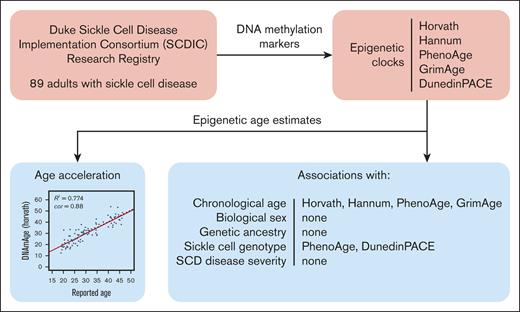
Chronological age and sickle cell genotype were associated with specific epigenetic age clocks in individuals with SCD.
Later generation epigenetic age clocks demonstrate age acceleration in SCD, whereas older clocks do not.
Visual Abstract
Sickle cell disease (SCD) affects ∼100 000 predominantly African American individuals in the United States, causing significant cellular damage, increased disease complications, and premature death. However, the contribution of epigenetic factors to SCD pathophysiology remains relatively unexplored. DNA methylation (DNAm), a primary epigenetic mechanism for regulating gene expression in response to the environment, is an important driver of normal cellular aging. Several DNAm epigenetic clocks have been developed to serve as a proxy for cellular aging. We calculated the epigenetic ages of 89 adults with SCD (mean age, 30.64 years; 60.64% female) using 5 published epigenetic clocks: Horvath, Hannum, PhenoAge, GrimAge, and DunedinPACE. We hypothesized that in chronic disease, such as SCD, individuals would demonstrate epigenetic age acceleration, but the results differed depending on the clock used. Recently developed clocks more consistently demonstrated acceleration (GrimAge, DunedinPACE). Additional demographic and clinical phenotypes were analyzed to explore their association with epigenetic age estimates. Chronological age was significantly correlated with epigenetic age in all clocks (Horvath, r = 0.88; Hannum, r = 0.89; PhenoAge, r = 0.85; GrimAge, r = 0.88; DunedinPACE, r = 0.34). The SCD genotype was associated with 2 clocks (PhenoAge, P = .02; DunedinPACE, P < .001). Genetic ancestry, biological sex, β-globin haplotypes, BCL11A rs11886868, and SCD severity were not associated. These findings, among the first to interrogate epigenetic aging in adults with SCD, demonstrate epigenetic age acceleration with recently developed epigenetic clocks but not older-generation clocks. Further development of epigenetic clocks may improve their predictive ability and utility for chronic diseases such as SCD.
Introduction
Sickle cell disease (SCD) is a progressive and complex chronic disease characterized by abnormally shaped red blood cells that adhere to blood vessel walls, ultimately resulting in vaso-occlusion of the microcirculation, subsequent ischemia, and organ damage.1 The complex pathophysiology of SCD includes elements of inflammation and adhesion that contribute to significant cellular damage over time,2 increased disease complications, and ultimately premature death (median age = 43 years).3 Previous reviews have highlighted the mechanisms of SCD pathophysiology through associations with genetic loci.4,5 However, the contribution of epigenetic factors to SCD pathophysiology is relatively unexplored despite the presence of age-related disparities in SCD, such as advanced clinical presentation and early mortality.6 The mean survival for individuals with SCD remains lower than that of individuals without SCD.7,8 These observed disparities, in combination with exposure to environmental stressors and the multisystem pathophysiologic involvement of SCD, suggest that accelerated epigenetic aging processes occur in this population. In particular, aging and early mortality in people with SCD may be informed by measuring epigenetic alterations.
One of the key hallmarks of normal biological aging is epigenetic alteration.9 DNA methylation (DNAm), a primary epigenetic mechanism for regulating gene expression,10 is an important driver of cellular aging.9,11 Epigenetic aging is determined by assessing the levels of DNAm at genome-wide Cytosine-phosphate-Guanine (CpG) loci.10 Using these CpG loci, several epigenetic clocks have been developed to calculate epigenetic age, including the Horvath,10 Hannum,12 PhenoAge,13 GrimAge,14 and DunedinPACE15 clocks. These clocks generate estimates of “epigenetic” age given a sample’s methylation profile. Epigenetic age acceleration or deceleration can then be estimated from the difference between the epigenetically predicted age and the chronological age (determined by the date of birth). Accelerated aging is associated with poor health outcomes and is a strong predictor of mortality.16-18 Putatively, this is a result of the accumulation of cellular damage and environmental exposure (eg, living conditions and chronic stress) that alter DNAm and accelerate the pace of cellular aging, ultimately causing molecular changes (eg, immune activation and inflammation, oxidative stress, and mitochondrial dysfunction).13,19,20 Global measures of epigenetic age and epigenetic age acceleration, rather than methylation at a single locus, provide a more comprehensive view of DNAm patterns, which are critical when investigating complex symptoms in complex conditions such as SCD.
