Somatic mutations were more frequently detected in patients with failure/resistance/progression than in optimal responders.
The presence of mutations, particularly in AS/MTF genes, is associated with worse outcomes.
Visual Abstract
Advancements in genomics are transforming the clinical management of chronic myeloid leukemia (CML) toward precision medicine. The impact of somatic mutations on treatment outcomes is still under debate. We studied the association of somatic mutations in epigenetic modifier genes and activated signaling/myeloid transcription factors (AS/MTFs) with disease progression and treatment failure in patients with CML after tyrosine kinase inhibitor (TKI) therapy. A total of 394 CML samples were sequenced, including 254 samples collected at initial diagnosis and 140 samples taken during follow-up. Single-molecule molecular inversion probe (smMIP)–based next-generation sequencing (NGS) was conducted targeting recurrently mutated loci in 40 genes, with a limit of detection of 0.2%. Seventy mutations were detected in 57 diagnostic samples (22.4%), whereas 64 mutations were detected in 39 of the follow-up samples (27.9%). Carrying any mutation at initial diagnosis was associated with worse outcomes after TKI therapy, particularly in AS/MTF genes. Patients having these mutations at initial diagnosis and treated with imatinib showed higher risks of treatment failure (hazard ratio, 2.53; 95% confidence interval, 1.13-5.66; P = .0239). The adverse prognostic impact of the mutations was not clear for patients treated with second-generation TKIs. The multivariate analysis affirmed that mutations in AS/MTF genes independently serve as adverse prognostic factors for molecular response, failure-free survival, and progression risk. Additionally, there was an observable nonsignificant trend indicating a heightened risk of progression to advanced disease and worse overall survival. In conclusion, mutations in the AS/MTF genes using smMIP-based NGS can help identify patients with a potential risk of both treatment failure and progression and may help upfront TKI selection.
Introduction
Increased genomics-based knowledge is currently transforming clinical practice in chronic myeloid leukemia (CML) into precision medicine–based management.1,2 Recently, several studies have characterized the genomic landscape of somatic mutations in CML, reporting that the presence of epigenetic modifier gene mutations is associated with poor treatment outcomes after tyrosine kinase inhibitor (TKI) therapy, particularly after imatinib treatment.3-6 A team of researchers indicated that there was a connection between somatic alterations in genes involved in epigenetic modifications, such as ASXL1, and the lack of response to imatinib treatment. However, this did not extend to second-generation TKI (2G-TKI) therapy,5 although there is a contradictory study reporting that ASXL1 mutation is associated with an inferior response to 2G-TKI, nilotinib.7
In terms of biological and functional relevance, it is not fully elucidated how epigenetic modifier gene mutations, such as an ASXL1 mutation, confer the development of TKI resistance in CML. Our group reported distinct patterns of mutation dynamics arising after TKI therapy in association with clinical outcomes. It was observed that after TKI therapy, the allele frequency of the ASXL1 mutation rapidly declined, but this did not correlate with clinical outcomes.3 Furthermore, we found that some patients showing optimal response did not experience any changes in the ASXL1 mutation burden, whereas other patients who failed TKI therapy demonstrated significant reductions in the ASXL1 mutation burden.3 This discrepant finding has raised the question of whether the ASXL1 mutation is a real driver of treatment failure in patients with CML or whether it is merely the detection of age-related mutations in epigenetic modifier genes that may predispose to the development of treatment failure.
Somatic mutations in activated signaling (AS) and myeloid transcription factor (MTF) genes have been associated with disease progression in diverse myeloid malignancies.8-14 Thus, it is plausible that those mutated genes are more potent drivers of treatment failure, resistance, and progression to the advanced phase in CML. Mutations in AS/MTF genes are strongly related with leukemic transformation of myelodysplastic syndrome (MDS).8,10-12,14 Particularly, preexisting or emerging RAS mutations are associated with a higher risk of blast progression in chronic myelomonocytic leukemia and myeloproliferative neoplasms.9,13,15 We hypothesized that somatic mutations in AS/MTF genes can provide prognostic value in the context of CML therapy. Thus, we have examined the impact of specific mutations on disease progression and treatment failure in patients with CML following TKI therapy.
Another question that has not been answered is whether the 2G-TKI therapy can overcome and abrogate adverse impact on treatment outcomes from somatic mutations in patients with CML.3,5,7 In this study, we evaluated the treatment outcomes of patients with CML according to the presence of any somatic mutations or mutations specifically in epigenetic modifier genes or in AS/MTF genes. By separately analyzing the outcomes between the imatinib-treated group vs the 2G-TKI–treated group, the impact of mutation profiles on treatment outcomes was determined according to the type of TKI drug administered.
Patients and methods
The study included 394 samples acquired from December 2000 to December 2020 from a cohort of 254 patients with CML who received treatment at 2 dedicated CML treatment facilities: the Princess Margaret Cancer Centre in Toronto, Canada, and University Hospital Brno in Brno, Czech Republic. Informed consent was obtained from all participating patients.
Broadly, patients’ samples can be categorized into groups 1 and 2 based on the time of sample collection. Group 1 includes 254 samples collected at initial diagnosis, whereas group 2 includes 140 samples taken during follow-up during TKI therapy, including 48 cases having pair or trio samples obtained serially from initial diagnosis and follow-up during TKI therapy. Frontline therapy included imatinib (n = 190 [75.3%]) or 2G-TKI (n = 64) including dasatinib (n = 15 [6.0%]), nilotinib (n = 37 [14.7%]), and bosutinib (n = 10 [4.0%]). The study was approved by the research ethics committees of the participant centers.
