novel treatment strategies in patients with MDS, especially those with high-risk disease.
Kinases are enzymes that catalyze the transfer of phosphate residues from phosphate donors to target proteins, a biological process known as phosphorylation.11 Collectively, the human kinome is composed of >500 kinases and comprises ∼1.7% of the coding regions of our genome. The human kinome can be classified into 9 typical and 13 atypical families.12 Kinases are critical players in various cellular processes, such as signal transduction, metabolism, proliferation, differentiation, and apoptosis.13 Due to their functional versatility, >85% of the kinases are found to be dysregulated in human diseases,13 and >70 small molecule kinase inhibitors have been approved by the US Food and Drug Administration (FDA) for therapeutic purposes.14 However, none of the kinase inhibitors have received regulatory approval for the treatment of MDS yet.
In this study, we hypothesized that the aberrant expression of kinases could exert an impact on the clinical prognosis and, moreover, indicate novel treatment options in patients with MDS. We first profiled the gene expressions of the human kinome to devise a highly prognostic kinase-based risk score that could refine the risk stratification of MDS and further explored novel therapeutic possibilities by mining the well-curated Genomics of Drug Sensitivity in Cancer (GDSC) database.
Materials and methods
Patients
From 1997 to 2019, a total of 341 adult patients diagnosed with primary MDS according to the 2016 World Health Organization classification criteria15 and who had adequate cryopreserved diagnostic bone marrow (BM) samples for DNA and RNA sequencing at the National Taiwan University Hospital (NTUH) were included in this study. These patients were further annotated according to the 2022 International Consensus Classification (ICC) classification of myeloid neoplasms and acute leukemias after the release of the updated criteria.16 Because allo-HSCT is a well-established disease course modifier in myeloid malignancies, including MDS, the 282 patients who did not receive allo-HSCT were designated as the NTUH-A cohort, whereas the other 59 patients who had received allo-HSCT were designated as the NTUH-B cohort. In addition, 19 healthy BM stem cell donors were recruited as healthy controls (HCs). This study was conducted in accordance with the Declaration of Helsinki and was approved by the Research Ethics Committee of the NTUH. All participants provided written informed consent.
RNA sequencing (RNA-seq) and raw data preprocessing
RNA was extracted from the diagnostic BM samples (without CD34+ cell isolation), and the sequencing libraries were constructed using the TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA) following the manufacturer’s recommendations. The libraries were then sequenced on an Illumina NovaSeq 6000 with 150 bp paired-end read mode. Adapter sequences and low-quality bases in the raw sequencing data were removed using Cutadapt (v 3.0), and the clean reads were then aligned to the human reference genome GRCh38 using STAR (v2.7.6a) with 2-pass mode.17 The raw count of each gene was calculated according to the GENCODE v28 annotation and was converted into to transcripts per million (TPMs) for further analysis.
Development of the KISS
The overall workflow for survival modeling and establishment of the kinase-based risk score is illustrated in supplemental Figure 1A. A total of 517 kinase genes were extracted from the KinHub database.18 We excluded 125 kinases with low expression values (<1 TPM) and then performed differential expression analysis between patients with MDS and HCs to identify 61 overexpressed kinases in MDS (adjusted P < .05; log2 fold-change > 0). For subsequent survival modeling, we transformed the original TPM values into the log2 (TPM + 1) scale, in which +1 term was to mitigate excessive variations of small values, and then performed z-transformation across all samples, so that the risk score calculated from different patient cohorts or different gene expression quantification methods (eg, RNA-seq or microarray) would be more comparable. Next, univariate Cox proportional hazards regression was used to select 24 of the 61 kinases that had significant impact on overall survival (OS; hazard ratio [HR] > 1.0; adjusted P < .05) in the NTUH-A cohort. To construct a parsimonious outcome prediction model, the least absolute shrinkage and selection operator (LASSO) Cox proportional hazards (PHs) regression modeling method was used. We fitted the LASSO Cox regression model with 10-fold crossvalidation to the 24 kinase genes (supplemental Figure 1B). Kinases with nonzero coefficients (n = 7) were then selected to construct the kinase stratification score (KISS), which was defined as the normalized gene expressions of component kinases weighted by their corresponding LASSO coefficients: (0.252 × protein tyrosine kinase 7 [PTK7]) + (0.145 × KIT) + (0.144 × microtubule associated serine/threonine kinase family member 4 [MAST4]) + (0.072 × neurotrophic receptor tyrosine kinase 1 [NTRK1]) + (0.065 × p21-activated kinase 6 [PAK6]) + (0.061 × calcium/calmodulin dependent protein kinase ID [CAMK1D]) + (0.003 × protein kinase C zeta [PRKCZ]). The expression levels of the 7 kinases were significantly higher in patients with MDS than HCs (supplemental Figure 1C).
Additional details on description of materials and methods are presented in the supplemental Methods.