Somatic mutation profiling using the smMIP panel
Peripheral blood samples were processed for DNA extraction from mononuclear cell fractions. We applied a barcoded sequencing approach using single-molecule molecular inversion probe (smMIP)–based next-generation sequencing (NGS) with high coverage (∼10 000×) to improve detection of low-frequency mutations,16,17 typically below the intrinsic sequencing error rate of conventional NGS technology.18 This in-house smMIP panel covers 35.3 kb across 40 genes (supplemental Table 1A), including 12 AS genes, 5 MTF genes, 3 chromatin modifiers genes, 4 DNA methylation, 3 spliceosome, 3 tumor suppressor genes, 4 cohesion genes, and 6 “miscellaneous” genes (supplemental Table 1B).17
Assessment of clinical outcomes and statistical endpoints
The initial assessment of CML included bone marrow examination to evaluate blast percentage in the marrow, determining disease stage based on the European Leukemia Net criteria19,20: chronic phase with blast counts <15%, accelerated phase with blast counts 15% to 29%, and blastic phase with blast counts >30%. Peripheral blood BCR::ABL1 transcript data were extracted from available charts, and in both centers, the standard of care involved evaluating BCR::ABL1 by polymerase chain reaction every 3 months. The quantification followed international recommendations for standardizing the procedure.21 The BCR::ABL1 transcript levels were measured and presented according to the international scale (%IS) .22 The ABL1 kinase domain mutations were also determined in any of the resistant patients, regardless of disease stage.23 Response criteria were considered as previously defined.24-32 Briefly, molecular response (MR) with 2 log reduction (MR2), which is equivalent to complete cytogenetic response, was defined as ≤1%IS of BCR::ABL1 fusion gene transcripts, whereas major molecular response (MMR) was defined as ≤0.1%IS of BCR::ABL1 fusion gene transcripts. Molecular response with 4 log reduction (MR4) was defined as <0.01%ISBCR::ABL1 transcript level.19,20
Disease characteristics and treatment outcomes were evaluated and compared according to the somatic mutation profiles and biological pathways of the mutations. The treatment outcomes were evaluated concerning MMR, MR2, MR4, failure-free survival (FFS), progression, and overall survival (OS). FFS was defined as the interval between the start of TKI therapy and the event of treatment failure based on the ELN 2013 criteria19 including (1) loss of hematologic response, (2) loss of complete cytogenetic response, (3) development of ABL1 kinase domain mutation, (4) acquisition of additional cytogenetic abnormality (ACA) in Ph+ clone, and (5) progression to advanced phase CML or death from any cause. The patients were classified into resistant/progressed group or as optimal responder according to the response milestone based on the ELN recommendation.19
Statistical analysis
Patients, disease characteristics, and treatment outcomes are presented in Table 1. Treatment outcomes were analyzed and compared using χ2, Fisher exact, or Kruskal-Wallis tests as appropriate.
Summary of patient characteristics and treatment outcomes according to the presence of any mutation
| Patient’s characteristics . | Overall . | Overall patients (n = 254) . | P value . | Imatinib (n = 190) . | P value . | 2G-TKI (n = 64) . | P value . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutated . | No mutation . | Mutated . | No mutation . | Mutated . | No mutation . | ||||||
| No. of patients (%) | 254 | 57 | 197 | - | 44 | 146 | - | 13 | 51 | ||
| Age, median (range), y | 54 (17-82) | 53.5 (21-82) | 53 (17-82) | .993 | 53.5 (21-77) | 57 (17-82) | .43 | 64.5 (47-82) | 48 (17-81) | .273 | |
| Gender (female:male) | 111/143 (43.7/56.3) | 25/32 (43.8/56.2) | 86/111 (22.5/55.5) | .065 | 17/27 (21.4/78.6) | 66/79 (34.7/65.3) | .097 | 5/8 (38.4/61.5) | 18/31 (35.3/64.7) | .505 | |
| Sokal risk group | Low | 61 (24.0) | 10 (17.5) | 51 (25.5) | .202 | 9 (20.4) | 35 (24.0) | .329 | 1 (0) | 16 (31.4) | |
| Int | 84 (33.1) | 15 (26.3) | 69 (34.5) | 11 (25) | 53 (36.3) | 3 (0) | 16 (31.4) | .329 | |||
| High | 74 (29.1) | 19 (33.3) | 55 (27.5) | 13 (29.5) | 40 (27.3) | 6 (100) | 14 (27.4) | ||||
| Advanced phase | 21 (8.3) | 2 (12.5) | 19 (8.0) | .629 | 2 (14.3) | 16 (9.1) | .627 | 0 (0) | 2 (3.3) | 1.000 | |
| ACAs∗ | 32 (15.7) | 2 (18.2) | 30 (15.5) | .844 | 2 (22.2) | 21 (15.6) | .636 | 0 (0) | 8 (14.0) | 1.000 | |
| Response and long-term outcomes | |||||||||||
| MR2 at 12 mo | 75.4 ± 5.35% | 68.8 (54.6-79.3) | 77.4 (70.7-82.7) | .0283 | 61.8 (45.3-74.6) | 71.6 (63.4-78.3) | .0234 | 92.3 (35.8-99.4) | 93.8 (80.1-98.1) | .672 | |
| MMR at 3 y | 80.9 ± 4.9% | 66.7 (52.1-77.7) | 84.9 (78.8-89.4) | .0101 | 58.6 (41.9-72.0) | 82.3 (74.7-87.8) | .00758 | 92.3 (35.8-99.4) | 92.4 (76.6-97.7) | .787 | |
| MR4 at 5 y | 72.1 ± 5.5% | 59.2 (44.2-71.4) | 75.8 (68.8-81.4) | .0178 | 56.1 (39.0-70.0) | 73.1 (64.7-79.8) | .0231 | 70.3 (29.6-90.3) | 82.9 (67.1-91.5) | .446 | |
| FFS at 3 y | 73.3 ± 1.7% | 61.6 (47.3-73.1) | 76.8 (69.9-82.2) | .00336 | 59.6 (43.2-72.7) | 71.6 (63.2-78.5) | .0257 | 68.4 (35.9-86.8) | 91.6 (79.2-96.8) | .05 | |
| Progression at 5 y | 7.1 ± 1.1% | 9.3 (3.4-19.1) | 6.3 (3.3-10.6) | .0225 | 12.0 (4.3-23.9) | 7.5 (3.8-12.9) | .0227 | 0 | 2.9 (0.2-13.0) | .672 | |
| OS at 10 y | 70.6 ± 6.8% | 59.9 (42.9-73.4) | 74.3 (66.1-80.8) | .0997 | 56.9 (39.3-71.2) | 69.1 (59.4-76.9) | .167 | 80.0 (20.4-96.9) | 92.0 (77.0-97.4) | .772 | |
| Patient’s characteristics . | Overall . | Overall patients (n = 254) . | P value . | Imatinib (n = 190) . | P value . | 2G-TKI (n = 64) . | P value . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutated . | No mutation . | Mutated . | No mutation . | Mutated . | No mutation . | ||||||
| No. of patients (%) | 254 | 57 | 197 | - | 44 | 146 | - | 13 | 51 | ||
| Age, median (range), y | 54 (17-82) | 53.5 (21-82) | 53 (17-82) | .993 | 53.5 (21-77) | 57 (17-82) | .43 | 64.5 (47-82) | 48 (17-81) | .273 | |
| Gender (female:male) | 111/143 (43.7/56.3) | 25/32 (43.8/56.2) | 86/111 (22.5/55.5) | .065 | 17/27 (21.4/78.6) | 66/79 (34.7/65.3) | .097 | 5/8 (38.4/61.5) | 18/31 (35.3/64.7) | .505 | |
| Sokal risk group | Low | 61 (24.0) | 10 (17.5) | 51 (25.5) | .202 | 9 (20.4) | 35 (24.0) | .329 | 1 (0) | 16 (31.4) | |
| Int | 84 (33.1) | 15 (26.3) | 69 (34.5) | 11 (25) | 53 (36.3) | 3 (0) | 16 (31.4) | .329 | |||
| High | 74 (29.1) | 19 (33.3) | 55 (27.5) | 13 (29.5) | 40 (27.3) | 6 (100) | 14 (27.4) | ||||
| Advanced phase | 21 (8.3) | 2 (12.5) | 19 (8.0) | .629 | 2 (14.3) | 16 (9.1) | .627 | 0 (0) | 2 (3.3) | 1.000 | |
| ACAs∗ | 32 (15.7) | 2 (18.2) | 30 (15.5) | .844 | 2 (22.2) | 21 (15.6) | .636 | 0 (0) | 8 (14.0) | 1.000 | |
| Response and long-term outcomes | |||||||||||
| MR2 at 12 mo | 75.4 ± 5.35% | 68.8 (54.6-79.3) | 77.4 (70.7-82.7) | .0283 | 61.8 (45.3-74.6) | 71.6 (63.4-78.3) | .0234 | 92.3 (35.8-99.4) | 93.8 (80.1-98.1) | .672 | |
| MMR at 3 y | 80.9 ± 4.9% | 66.7 (52.1-77.7) | 84.9 (78.8-89.4) | .0101 | 58.6 (41.9-72.0) | 82.3 (74.7-87.8) | .00758 | 92.3 (35.8-99.4) | 92.4 (76.6-97.7) | .787 | |
| MR4 at 5 y | 72.1 ± 5.5% | 59.2 (44.2-71.4) | 75.8 (68.8-81.4) | .0178 | 56.1 (39.0-70.0) | 73.1 (64.7-79.8) | .0231 | 70.3 (29.6-90.3) | 82.9 (67.1-91.5) | .446 | |
| FFS at 3 y | 73.3 ± 1.7% | 61.6 (47.3-73.1) | 76.8 (69.9-82.2) | .00336 | 59.6 (43.2-72.7) | 71.6 (63.2-78.5) | .0257 | 68.4 (35.9-86.8) | 91.6 (79.2-96.8) | .05 | |
| Progression at 5 y | 7.1 ± 1.1% | 9.3 (3.4-19.1) | 6.3 (3.3-10.6) | .0225 | 12.0 (4.3-23.9) | 7.5 (3.8-12.9) | .0227 | 0 | 2.9 (0.2-13.0) | .672 | |
| OS at 10 y | 70.6 ± 6.8% | 59.9 (42.9-73.4) | 74.3 (66.1-80.8) | .0997 | 56.9 (39.3-71.2) | 69.1 (59.4-76.9) | .167 | 80.0 (20.4-96.9) | 92.0 (77.0-97.4) | .772 | |
Int, intermediate.
The mutation frequency was compared between patients with failure/progression vs other patients who achieved optimal response to TKI therapy using the χ2 test. The cumulative incidences of MMR, MR2, MR4, and progression were estimated using the cumulative incidence method considering competing risks,33 whereas FFS and OS were calculated using the Kaplan-Meier method. The cumulative incidences of MMR, MR2, and MR4 were calculated using dates from the start of frontline TKI therapy until the first day of MMR, MR2, and MR4 achievement throughout the TKI therapies; death or permanent TKI discontinuation due to intolerance/resistance were accounted for as competing events. Progression was defined as the interval between TKI therapy start and confirmation of progression to advanced phase (ie, accelerated or blastic phase); death from non-CML–related causes was accounted as a competing event. The OS was calculated from the TKI therapy start date until the date of death from any cause or date of last follow-up. Hazard ratio (HR) with a 95% confidence interval (CI) was calculated for risk factors using Cox proportional hazard regression models for FFS and OS or using the Fine-Gray model for MMR, MR2, MR4, and progression to advanced disease.
Logistic regression analysis was performed to evaluate the risk factors between patients who experienced failure/progression vs others with optimal response, whereas the odds ratio (OR) with 95% CI was calculated for risk factors including any mutations, mutations in the epigenetic modification pathway, mutations in AS/MTF, or other potential clinical risk factors.
The univariate analysis incorporated additional covariates alongside the somatic mutation profile, encompassing the Sokal risk group during initial diagnosis, disease stage (chronic phase vs advanced phase), type of TKI (imatinib vs 2G-TKI), and presence of ACAs at the outset of diagnosis. In conducting the multivariate analysis, 2 models were used: 1 encompassing the entire population regardless of disease stage, incorporating disease stage, TKI type, ACAs, and the mutational profile during initial diagnosis; the second model focused solely on chronic phase CML, comprising the Sokal risk group, TKI type, ACAs, and the mutational profile during initial diagnosis. The final model construction involved applying both enter mode and stepwise selection procedures, with only variables demonstrating a P value <.1 being included in the ultimate model. A statistical significance level of .05 was applied throughout the study. All statistical analyses were performed using the EZR software (Saitama Medical Center, Jichi Medical University, Saitama, Japan). EZR is a modified version of R commander (version 1.5.4), which is based on the R package.34
Results
Summary of patient characteristics, treatment, and clinical outcomes
The full description of patients' characteristics is summarized in Table 1. The median age of patients at initial diagnosis was 54 years, with a male-to-female ratio of 143:111 (56.3%:43.7%). The disease status at presentation included chronic (n = 233 [91.7%]), accelerated (n = 15 [5.9%]), blastic phase (n = 4 [1.6%]), or not available (n = 2 [0.8%]). The Sokal risk groups were categorized as low (n = 61 [24.0%]), intermediate (n = 84 [33.1%]), high (n = 74 [29.1%]), or not available (n = 35 [13.8%]). Frontline treatment consisted of imatinib (n = 190 [74.8%]) or 2G-TKI (n = 64 [25.2%]).
The treatment outcomes in the overall study population were as follows: the MR2 rate at 12 months was 75.4% (95% CI, 69.6-80.3); the MMR rate at 3 years, 80.9% (95% CI, 75.2-85.3); the MR4 rate at 5 years, 72.1% (95% CI, 65.9-77.5), the FFS rate at 3 years, 73.3% (95% CI, 67.1-78.4); the incidence of progression at 5 years, 7.1% (95% CI, 4.2-10.9); and OS rate at 10 years reached 70.6% (95% CI, 63.2-76.8). Of the 254 patients whose samples were obtained at the time of diagnosis, 89 patients had to switch TKI, with a total of 46 switching from those who initially received imatinib and 43 from those who were initially on 2G-TKI. The median duration until the first switch occurred was 304 days (range, 14-1870). The primary motivations for switching to a different TKI were identified as resistance in 46 cases and intolerance in 25 cases, whereas the reasons for the remaining cases were unknown.