Results
Patient characteristics
The demographic and clinical characteristics of the overall 341 NTUH patients with MDS had been described previously.19 Briefly, the cohort consisted of 36.7% females and 63.3% males, with a median age of 68.3 years. A total of 59 of the 341 patients (17.3%) received allo-HSCT as part of their treatment. In addition, 332 patients (97.4% of the cohort) had diagnostic cytogenetic information, and of these, 4.5%, 24.1%, 23.5%, 24.4%, and 23.5% could be classified into very low-, low-, intermediate-, high-, and very high-risk categories in the IPSS-R scheme, respectively.
The KISS could risk stratify patient outcomes across multiple MDS cohorts
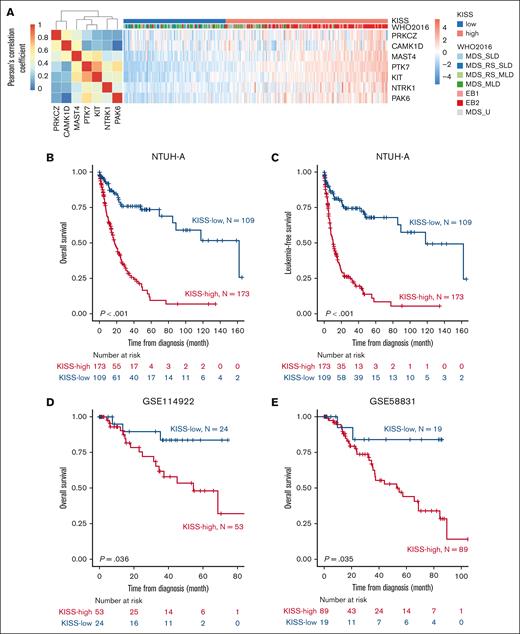
Because dysregulation of kinases may play an important role in the pathogenesis and progression of MDS, we modeled the kinase expressions in MDS (detailed in the “Materials and Methods”) and identified 7 kinases, namely PTK7, KIT, MAST4, NTRK1, PAK6, CAMK1D, and PRKCZ, whose expressions were significantly associated with compromised survival in MDS. Coexpression analysis of the 7 members of KISS revealed that PRKCZ and CAMK1D formed a coexpression module, whereas MATS4, PTK7, KIT, NTRK1, and PAK6 formed another one (Figure 1A). We further integrated the weighted sum of these 7 kinase into the KISS prognostic score and used the maximally selected log-rank statistics method to identify the optimal threshold dichotomizing patients with MDS into KISS-high and KISS-low (ie, prognostically unfavorable vs favorable) subgroups (supplemental Figure 1D).
Establishment of the KISS for MDS risk stratification. (A) Pearson correlation matrix of the 7 kinases selected into the KISS model, and heat map illustrating the normalized expression of the kinases, across the patients in the NTUH-A cohort. Patients with higher-risk MDS (EB1 and EB2) were notably clustered within the KISS-high subgroup. OS (B) and LFS (C) of patients with MDS in the NTUH-A cohort, stratified by the KISS. (D) OS of patients with MDS in the GSE114922 data set, stratified by the KISS. (E) OS of patients with MDS in the GSE58831 data set, stratified by the KISS.
Establishment of the KISS for MDS risk stratification. (A) Pearson correlation matrix of the 7 kinases selected into the KISS model, and heat map illustrating the normalized expression of the kinases, across the patients in the NTUH-A cohort. Patients with higher-risk MDS (EB1 and EB2) were notably clustered within the KISS-high subgroup. OS (B) and LFS (C) of patients with MDS in the NTUH-A cohort, stratified by the KISS. (D) OS of patients with MDS in the GSE114922 data set, stratified by the KISS. (E) OS of patients with MDS in the GSE58831 data set, stratified by the KISS.
We first demonstrated that KISS-high patients had significantly shorter OS (Figure 1B; median, 17.7 vs 162.1 months; P < .001) and leukemia-free survival (LFS; Figure 1C; median, 10.3 vs 118.1 months; P < .001) than KISS-low patients in the NTUH-A cohort. To assess the robustness of our scoring method, we also evaluated the prognostic capability of KISS in 2 external MDS cohorts. Using the same cutoff value as defined in the NTUH-A cohort, KISS-high patients consistently had significantly worse OS than KISS-low patients in both the external validation cohorts, namely GSE114922 (Figure 1D; median OS, 54.5 months vs not reached [NR]; P = .036) and GSE58831 (Figure 1E; median OS, 54.5 months vs NR; P = .035), demonstrating the robustness of our KISS for outcome prediction in MDS.