Mutation profiles of newly diagnosed CML using the smMIP-based NGS
A total of 70 mutations were identified in 57 of the 254 samples (22.4%) at the initial diagnosis of CML. The most common mutations involved epigenetic modifier genes, including ASXL1 (n = 20 [7.9%]), followed by TET2 (n = 11 [4.3%]), DNMT3A (n = 6 [2.4%]), PHF6 (n = 4 [1.6%]), TP53 (n = 4 [1.6%]), FLT3 (n = 3 [1.2%]), IKZF1 (n = 3 [1.2%]), and WT1 (n = 2 [0.8%]). RUNX1 and ABL1 mutations that have known implications in disease progression and treatment resistance, respectively, were noted in 1 patient. When mutations were grouped according to the relevant biological pathways of the genes, genes in chromatin modifiers were the most frequently mutated (n = 23 [9.1%]), followed by DNA methylation (n = 17 [6.7%]), tumor suppressors (n = 10 [3.9%]), AS (n = 9 [3.5%]), MTF (n = 5 [2.0%]), and splicing (n = 1 [0.4%]).
Mutation profiles at initial diagnosis between failure/progression group and optimal responders
We have evaluated associations between patterns of mutations at initial diagnosis and TKI response. Of the 254 patients from whom we collected samples at diagnosis, 165 patients (65.0%) exhibited an optimal response, whereas 55 patients (21.7%) were resistant to TKI therapy, and 34 patients (13.4%) experienced progression. Among the 57 patients with at least 1 mutation, 29 patients were within the resistant/progression group (resistance, n = 17; progressed, n = 12), whereas 28 patients were optimal responders. The resistant/progression group showed a higher proportion of patients having at least 1 mutation (32.6%) than the optimal responders (10.3%; P = .007, by Fisher exact test; Figure 1A).
Frequency of mutations in patients with CML by TKI therapy response. Summary of mutation frequency at diagnosis (A) and follow-up sample (B). The mutations were classified according to their biological pathway.
Frequency of mutations in patients with CML by TKI therapy response. Summary of mutation frequency at diagnosis (A) and follow-up sample (B). The mutations were classified according to their biological pathway.
For the genes that were mutated in at least 5 patients (ie, ASXL1 [n = 20], TET2 [n = 11], and DNMT3A [n = 6]), we did not find significant differences in their mutation frequencies between the failure/progression group and the optimal responders (P = .632, 1.0, and .425, respectively). Next, we conducted the same analysis according to the following biological category designated to each gene (Figure 1B): frequency of mutations in epigenetic modifier genes including chromatin modifiers and DNA methylation genes were not significantly different between the patients in our cohort who experienced failure/progression vs optimal responders (P = .65 and .60, respectively). Due to the relatively small number of patients with AS and MTF genes (n = 9 and 5, respectively), these 2 groups were combined. Mutations in those 2 pathways were more frequently noted in the failure/progression group than in optimal responders (9/89 [10%] vs 4/165 [2.4%]; P = .01, by Fisher exact test). When these 2 mutations in AS and MTF genes were separately analyzed, it still showed a statistical trend toward higher frequency of these mutations in the failure/progression group (P = .07 and .05, respectively).
As shown in Table 2 and supplemental Figure 1, logistic regression analysis showed that regardless of the mutations’ categories, any mutations were 2.78-times more frequently detected in the failure/progression group than in optimal responders (P = .00861; OR, 2.78; 95% CI, 1.3-5.97). Although the frequency of mutations in the epigenetic modification genes was not different between the 2 groups, mutations in the AS/MTFs were 4.04-times more frequently detected in the failure/progression group than the other group (P = .034; OR, 4.04; 95% CI, 1.11-14.7). In addition, the adverse impacts of mutations in AS/MTFs were significant in patients treated with imatinib (P = .036; OR, 6.02; 95% CI, 1.12-32.4). A similar trend was noted when the analysis was restricted to patients in chronic phase (n = 190; supplemental Figure 2).
Univariate and multivariate logistic regression analysis for the impact of somatic mutations between progression/treatment failure group vs optimal responder to TKI therapy in overall population of 254 patients with CML
| Any mutation . | Univariate . | Multivariate, enter . | Multivariate, stepwise . | |||
|---|---|---|---|---|---|---|
| P value . | OR . | P value . | OR . | P value . | OR . | |
| Any mutation∗ | ||||||
| Mutation∗ | .00503 | 2.36 (1.30-4.32) | .00861 | 2.78 (1.300-5.970) | .0173 | 2.36 (1.160-4.800) |
| ACA | .0898 | 1.95 (0.901-4.23) | .416 | 1.57 (0.526-4.717) | - | |
| Disease phase | .983 | 1.03 × 108 (0.000-Inf) | .985 | 1.67 × 108 (0.000-Inf) | - | |
| TKI type | .00214 | 0.339 (0.170-0.676) | .0101 | 0.328 (0.141-0.767) | .00508 | 0.333 (0.154-0.718) |
| Mutation in epigenetic modification pathway† | ||||||
| Mutation† | .535 | 1.25 (0.616-2.54) | .541 | 1.34 (0.527-3.390) | - | |
| ACA | .0898 | 1.95 (0.901-4.23) | .544 | 1.40 (0.476-4.10) | - | |
| Disease phase | .983 | 1.03 × 108 (0.000-Inf) | .986 | 1.43 × 108 (0.000-Inf) | - | |
| TKI type | .00214 | 0.339 (0.170-0.676) | .012 | 0.344 (0.150-0.791) | .00576 | 0.343 (0.161-0.733) |
| Mutation in AS and MTF‡ | ||||||
| Mutation‡ | .0235 | 3.35 (1.18-9.56) | .0646 | 3.70 (0.924-14.8) | .0345 | 4.04 (1.11-14.7) |
| ACA | .0898 | 1.95 (0.901-4.23) | .598 | 1.34 (0.455-3.92) | — | |
| Disease phase | .983 | 1.03 × 108 (0.000-Inf) | .986 | 1.29 × 108 (0.000-Inf) | — | |
| TKI type | .00214 | 0.339 (0.170-0.676) | .014 | 0.349 (0.151-0.808) | .00754 | 0.352 (0.164-0.757) |
| Any mutation . | Univariate . | Multivariate, enter . | Multivariate, stepwise . | |||
|---|---|---|---|---|---|---|
| P value . | OR . | P value . | OR . | P value . | OR . | |
| Any mutation∗ | ||||||
| Mutation∗ | .00503 | 2.36 (1.30-4.32) | .00861 | 2.78 (1.300-5.970) | .0173 | 2.36 (1.160-4.800) |
| ACA | .0898 | 1.95 (0.901-4.23) | .416 | 1.57 (0.526-4.717) | - | |
| Disease phase | .983 | 1.03 × 108 (0.000-Inf) | .985 | 1.67 × 108 (0.000-Inf) | - | |
| TKI type | .00214 | 0.339 (0.170-0.676) | .0101 | 0.328 (0.141-0.767) | .00508 | 0.333 (0.154-0.718) |
| Mutation in epigenetic modification pathway† | ||||||
| Mutation† | .535 | 1.25 (0.616-2.54) | .541 | 1.34 (0.527-3.390) | - | |
| ACA | .0898 | 1.95 (0.901-4.23) | .544 | 1.40 (0.476-4.10) | - | |
| Disease phase | .983 | 1.03 × 108 (0.000-Inf) | .986 | 1.43 × 108 (0.000-Inf) | - | |
| TKI type | .00214 | 0.339 (0.170-0.676) | .012 | 0.344 (0.150-0.791) | .00576 | 0.343 (0.161-0.733) |
| Mutation in AS and MTF‡ | ||||||
| Mutation‡ | .0235 | 3.35 (1.18-9.56) | .0646 | 3.70 (0.924-14.8) | .0345 | 4.04 (1.11-14.7) |
| ACA | .0898 | 1.95 (0.901-4.23) | .598 | 1.34 (0.455-3.92) | — | |
| Disease phase | .983 | 1.03 × 108 (0.000-Inf) | .986 | 1.29 × 108 (0.000-Inf) | — | |
| TKI type | .00214 | 0.339 (0.170-0.676) | .014 | 0.349 (0.151-0.808) | .00754 | 0.352 (0.164-0.757) |
Inf, infinite.
Any mutation.
Mutation in epigenetic modification pathway.
Mutation in AS and myeloid transcription factor.
In summary, the presence of somatic mutations at initial diagnosis, particularly in AS/MTFs, is associated with a higher risk of treatment failure/resistance and progression in patients with CML, particularly after imatinib therapy (supplemental Tables 2A and 2B).
Treatment response and long-term outcomes after TKI therapy according to mutation profiles at initial diagnosis
We have evaluated the prognostic impact of the mutation profiles at initial diagnosis concerning 6 different treatment outcome parameters including response parameters (ie, MR, MMR, and MR4), treatment failure (ie, FFS and progression), and OS, across all lines of TKI therapies (Table 1).
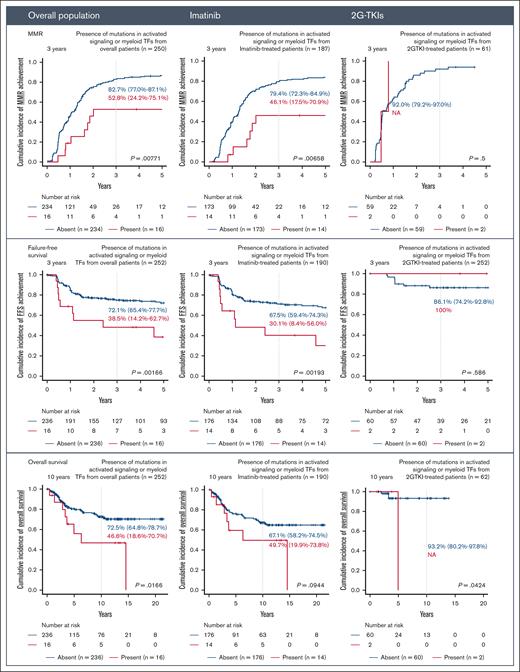
Carrying any mutations at the time of initial diagnosis was associated with worse outcomes after TKI therapy, particularly in those treated with imatinib but not in those who received 2G-TKI therapy as shown in Figure 2. When treated with imatinib, patients with mutations showed lower incidence of FFS (3 years: 59.5% [95% CI, 43.2-72.7] vs 71.6% [95% CI, 63.2-78.5]; P = .0257), lower incidence of MMR achievement (3 years: 58.6% [95% CI, 41.9-73] vs 82.3% [95% CI, 74.7-87.8]; P = .00758), and lower OS (3 years: 58.5% [95% CI, 41.9-72] vs 82.3% [95% CI, 74.7-87.8]; P = .00758). The adverse impacts of somatic mutations were mainly derived from mutations in AS/MTF genes (Figure 3). The adverse prognostic impact of these mutations was not confirmed in patients who received 2G-TKI (supplemental Figure 3).
Impact of the presence of any somatic mutation on treatment outcomes after TKI therapy. Treatment outcomes include MMR, FFS, and OS. Results are shown for the overall population and imatinib-treated and 2G-TKI–treated groups.
Impact of the presence of any somatic mutation on treatment outcomes after TKI therapy. Treatment outcomes include MMR, FFS, and OS. Results are shown for the overall population and imatinib-treated and 2G-TKI–treated groups.
Impact of the presence of mutations in AS/MTF genes on treatment outcomes after TKI therapy. Treatment outcomes include MMR, FFS, and OS. Results are shown for the overall population and imatinib-treated and 2G-TKI–treated groups.
Impact of the presence of mutations in AS/MTF genes on treatment outcomes after TKI therapy. Treatment outcomes include MMR, FFS, and OS. Results are shown for the overall population and imatinib-treated and 2G-TKI–treated groups.
After considering other confounding covariates, multivariate analyses were conducted to validate that the mutation profile is an independent predictive and prognostic factor for treatment outcomes. Any mutations detected at initial diagnosis showed a statistical trend toward a higher risk of FFS (P = .06; HR, 1.69; 95% CI, 0.977-2.94), although it was not confirmed to be an independent risk factor for MR2 (P = .19), MMR (P = .32), MR4 (P = .37), progression (P = .57), and OS (P = .86; supplemental Figure 4).
When the analysis was restricted to the AS/MTF mutations (Table 3; supplemental Figure 5), the presence of such mutations was confirmed to be an independent risk factor for decreased MR2 (P = .039; HR, 0.449; 95% CI, 0.210-0.963), MMR (P = .023; HR, 0.416; 95% CI, 0.195-0.888), MR4 (P = .024; HR, 0.447; 95% CI, 0.222-0.900), and worse FFS (P = .0239; HR, 2.53; 95% CI, 1.13-5.66), while showing a trend toward a higher risk of progression to advanced disease (P = .27; HR, 2.05; 95% CI, 0.57-7.39) with worse OS (P = .235; HR, 1.77; 95% CI, 0.689-4.58).