The KISS-high subgroup was associated with high-risk clinical and genetic features
The KISS as a continuous variable was significantly higher in the clinically defined high-risk MDS subclasses, such as MDS with excess blasts 1 (MDS-EB1) and MDS-EB2 (supplemental Figure 1E; Kruskal-Wallis P < .001). The KISS also appeared to elevate progressively ranging from IPSS-R very low– to very high–risk subgroups (supplemental Figure 1F; Kruskal-Wallis P < .001). The comparison of clinical characteristics and genetic alterations between the KISS-high and KISS-low patients of the NTUH-A cohort is presented in Table 1 (the annotation by the 2022 ICC classification is presented in supplemental Table 2). The KISS-high subgroup had more advanced age (P = .020), higher peripheral blood and BM (both P < .001) blast percentages, and lower platelet levels (P = .003) at diagnosis than the KISS-low subgroup. According to the 2016 World Health Organization classification of MDS, the KISS-high subgroup had less frequent MDS with single-lineage dysplasia (MDS-SLD; P < .001), MDS with multilineage dysplasia (MDS-MLD; P < .001), and MDS with ring sideroblasts and SLD (P < .001) or MLD (P = .001) subtypes but more frequent MDS-EB1 and MDS-EB2 (both P < .001) subtypes. According to the 2022 ICC classification, the KISS-high subgroup had lower proportions of MDS with mutated SF3B1 (P < .001), MDS, not otherwise specified, with SLD (P < .001), and MDS, not otherwise specified, with MLD (P < .001) but higher proportions of MDS with mutated TP53 (P = .008), MDS-EB (P < .001), MDS/AML with mutated TP53 (P = .003), and MDS/AML with myelodysplasia-related gene mutations or cytogenetics abnormalities (P < .001). In addition, patients in the KISS-high subgroup were more likely to harbor complex cytogenetics (P < .001) but had less frequent normal karyotype (P < .001). In terms of IPSS-R, the KISS-high subgroup was enriched for very high and high risks (both P < .001), while inversely correlated with low (P < .001) and very low risks (P = .007). The KISS-high subgroup was significantly enriched for mutations in ASXL1, EZH2, NPM1, RUNX1, STAG2, and TP53 (P < .001; P = .020; P = .027; P = .001; P = .029; and P < .001, respectively), whereas the KISS-low subgroup had significantly higher frequency of the SF3B1 mutation (P = .004; supplemental Table 3). Collectively, these results showed that the KISS was able to identify a subset of prognostically unfavorable patients with high-risk clinical and genetic features.
Clinical and laboratory characteristics according to the KISS strata
| Variable . | KISS-high (n = 173) . | KISS-low (n = 109) . | P value . |
|---|---|---|---|
| Sex, n (%) | .434 | ||
| Female | 59 (34.1) | 43 (39.4) | |
| Male | 114 (65.9) | 66 (60.6) | |
| Age, median (range) | 72.1 (17.8-94.2) | 67.6 (20.0-94.2) | .020 |
| Laboratory data, median (range) | |||
| WBC, × 109/L | 3.8 (0.3-56.3) | 4.3 (0.8-25.5) | .311 |
| Hb, g/dL | 8.4 (3.5-15.3) | 8.2 (4.2-16.9) | .535 |
| Platelet, × 109/L | 75 (1-721) | 111 (3-405) | .003 |
| PB blast, % | 1.0 (0-18.0) | 0 (0-4.0) | <.001 |
| BM blast, % | 9.2 (0-19.0) | 2 (0-17.8) | <.001 |
| 2016 WHO classification, n (%) | |||
| MDS-SLD | 5 (2.9) | 26 (23.9) | <.001 |
| MDS-MLD | 19 (11.0) | 37 (33.9) | <.001 |
| MDS-RS-SLD | 2 (1.2) | 17 (15.6) | <.001 |
| MDS-RS-MLD | 4 (2.3) | 11 (10.1) | .010 |
| MDS-del(5q) | 1 (0.6) | 1 (0.9) | >.999 |
| MDS-EB1 | 55 (31.8) | 7 (6.4) | <.001 |
| MDS-EB2 | 83 (48.0) | 9 (8.3) | <.001 |
| MDS-U | 4 (2.3) | 1 (0.9) | .689 |
| IPSS-R, n (%) | |||
| Very low | 3 (1.8) | 10 (9.7) | .007 |
| Low | 21 (12.4) | 53 (51.5) | <.001 |
| Int | 32 (18.8) | 29 (28.2) | .1 |
| High | 57 (33.5) | 8 (7.8) | <.001 |
| Very high | 57 (33.5) | 3 (2.9) | <.001 |
| Karyotype, n (%) | |||
| Normal karyotype | 84 (49.4) | 78 (75.7) | <.001 |
| Complex karyotype | 35 (20.6) | 1 (1.0) | <.001 |
| Variable . | KISS-high (n = 173) . | KISS-low (n = 109) . | P value . |
|---|---|---|---|
| Sex, n (%) | .434 | ||
| Female | 59 (34.1) | 43 (39.4) | |
| Male | 114 (65.9) | 66 (60.6) | |
| Age, median (range) | 72.1 (17.8-94.2) | 67.6 (20.0-94.2) | .020 |
| Laboratory data, median (range) | |||
| WBC, × 109/L | 3.8 (0.3-56.3) | 4.3 (0.8-25.5) | .311 |
| Hb, g/dL | 8.4 (3.5-15.3) | 8.2 (4.2-16.9) | .535 |
| Platelet, × 109/L | 75 (1-721) | 111 (3-405) | .003 |
| PB blast, % | 1.0 (0-18.0) | 0 (0-4.0) | <.001 |
| BM blast, % | 9.2 (0-19.0) | 2 (0-17.8) | <.001 |
| 2016 WHO classification, n (%) | |||
| MDS-SLD | 5 (2.9) | 26 (23.9) | <.001 |
| MDS-MLD | 19 (11.0) | 37 (33.9) | <.001 |
| MDS-RS-SLD | 2 (1.2) | 17 (15.6) | <.001 |
| MDS-RS-MLD | 4 (2.3) | 11 (10.1) | .010 |
| MDS-del(5q) | 1 (0.6) | 1 (0.9) | >.999 |
| MDS-EB1 | 55 (31.8) | 7 (6.4) | <.001 |
| MDS-EB2 | 83 (48.0) | 9 (8.3) | <.001 |
| MDS-U | 4 (2.3) | 1 (0.9) | .689 |
| IPSS-R, n (%) | |||
| Very low | 3 (1.8) | 10 (9.7) | .007 |
| Low | 21 (12.4) | 53 (51.5) | <.001 |
| Int | 32 (18.8) | 29 (28.2) | .1 |
| High | 57 (33.5) | 8 (7.8) | <.001 |
| Very high | 57 (33.5) | 3 (2.9) | <.001 |
| Karyotype, n (%) | |||
| Normal karyotype | 84 (49.4) | 78 (75.7) | <.001 |
| Complex karyotype | 35 (20.6) | 1 (1.0) | <.001 |
Hb, hemoglobin; Int, intermediate; MDS-RS-SLD, MDS with ring sideroblasts and single-lineage dysplasia; MDS-RS-MLD, MDS with ring sideroblasts and multilineage dysplasia; MDS-U, MDS, unclassifiable.
Multivariable Cox analysis of the prognostic impact of KISS and other clinically relevant variables
| Variable . | OS . | LFS . | ||||||
|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI lower . | 95% CI upper . | P value . | HR . | 95% CI lower . | 95% CI upper . | P value . | |
| Age∗ | 1.032 | 1.019 | 1.045 | <.001 | 1.020 | 1.009 | 1.031 | .001 |
| IPSS-R† | 1.603 | 1.341 | 1.916 | <.001 | 1.502 | 1.266 | 1.783 | < .001 |
| Mutation | ||||||||
| ASXL1 | 0.819 | 0.510 | 1.316 | .409 | 0.811 | 0.516 | 1.274 | .363 |
| EZH2 | 2.754 | 1.450 | 5.233 | .002 | 1.835 | 0.977 | 3.447 | .059 |
| RUNX1 | 1.066 | 0.692 | 1.641 | .772 | 1.036 | 0.679 | 1.581 | .870 |
| SF3B1 | 0.561 | 0.302 | 1.041 | .067 | 0.560 | 0.311 | 1.010 | .054 |
| SRSF2 | 1.084 | 0.632 | 1.861 | .769 | 1.294 | 0.778 | 2.150 | .321 |
| STAG2 | 1.139 | 0.682 | 1.903 | .619 | 1.380 | 0.847 | 2.247 | .196 |
| TET2 | 1.646 | 1.033 | 2.623 | .036 | 1.518 | 0.975 | 2.362 | .065 |
| TP53 | 4.007 | 2.428 | 6.613 | <.001 | 2.144 | 1.349 | 3.408 | .001 |
| KISS∗ | 1.692 | 1.151 | 2.488 | .008 | 1.797 | 1.238 | 2.608 | .002 |
| Variable . | OS . | LFS . | ||||||
|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI lower . | 95% CI upper . | P value . | HR . | 95% CI lower . | 95% CI upper . | P value . | |
| Age∗ | 1.032 | 1.019 | 1.045 | <.001 | 1.020 | 1.009 | 1.031 | .001 |
| IPSS-R† | 1.603 | 1.341 | 1.916 | <.001 | 1.502 | 1.266 | 1.783 | < .001 |
| Mutation | ||||||||
| ASXL1 | 0.819 | 0.510 | 1.316 | .409 | 0.811 | 0.516 | 1.274 | .363 |
| EZH2 | 2.754 | 1.450 | 5.233 | .002 | 1.835 | 0.977 | 3.447 | .059 |
| RUNX1 | 1.066 | 0.692 | 1.641 | .772 | 1.036 | 0.679 | 1.581 | .870 |
| SF3B1 | 0.561 | 0.302 | 1.041 | .067 | 0.560 | 0.311 | 1.010 | .054 |
| SRSF2 | 1.084 | 0.632 | 1.861 | .769 | 1.294 | 0.778 | 2.150 | .321 |
| STAG2 | 1.139 | 0.682 | 1.903 | .619 | 1.380 | 0.847 | 2.247 | .196 |
| TET2 | 1.646 | 1.033 | 2.623 | .036 | 1.518 | 0.975 | 2.362 | .065 |
| TP53 | 4.007 | 2.428 | 6.613 | <.001 | 2.144 | 1.349 | 3.408 | .001 |
| KISS∗ | 1.692 | 1.151 | 2.488 | .008 | 1.797 | 1.238 | 2.608 | .002 |
CI, confidence interval.
As continuous variable.