Univariate and multivariate Cox or Fine-gray analysis for the impact of mutations in AS and MTF on the treatment outcomes with respect to MMR, MR2, MR4, FFS, progression, and OS after TKI therapy in patients with CML
| . | Univariate . | Multivariate, enter . | Multivariate, stepwise . | |||
|---|---|---|---|---|---|---|
| P value . | HR . | P value . | HR . | P value . | HR . | |
| MR2 | ||||||
| Mutation | .043 | 0.485 (0.241-0.976) | .04 | 0.449 (0.210-0.963) | .039 | 0.449 (0.210-0.960) |
| ACA | .0015 | 0.478 (0.304-0.754) | .024 | 0.603 (0.390-0.934) | .0025 | 0.522 (0.342-0.795) |
| Disease phase | .00011 | 0.307 (0.169-0.560) | .12 | 0.603 (0.318-1.14) | - | |
| TKI type | .0000019 | 2.34 (1.65-3.33) | .003 | 1.81 (1.23-2.68) | .0026 | 1.82 (1.23-2.68) |
| MMR | ||||||
| Mutation | .0047 | 0.364 (0.180-0.733) | .023 | 0.416 (0.195-0.888) | .03 | 0.421 (0.193-0.919) |
| ACA | .0021 | 0.484 (0.304-0.769) | .068 | 0.654 (0.414-1.03) | - | |
| Disease phase | .000052 | 0.274 (0.146-0.513) | .02 | 0.440 (0.220-0.881) | .002 | 0.341 (0.172-0.674) |
| TKI type | .000056 | 1.97 (1.42-2.74) | .045 | 1.46 (1.01-2.12) | .021 | 1.53 (1.07-2.19) |
| MR4 | ||||||
| Mutation | .012 | 0.403 (0.198-0.819) | .024 | 0.447 (0.222-0.900) | .018 | 0.429 (0.212-0.866) |
| ACA | .016 | 0.516 (0.302-0.882) | .061 | 0.575 (0.322-1.03) | .017 | 0.522 (0.306-0.889) |
| Disease phase | .0052 | 0.367 (0.181-0.741) | .44 | 0.736 (0.340-1.60) | - | |
| TKI type | .026 | 1.46 (1.05-2.02) | .77 | 1.06 (0.734-1.52) | - | |
| FFS | ||||||
| Mutation | .00261 | 2.77 (1.43-5.39) | .0239 | 2.53 (1.13-5.66) | .0238 | 2.53 (1.13-5.66) |
| ACA | .278 | 1.43 (0.749-2.74) | .984 | 0.993 (0.493-2.00) | - | |
| Disease phase | .00000321 | 3.99 (2.23-7.13) | .00279 | 3.34 (1.51-7.35) | .00132 | 3.33 (1.60-6.92) |
| TKI type | .00163 | 0.309 (0.149-0.641) | .00675 | 0.334 (0.151-0.738) | .00675 | 0.334 (0.151-0.738) |
| Progression | ||||||
| Mutation | .012 | 2.481 (1.221-5.050) | .27 | 2.049 (0.571-7.407) | - | |
| ACA | .016 | 0.516 (0.302-0.882) | .021 | 3.05 (1.19-7.85) | .035 | 2.79 (1.07-7.24) |
| Disease phase | .0052 | 0.367 (0.181-0.741) | .000012 | 7.17 (2.97-17.3) | .0000087 | 8.13 (3.23-20.5) |
| TKI type | .026 | 1.46 (1.05-2.02) | .20 | 0.261 (0.0342-2.00) | - | |
| OS | ||||||
| Mutation | .0204 | 2.42 (1.15-5.11) | .235 | 1.78 (0.689-4.58) | - | |
| ACA | .0865 | 1.87 (0.914-3.83) | .434 | 1.36 (0.630-2.94) | - | |
| Disease phase | .0000508 | 3.73 (1.97-7.06) | .0211 | 2.82 (1.17-6.79) | .00424 | 3.30 (1.46-7.47) |
| TKI type | .00557 | 0.237 (0.0856-0.656) | .0076 | 0.143 (0.0343-0.597) | .00648 | 0.138 (0.0332-0.574) |
| . | Univariate . | Multivariate, enter . | Multivariate, stepwise . | |||
|---|---|---|---|---|---|---|
| P value . | HR . | P value . | HR . | P value . | HR . | |
| MR2 | ||||||
| Mutation | .043 | 0.485 (0.241-0.976) | .04 | 0.449 (0.210-0.963) | .039 | 0.449 (0.210-0.960) |
| ACA | .0015 | 0.478 (0.304-0.754) | .024 | 0.603 (0.390-0.934) | .0025 | 0.522 (0.342-0.795) |
| Disease phase | .00011 | 0.307 (0.169-0.560) | .12 | 0.603 (0.318-1.14) | - | |
| TKI type | .0000019 | 2.34 (1.65-3.33) | .003 | 1.81 (1.23-2.68) | .0026 | 1.82 (1.23-2.68) |
| MMR | ||||||
| Mutation | .0047 | 0.364 (0.180-0.733) | .023 | 0.416 (0.195-0.888) | .03 | 0.421 (0.193-0.919) |
| ACA | .0021 | 0.484 (0.304-0.769) | .068 | 0.654 (0.414-1.03) | - | |
| Disease phase | .000052 | 0.274 (0.146-0.513) | .02 | 0.440 (0.220-0.881) | .002 | 0.341 (0.172-0.674) |
| TKI type | .000056 | 1.97 (1.42-2.74) | .045 | 1.46 (1.01-2.12) | .021 | 1.53 (1.07-2.19) |
| MR4 | ||||||
| Mutation | .012 | 0.403 (0.198-0.819) | .024 | 0.447 (0.222-0.900) | .018 | 0.429 (0.212-0.866) |
| ACA | .016 | 0.516 (0.302-0.882) | .061 | 0.575 (0.322-1.03) | .017 | 0.522 (0.306-0.889) |
| Disease phase | .0052 | 0.367 (0.181-0.741) | .44 | 0.736 (0.340-1.60) | - | |
| TKI type | .026 | 1.46 (1.05-2.02) | .77 | 1.06 (0.734-1.52) | - | |
| FFS | ||||||
| Mutation | .00261 | 2.77 (1.43-5.39) | .0239 | 2.53 (1.13-5.66) | .0238 | 2.53 (1.13-5.66) |
| ACA | .278 | 1.43 (0.749-2.74) | .984 | 0.993 (0.493-2.00) | - | |
| Disease phase | .00000321 | 3.99 (2.23-7.13) | .00279 | 3.34 (1.51-7.35) | .00132 | 3.33 (1.60-6.92) |
| TKI type | .00163 | 0.309 (0.149-0.641) | .00675 | 0.334 (0.151-0.738) | .00675 | 0.334 (0.151-0.738) |
| Progression | ||||||
| Mutation | .012 | 2.481 (1.221-5.050) | .27 | 2.049 (0.571-7.407) | - | |
| ACA | .016 | 0.516 (0.302-0.882) | .021 | 3.05 (1.19-7.85) | .035 | 2.79 (1.07-7.24) |
| Disease phase | .0052 | 0.367 (0.181-0.741) | .000012 | 7.17 (2.97-17.3) | .0000087 | 8.13 (3.23-20.5) |
| TKI type | .026 | 1.46 (1.05-2.02) | .20 | 0.261 (0.0342-2.00) | - | |
| OS | ||||||
| Mutation | .0204 | 2.42 (1.15-5.11) | .235 | 1.78 (0.689-4.58) | - | |
| ACA | .0865 | 1.87 (0.914-3.83) | .434 | 1.36 (0.630-2.94) | - | |
| Disease phase | .0000508 | 3.73 (1.97-7.06) | .0211 | 2.82 (1.17-6.79) | .00424 | 3.30 (1.46-7.47) |
| TKI type | .00557 | 0.237 (0.0856-0.656) | .0076 | 0.143 (0.0343-0.597) | .00648 | 0.138 (0.0332-0.574) |
Overall, samples from 254 patients were analyzed.