IPSS-R risk groups: very low, low, intermediate, high, and very high.
KISS as an independent prognostic factor for OS and LFS in patients with MDS
Given that several conventional clinical factors also influence the survival of patients with MDS, we conducted further subgroup analyses to explore the prognostic significance of KISS across various clinical scenarios. We found that the adverse prognostic impact of KISS on OS and LFS remained significant in both lower-risk (IPSS-R very low, low, and intermediate risks merged; Figure 2A; median OS, 45.1 vs 162.1 months; P = .001; Figure 2B; median LFS, 32.5 vs 162.1 months; P < .001) and higher-risk (IPSS-R high and very high risks merged; Figure 2C; median OS, 13.0 vs 85.2 months; P = .009; Figure 2D; median LFS, 7.0 vs 43.0 months; P = .004) subsets of patients with MDS. KISS was also able to stratify risk in patients with MDS harboring either normal karyotype (supplemental Figure 2A) or complex cytogenetics (supplemental Figure 2B). Furthermore, in the multivariate Cox PH regression analysis considering KISS and those statistically significant clinical variables and genetic mutations in the univariate Cox PH regression analysis (supplemental Table 4), the KISS remained an independent adverse prognostic factor for OS (HR 1.692; 95% confidence interval, 1.151-2.488; P = .008) and LFS (HR, 1.797; 95% confidence interval, 1.238-2.608; P = .002; Table 2) Taken together, higher KISS predicted worse clinical outcomes not only in the overall MDS cohort but also in various clinically relevant subgroups, and this adverse prognostic impact appeared to be independent of other conventional risk factors, such as age, IPSS-R, and genetic mutations.
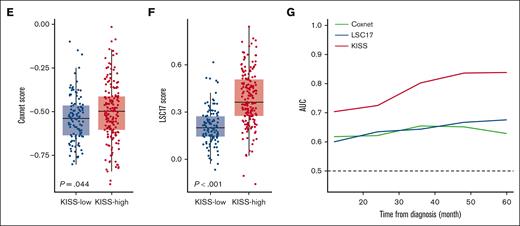
The KISS can refine the current risk-stratification scheme for MDS. OS (A) and LFS (B) of patients with lower-risk IPSS-R (very low, low, and intermediate risks) MDS, stratified by the KISS. OS (C) and LFS (D) of patients with higher-risk IPSS-R (high and very high risks) MDS, stratified by the KISS. The Coxnet predictor (E) and LSC17 scores (F) were significantly higher in the KISS-high subgroup. (G) The time-dependent ROC analysis of the KISS, Coxnet, and LSC17 scores demonstrated the superior prognostic performance of KISS.
The KISS can refine the current risk-stratification scheme for MDS. OS (A) and LFS (B) of patients with lower-risk IPSS-R (very low, low, and intermediate risks) MDS, stratified by the KISS. OS (C) and LFS (D) of patients with higher-risk IPSS-R (high and very high risks) MDS, stratified by the KISS. The Coxnet predictor (E) and LSC17 scores (F) were significantly higher in the KISS-high subgroup. (G) The time-dependent ROC analysis of the KISS, Coxnet, and LSC17 scores demonstrated the superior prognostic performance of KISS.
Additionally, to compare the prognostic performance of the KISS with other reported gene expression–based predictors in MDS, we calculated the Coxnet predictor and the LSC17 scores in our MDS cohort.8,9 Although the LSC17 score, composed of the weighted expression of 17 LSC-related genes, was initially developed for risk determination in AML, because MDS is also a stem cell disease,20,21 we reasoned that the LSC17 score may also play a role in MDS risk stratification. Indeed, both the Coxnet predictor and the LSC17 scores could risk stratify patients in the NTUH-A cohort (supplemental Figure 3A-B), and both scores were significantly higher in the KISS-high patients (Figure 2E-F; Wilcoxon rank-sum P = .044 and P < 0.001, respectively). Nonetheless, we found that the KISS had superior predictive power for patient outcomes compared with the Coxnet or LSC17 scores, assessed by the time-dependent receiver operating characteristic (ROC) curves (Figure 2G).
KISS could help identify appropriate candidates for allo-HSCT
Because we demonstrated the adverse prognostic impact of KISS on patients with MDS, we were interested in whether the adverse prognostic impact of higher KISS could be mitigated by allo-HSCT. We therefore examined KISS-high patients in the entire MDS cohort (NTUH-A and NTUH-B) and found that allo-HSCT could significantly improve both OS and LFS in these patients at high risk (supplemental Figure 3C; median OS, 37.7 vs 17.7 months; P < .001; median LFS, 32.9 vs 10.3 months; P < .001). Conversely, patients categorized as KISS-low did not derive survival benefit from allo-HST (supplemental Figure 3D). These findings suggested that the KISS could mitigate the adverse prognostic effect of the KISS-high status and thus holds the potential for selecting patients with MDS who would benefit from the allo-HSCT procedure.
Functional analysis revealed an enhanced stem cell signature in KISS-high patients
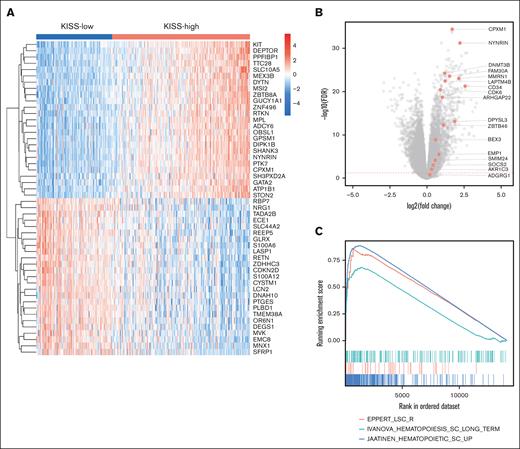
To elucidate the possible biological mechanisms underlying the adverse prognostic effect of higher KISS, we analyzed the RNA-seq data of BM samples in our MDS patient cohort (NTUH-A and NTUH-B merged). The differential expression analysis between the KISS-high vs KISS-low patients was performed. The most significant overexpressed and underexpressed genes are illustrated in Figure 3A (the details of the top 300 differentially expressed (DE) genes were listed in supplemental Table 5). Notably, the genes involved in the regulation of HSC proliferation and maintenance were significantly overexpressed in the KISS-high subgroup (false discovery rate (FDR) < 0.05), such as CPXM1, PTK7, KIT, NYNRIN, and MSI2.9,22 The LSC17 score–associated genes and HOX family genes were also significantly upregulated in the KISS-high subgroup (Figure 3B; supplemental Figure 4A). The results of GSEA (detailed in supplemental Table 6) revealed that many of the pathways associated with HSCs or LSCs were positively enriched (Figure 3C),23 whereas pathways encompassing genes downregulated in HSCs or LSCs were negatively correlated (supplemental Figure 4B). Together, the transcriptomic data analysis suggested that an enhanced stem cell transcriptional program may contribute to disease aggressiveness in the KISS-high patients.
Functional analysis of KISS-high vs KISS-low patients with MDS. (A) Heat map illustrating the most significant DE genes (25 most upregulated and 25 most downregulated genes). (B) The LSC17 genes were significantly upregulated in the KISS-high subgroup. (C) GSEA highlighted the enrichment of HSC and LSC-associated gene sets in the KISS-high subgroup. GSEA, gene set enrichment analysis.
Functional analysis of KISS-high vs KISS-low patients with MDS. (A) Heat map illustrating the most significant DE genes (25 most upregulated and 25 most downregulated genes). (B) The LSC17 genes were significantly upregulated in the KISS-high subgroup. (C) GSEA highlighted the enrichment of HSC and LSC-associated gene sets in the KISS-high subgroup. GSEA, gene set enrichment analysis.
Exploring novel therapeutic opportunities for the KISS-high patients
Several kinase inhibitors have been approved for the treatment of solid cancers but not yet for MDS. Because we demonstrated that the KISS-high patients with MDS had significantly worse survival, we sought to investigate whether these patients would benefit from kinase inhibitor treatments. To this end, we queried the Genomics of Drug Sensitivity in Cancer (GDSC) and Cancer Cell Line Encyclopedia (CCLE) data sets to model drug sensitivities with regard to the KISS.24,25 Although the GDSC collection did not include MDS cell lines, because MDS and AML are closely related clonal myeloid diseases, and the diagnostic boundary for high-risk MDS and AML is gradually being blurred in recent years,16 we selected the AML cell lines curated in the GDSC project as in vitro surrogates for drug response investigation.
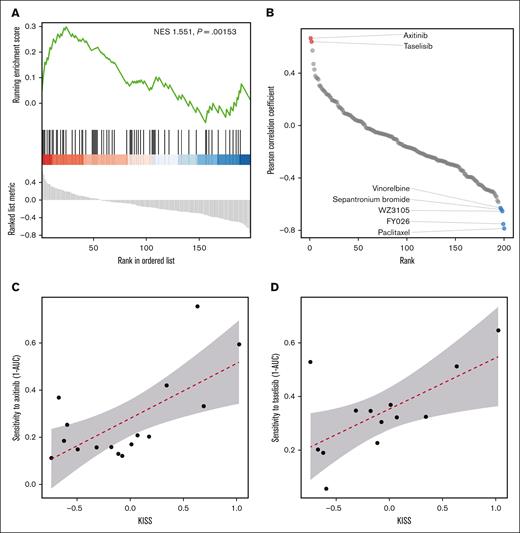
The Pearson correlation coefficients between the drug sensitivity score (we used 1 minus area under the drug response curve (1 – AUC) as the surrogate for drug sensitivity) and the KISS across all AML cell lines were calculated. Intriguingly, we observed that kinase inhibitors were positively enriched at the top of the compound list, ordered by descending Pearson correlation coefficients (Figure 4A-B), whereas the cytotoxic agents (such as paclitaxel and vinorelbine, etc) were mostly at the bottom of the list, indicating that our analysis did prioritize kinase inhibitors for the treatment of KISS-high cell lines. Importantly, we observed that axitinib and taselisib represented the most effective compounds (Pearson correlation coefficient > 0.6; supplemental Table 7) against KISS-high cell lines (Figure 4C-D).