Similar findings were also consistently observed when the analysis was restricted to patients treated with imatinib: MR2 (P = .019; HR, 0.325; 95% CI, 0.127-0.829), MMR (P = .0057; HR, 0.286; 95% CI, 0.118-0.694), MR4 (P = .0074; HR, 0.325; 95% CI, 0.143-0.740), and FFS (P = .0146; HR, 2.76; 95% CI, 1.22-6.24). However, such trends were not noted in patients treated with 2G-TKIs.
Mutation profiles at follow-up and its comparison between the failure/progression group and others
A total of 140 samples taken during TKI treatment were sequenced. Of the 140 samples taken after TKI therapy, 39 samples (n = 39 [27.9%]) were obtained from patients who had progressed or were resistant to TKI therapy, and the remaining 101 samples (n = 101 [72.1%]) were obtained from optimal responders. Overall, 64 mutations were identified (Figure 1B). The most frequently mutated gene at follow-up was DNMT3A (n = 13 [9.3%]), followed by TET2 (n = 12 [8.6%]), ASXL1 (n = 9 [6.4%]), RUNX1 (n = 6 [4.3%]), and ABL1 (n = 5 [3.6%]). After regrouping according to the previously mentioned biological category, 21 patients carried mutations in DNA methylation category and 11 patients in chromatin modifier genes, and the frequency of patients carrying mutation in AS/MTF genes was 12 and 6 (8.6% and 4.3%), respectively.
When evaluating the occurrences of mutations according to responses to TKI therapy, a significantly higher number of mutations were detected in the failure/progression group than in optimal responders (17/39 [44%] vs 22/101 [22%]; P = .01, Fisher exact test). This result is in line with the result obtained through the analysis of diagnostic samples. As evaluated in the diagnostic samples, in these follow-up samples, there was no statistical significance regarding mutation frequencies in genes associated with epigenetic modifier genes, including between the failure/progression group and optimal responders (P = 1.0, .74, and .27, respectively). On the contrary, ABL1 mutations were carried by 5 patients with failure/progression, whereas ABL1 mutations were not found in any of the optimal responders (5/39 [13%] vs 0/101 [0%]; P = .001, Fisher exact test). RUNX1 mutations were also observed nearly exclusively in the failure/progression group (5/39 [13%] vs 1/101 [1%]). We observed significantly higher occurrences of mutations in the AS/MTF genes (12/39 [31%] vs 4/101 [4%]; P < .001, Fisher exact test), similar to the mutation pattern observed in the diagnostic samples.
Changes in the allelic burden from initial diagnosis to follow-up
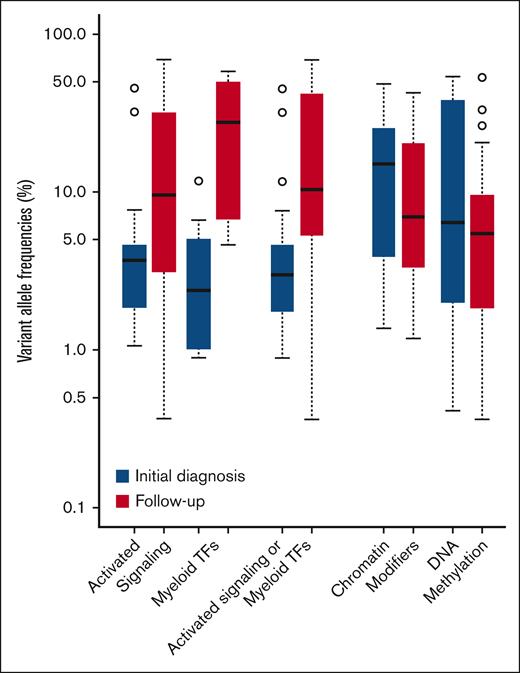
We compared variants alleles frequencies (VAFs) of mutations samples obtained either before or after TKI therapy. Mutations in AS/MTFs were found at significantly higher VAF in patients’ follow-up samples than in the samples collected at initial diagnosis (P = .003, by t test). The mean VAF was elevated to 22.1% at follow-up (range, 0.4%-69.2%) from 6.8% at diagnosis (range, 0.8%-45.6%; Figure 4). On the contrary, VAFs between these 2 time points were not statistically significant for mutations in chromatin modifiers or DNA methylation pathways (P = .35 and .09, respectively, by t test). For mutations in the chromatin modifier pathway, mean allele frequencies at diagnosis and follow-up were 17.3% (range, 1.4%-49.0%) and 12.0% (range, 1.2%-42.6%), respectively. For mutations in DNA methylation, mean allele frequencies at diagnosis and follow-up were 20.0% (range, 0.4%-54.6%) and 9.1% (range, 0.3%-53.7%), respectively. Despite being based on nonpaired samples, we observed significantly higher VAFs of mutations in AS/MTFs in patients’ follow-up samples, whereas VAFs in the epigenetic modifier gene mutations remained stable after TKI therapy. These results further implicate AS/MTF mutations as drivers of poor outcomes of patients with CML.
Longitudinal comparison of allele burdens change according to the biologic pathway of somatic mutation between diagnosis and follow-up samples.
Longitudinal comparison of allele burdens change according to the biologic pathway of somatic mutation between diagnosis and follow-up samples.