Pharmacogenomic investigation identified compounds that could specifically target the KISS-high myeloblasts. (A) Kinase inhibitors were positively enriched at the top of the compound list ordered by the Pearson correlation coefficients between the KISS and drug sensitivity. (B) Pearson correlation of the KISS and drug sensitivity; a positive Pearson correlation coefficient implies that a compound is more effective against the KISS-high AML cell lines. (C) The correlation between axitinib sensitivity and KISS. (D) The correlation between taselisib sensitivity and KISS. NES, net enrichment score.
Pharmacogenomic investigation identified compounds that could specifically target the KISS-high myeloblasts. (A) Kinase inhibitors were positively enriched at the top of the compound list ordered by the Pearson correlation coefficients between the KISS and drug sensitivity. (B) Pearson correlation of the KISS and drug sensitivity; a positive Pearson correlation coefficient implies that a compound is more effective against the KISS-high AML cell lines. (C) The correlation between axitinib sensitivity and KISS. (D) The correlation between taselisib sensitivity and KISS. NES, net enrichment score.
Discussion
MDS are clonal stem cell diseases with a genomic landscape dominated by genetic mutations in the epigenetic regulators and splicing machinery, whereas variants of the transcription factors, DNA repair proteins, and signaling pathways constitute a minor proportion.26 The prognostic significance and therapeutic implications of kinome profiling in MDS have not been extensively explored. Although a wide array of kinase inhibitors have been approved by the FDA for the treatment of several solid cancers and hematological malignancies,27 and others are entering various phases of clinical trials, none of them have been approved specifically for MDS yet. Therefore, we reason that the investigation of the kinome expression in MDS addresses this unmet medical need.
In this study, we discovered that the expression levels of 7 kinases were significantly linked with the clinical outcomes of patients with MDS through rigorous statistical modeling. We further integrated the weighted expression of these kinases into the KISS, a concise yet powerful risk score, and validated its prognostic impact in 2 external MDS cohorts. Even though the KISS-high subgroup was associated with distinct clinical characteristics and more frequent deleterious mutations in ASXL1, EZH2, NPM1, RUNX1, STAG2, and TP53, our multivariate analysis attested the independent prognostic significance of the KISS. Notably, the KISS outperformed previously published Coxnet and LSC17 scores9,28 and held the ability to identify prognostically unfavorable patients even within IPSS-R lower-risk or normal karyotype subgroups.
The components of the KISS include PTK7, KIT, MAST4, NTRK1, PAK6, CAMK1D, and PRKCZ, which are all overexpressed in patients with MDS compared with the HCs and associated with inferior clinical outcomes. PTK7 is an evolutionarily conserved transmembrane receptor tyrosine kinase, which was originally found to be involved in the canonical and noncanonical Wnt pathways of epithelial cells. More recently, PTK7 was also found to be expressed by the HSPCs, with the highest expression level observed in the HSCs,29 and its deficiency in the mouse model led to a diminished HSC pool.30KIT is a type III receptor tyrosine kinase, which is strongly expressed in the HSPCs in which it plays a role in the maintenance of balance between self-renewal and lineage commitment.31,32MAST4 is widely expressed in the nervous system, especially in the cerebellar Purkinje cells and hippocampus,33 and also determines the mesenchymal stromal cell commitment toward the chondro-osteogenic fate by hampering the Sox9 transcriptional activity.34 However, its role in hematopoiesis and HSC biology has been less reported. NTRK1 fusions have been implicated in a number of solid tumor malignancies,35-38 representing an important therapeutic target.39 In normal hematopoiesis, NTRK1 is expressed at the highest levels in the common myeloid progenitors and early monocytes, whereas in AML, NTRK1 is overexpressed mainly in core-binding factor AML.40,41 In RUNX1::RUNX1T1 rearranged AML, BM stromal cells express nerve growth factor, which can bind to TRKA (encoded by the NTRK1 gene), leading to leukemogenesis.41PAK6 is implicated in tyrosine kinase inhibitor resistance in chronic myeloid leukemia by interacting with the tumor suppressor miR-185.42-44CAMK1D can network with the inhibitory leukocyte immunoglobulin-like receptor signaling pathway and plays an essential role in AML development and maintenance.45,46PRKCZ was found to be upregulated in MDS CD34+ BM cells and participate in the thrombopoietin signaling axis and HSC self-renewal.47,48 Overall, we provide evidence that the kinases selected by our KISS model are closely connected to the pathogenesis of myeloid malignancies. This is in parallel with the findings of our transcriptomic analysis that KISS-high patients had overexpressed HOX genes and a positive enrichment for HSC and LSC gene signatures.
Having demonstrated that KISS-high patients fared worse clinically, we were interested in whether certain treatment modalities could reverse the adverse prognostic impact of higher KISS. We found that allo-HSCT could indeed improve survival in the KISS-high subgroup; whereas on the contrary, the survival benefit of allo-HCST was not observed in the KISS-low subgroup, indicating that such intensive treatment option mainly benefits for those with truly high-risk diseases. Next, we attempted to search for compounds that could specifically target the KISS-high leukemic myeloblasts. By interrogating the GDSC project data sets, we identified axitinib and taselisib as our top hits. Axitinib is a selective inhibitor of vascular endothelial growth factor receptors 1, 2, and 3. It can bind to the intracellular tyrosine kinase domains of vascular endothelial growth factor receptors and subsequently reduce the downstream phosphorylation of AKT and ERK1/2.49,50 Although its first FDA approved indication was for the treatment of advanced renal cell carcinoma,51 axitinib has also demonstrated antitumor activities in hematological malignancies. In the case of chronic myeloid leukemia, axitinib can selectively target the BCR::ABL1 fusion transcript that harbors the T315I gatekeeper mutation through a mutation-selective mechanism and potently reduce the downstream phosphorylation targets in the leukemic cells.52 Moreover, combining axitinib with sorafenib can also overcome the tyrosine kinase inhibitor resistance caused by T315I, by inhibiting the Bcr-Abl/Grb2/Gab2 axis.53 Although in a previous study by Giles et al, axitinib had only minimal activity in AML or MDS,54 the sample size in this study was rather limited, and it is possible that only patient with higher KISS might derive greater treatment benefits. Taselisib is a PI3K inhibitor that potently inhibits the p110-alpha, delta, and gamma isoforms.55 When combined with antimicrotubule chemotherapy, such as vinorelbine or paclitaxel, taselisib induced antiproliferative, proapoptotic, and antimetastatic effects in human breast cancer cells, via the inhibition of downstream PI3K and MAPK pathway activities.56 Furthermore, when combined with the BCL2 inhibitor venetoclax, taselisib could effectively inhibit leukemic cell growth in AML cell lines or xenograft models derived from patients with AML.57 Overall, our pharmaco-transcriptomic investigation provided evidence that axitinib and taselisib may be potential novel treatment options for KISS-high patients with MDS.
We acknowledge there are some limitations in this study. First, our study population consisted of a retrospective MDS cohort with nonhomogeneous treatments that might have been influenced primarily by the individual patient’s comorbidities, quality-of-life expectations, as well as local reimbursement policies. Second, the patients included in this study were diagnosed with primary MDS and were from Asian ethnicity background, which needs to be considered when extrapolating the findings of this study to therapy-related MDS or other MDS patient populations. Third, although MDS and AML are closely related disease entities, the exact pathogenic mechanisms are not identical after all. Although we were only able to explore drug sensitivity data collected from the AML cell lines in the publicly accessible databases, we recognize that drug sensitivity experiments performed on BM hematopoietic cells derived from patients with MDS or MDS-derived cell lines would reflect the real-life treatment efficacy in patients with MDS more rigorously.
In conclusion, we performed, to our knowledge, the first extensive kinome expression analysis in MDS and constructed the kinase-based risk score, KISS, that could not only robustly risk stratify patients with MDS but also infer novel therapeutic approaches. We also provided evidence that the KISS can be externally validated in 2 additional MDS cohorts, profiled by different gene expression quantification techniques (both RNA-seq and microarray). The KISS can serve as an independent adverse prognostic factor in MDS, outcompeting the previously reported Coxnet and LSC17 signatures. Prospective validation in larger MDS cohorts is anticipated to further establish the applicability of KISS in a wider population of patients with MDS.
Acknowledgments
The authors acknowledge service provided by the Department of Laboratory Medicine, Department of Medical Research, and Division of Hematology, Department of Internal Medicine, National Taiwan University Hospital.
This research was supported by National Taiwan University Hospital (111-L2005 & 112-L3005; C.-L.H.) and National Science and Technology Council (110-2221-E-002 -129 -MY3; C.-L.H.).
Authorship
Contribution: C.-Y.Y. was responsible for data collection and management, statistical and bioinformatic analysis, result interpretation, literature research, and manuscript writing; C.-C.L. and Y.-H.W. were responsible for data management and statistical analysis; C.-J.K., C.-H.T., H.-A.H., and H.-F.T. were responsible for data collection and management; C.-L.H., and W.-C.C. planned, designed, and coordinated the study over the entire period and wrote the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wen-Chien Chou, Department of Laboratory Medicine, National Taiwan University Hospital, No. 7, Chung-Shan South Rd, Taipei City 10002, Taiwan; email: wchou@ntu.edu.tw; and Chia-Lang Hsu, Department of Medical Research, National Taiwan University Hospital, No. 7, Chung Shan South Rd, Taipei City 10002, Taiwan; email: chialanghsu@ntuh.gov.tw.
References
Author notes
C.-L.H. and W.-C.C. are joint senior authors and contributed equally to this work.
The fastq files will be uploaded to the Gene Expression Omnibus database (accession number GSE223305).
The data reported in this article are available upon reasonable request from the corresponding author, Wen-Chien Chou (wchou@ntu.edu.tw).
The full-text version of this article contains a data supplement.