Discussion
This study reports that (1) somatic mutations were more frequently detected in patients with failure/resistance/progression than in optimal responders both at initial diagnosis and follow-up; and (2) mutations in AS/MTFs greatly contributed to the adverse impact on the outcomes after TKI therapy, particularly with imatinib.
Clonal evolution is responsible for the progression to advanced disease phase and treatment failure, which include the acquisition of new, additional mutations in the leukemic clone.35,36 When clonal evolution occurs, the leukemic clone with a growth advantage accelerates its survival pathway and repopulates to become the predominant clone. During this process, the clone becomes more resistant to treatment and biologically more aggressive.37 Accordingly, the acquisition of an additional mutation in the leukemic clone implies that the treatment outcome would be worse than in those without such a mutation. However, different types of gene mutations would have different biological and functional relevance on clonal evolution and treatment outcomes. It is still not fully elucidated which biological pathways and gene mutations are responsible for the development of treatment failure and disease progression in CML. Mutations in AS/MTFs would be a strong candidate as the driver of treatment failure, resistance, and progression to the advanced phase. We hypothesized that mutations in the AS/MTFs would be associated with treatment failure/resistance/progression, and the clone carrying a mutation in the AS/MTFs could expand in those who failed TKI therapy, progressed to advanced disease, or both. Although a paired analysis was not performed, mutations in the epigenetic pathway did not show any significant changes in allele frequency. Single-cell sequencing study in paired samples would help reach a clearer conclusion to this question.
Progression of leukemia requires an acceleration in cellular growth and differentiation processes. This cellular pathway includes those involved in DNA transcription or signal transduction. For the AS pathway, several permutations of RAS (ie, N-RAS, K-RAS, and H-RAS) were reported to be mutated in MDS and acute myeloid leukemia.38 Normal cellular growth and differentiation are regulated by other important signaling pathways, which can be also mutated. For instance, mutations leading to abnormal and overactive tyrosine kinase JAK/STAT signaling and ensuing myelopoiesis have been implicated in a small subset of MDS,39 similar to the mitogen-activated protein and phosphatidylinositol-3 kinase (PI3K)/AKT/mTOR pathways. As was in MDS, this study observed clues indicating that mutations in the AS pathway could be associated with worse treatment outcomes and the progression of chronic phase CML into a more advanced disease phase.
Transcriptional dysregulation arises in cancer from disease-defining genetic alterations via mutation of signaling factors converging on transcriptional control or via genetic alterations to control transcription.40-42 It has been known that genes regulated by transcriptional regulators, such as HNF4A, RICTOR, E2F1, MYC, MYCN, and RB1, all major controllers of cell growth/cell cycle,43,44 showed significant enrichment for aberrantly spliced genes associated with splicing factor gene mutations in MDS.45 Interestingly, HNF4A inhibition promotes tumorigenesis in solid cancers as well.46 In this study, a strong signal alarming the association of mutations in myeloid MTF toward worse treatment outcomes in patients with CML was also captured.
In contrast to our previous report and other results that suggest the epigenetic modification pathway mutations would correlate with poor outcomes,3 we could not find any statistically significant association between the epigenetic modification pathway mutations and poor treatment outcomes. Different from our previous study or from others, this study adopted a barcoded, error-corrected sequencing method that can detect any mutations with very low allele frequencies down to 0.2%,16,17 whereas conventional NGS has about a 5% limit of detection. At the time of initial diagnosis of CML, mutations in AS/MTFs may not be a predominant clone and would be too minor of a clone to be captured with conventional NGS. While TKI therapy continues, this clone can expand due to its survival advantage over other clones, thus becoming a predominant clone and resulting in treatment failure or progression to advanced disease stage.37 Accordingly, more sensitive and accurate NGS technology such as our smMIP panel can help capture such low allele frequency mutations even at the time of initial diagnosis of CML.17 Therefore, we suggest applying barcoded, error-corrected, NGS methodology for molecular diagnostics in CML, which can capture very low allele frequency mutations at initial diagnosis that could be easily missed by conventional NGS.
In summary, this study demonstrated that mutation profiling using smMIP-based NGS can help identify patients with a potential adverse prognosis from initial diagnosis and may help upfront TKI selection. Overall, our data and analyses suggest that the presence/emergence of any mutations, particularly in MTFs and AS pathways, need to be monitored closely in clinical practice. Further follow-up analysis is ongoing to evaluate the effect of TKI therapy on the longitudinal kinetics of mutations in serially collected samples.
Acknowledgments
This study was supported by the Princess Margaret Cancer Foundation, Toronto, ON, Canada, a Pfizer quality improvement grant, and a research grant from Paladin, Canada.
Authorship
Contribution: D.D.H.K. contributed to study conceptualization, study design, data collection, data analysis and interpretation, writing of the manuscript, and administrative support; M.A.P. contributed to data analysis and interpretation, writing of the manuscript, and administrative support; D.Ž. also contributed to the study design, data collection and analysis, critical revision of the manuscript, and administrative support; T.K. was involved in data analysis, data interpretation, and the manuscript's critical revision; K.P. and C.P. were involved in the study design, data analysis, critical revision of the manuscript, and data collection; I.J., A.K., T.J., J.K., Y.Y., S.Y., H.L., K.K., J.-M.C.-C., A.A., M.M., and M.C. were involved in data analysis, data interpretation, and the manuscript's critical revision; Z.Z. contributed to data analysis, interpretation of data, and critical revision of the manuscript; J.M. contributed to study design, data collection, data analysis and interpretation, writing of the manuscript, and administrative support; and J.J.F.M. and S.A. were involved in sequencing using the smMIP panel, data analysis, interpretation of data, and critical revision of the manuscript.
Conflict-of-interest disclosure: D.D.H.K. received honoraria from Bristol Myers Squibb and Novartis, and is involved in consultancy for Novartis, Pfizer, and Paladin. The remaining authors declare no competing financial interests.
Correspondence: Dennis Dong Hwan Kim, Princess Margaret Cancer Centre, University of Toronto, 610 University Ave, OPG Room 6-222, Toronto, ON, Canada M5G 2M9; email: dr.dennis.kim@uhn.ca.
References
Author notes
M.A.P. and D.Ž. contributed to the work equally as joint first authors.
J.M., S.A., and D.D.H.K. contributed to the work equally as joint senior authors.
The data have been deposited in the European Nucleotide Archive under the accession project number PRJEB73516 and the submission number ERA29258610.
Data are available for sharing upon reasonable request from the corresponding author, Dennis Dong Hwan Kim (dr.dennis.kim@uhn.ca).
The full-text version of this article contains a data supplement.