Each epigenetic clock has been developed for different purposes and outcomes. The “first-generation” clocks, Horvath and Hannum, estimate chronological age.10,12 The “second-generation” clocks, PhenoAge and GrimAge, estimate composite measures that incorporate biomarkers alongside chronological age.13,14 Notably, GrimAge incorporates cystatin-C, which is associated with SCD nephropathy and early mortality.21 The “third-generation” clock, DunedinPACE, uses longitudinal methylation data to produce an age-adjusted rate of aging as opposed to epigenetic age.15 These clocks assess methylation at disparate age-specific CpG loci, likely capturing different components of aging.18,22
Overall, the baseline for the evaluation of epigenetic age acceleration is healthy and nonage-accelerated individuals. These clocks are calibrated using methylation data from healthy individuals without chronic disease. Analyses of epigenetic aging in nonhealthy individuals have been performed in other disease contexts, yielding accelerated epigenetic age estimates.16 These observations underpin our hypothesis that individuals with SCD would demonstrate age acceleration. Although SCD is an age-dependent chronic disease, its effects on global methylation patterns remain unknown, motivating this exploratory analysis of an SCD cohort.
Importantly, the cohorts used to develop these clocks did not match the genetic and ancestral backgrounds of the SCD cohorts. Of the 82 cohorts used in the Horvath clock, only 2 contained individuals of African ancestry.10 The Hannum clock used a mixed Caucasian and Hispanic cohort,12 PhenoAge used an Italian population,13 GrimAge used the Framingham Heart Study (majority European descent),14 and DunedinPACE used the Dunedin study (European descent).15 Moreover, genetic ancestry is known to be associated with epigenetic age estimates.23 This highlights the need to assess individuals that closely match the epigenetic background of people with SCD. A few studies have characterized methylation data in African Americans, but such studies are rare compared with those in European populations.24
To assess epigenetic aging in individuals with SCD, we calculated the epigenetic ages of an adult cohort of individuals with SCD (from the Sickle Cell Disease Implementation Consortium; SCDIC)25 across 5 epigenetic clocks: Horvath,10 Hannum,12 PhenoAge,13 GrimAge,14 and DunedinPACE.15 We examined (1) the correlations between epigenetic age and reported chronological age to identify instances of accelerated epigenetic aging, (2) epigenetic age acceleration or deceleration beyond chronological age contributions, and (3) the effects of various demographic and clinical characteristics on epigenetic age. We also compared CpG probes used in the 5 clocks. In this study, we present the results of one of the first exploratory assessments of epigenetic aging in adult individuals with SCD and lay the groundwork for interpreting epigenetic age estimates in SCD cohorts of African ancestry.
Methods
Design and participants
Data included in this cross-sectional, epigenetic study were obtained from the SCDIC Research Registry at Duke University (5U01HL133964). The SCDIC was a multisite program that consisted of a coordinating center and 8 clinical centers, including Duke University. One of the goals of this consortium was to develop and maintain a comprehensive research registry that included patient-reported outcomes and clinical data collected through participant self-reports or obtained through medical records.25,26 At Duke, blood specimens were obtained from a subset of participants and banked for future analyses.
The inclusion criteria for the Duke SCDIC Research Registry included individuals who were 15 years or older, lived in North Carolina, had laboratory documentation of a genetically confirmed SCD diagnosis (confirmed in the electronic health record or a genetic laboratory report obtained from a health care provider), were literate in English, and completed informed consent procedures. Additional inclusion criteria for this study included individuals who consented to and provided a blood sample. A total of 91 participants met the inclusion criteria for this study. The study was approved by the Duke University Institutional Review Board.
Sociodemographic and clinical data
Self-reported sociodemographic data collected through the registry included chronological age, biological sex, race, ethnicity, marital status, and the highest level of education completed. SCD genotype (SS/sickle cell anemia, SC disease, S β0 thalassemia, and S β+ thalassemia) data were collected from medical records. SCD disease severity was collected using the Adult Sickle Cell Quality of Life Measurement (ASCQ-Me) SCD Medical History Checklist, a 9-item survey that captures common treatments and complications associated with the disease, including avascular necrosis, lung damage, leg ulcers, stroke, spleen damage, kidney disease, retinopathy, regular blood transfusions, and daily pain medication use.27 Each “yes” response (indicating the presence of treatment or complication) was tallied, and the total score was calculated. Higher scores (0-9 range) indicate greater disease severity.
TOPMed sequence data generation
Whole genome sequencing (WGS) data from 91 participants of the Duke SCDIC Research Registry were generated using the National Heart, Lung, and Blood Institute TOPMed program as part of its Freeze 11 release.28 Information on the latest published sequencing, variant calling, and quality control methodology can be found on the TOPMed website (https://topmed.nhlbi.nih.gov/topmed-whole-genome-sequencing-methods-freeze-9).
Methylation data processing
DNA was previously extracted from 96 blood samples from the Duke SCDIC cohort, representing 91 unique subjects. From these DNA samples, DNAm data were generated in the Duke Molecular Physiology Institute Core laboratory using the Infinium MethylationEPIC Beadchip (Illumina, San Diego, CA). A total of 250 ng of DNA was plated in 96-well plates and bisulfite treated using the EZ DNA Methylation-Direct™ Kit (Zymo). Five samples were repeated for use in assessing replicability of the experiment. Genome-wide DNA methylation was measured on a beadchip from bisulfite-converted DNA samples. These data were preprocessed using Illumina GenomeStudio software. Sample and probe quality control (QC) was performed using the minfi29 and ChAMP30 R packages. The relative levels of methylation (β) were calculated as the ratio of the methylated probe signal to total locus signal intensity. Probe QC and data normalization were performed within each batch using R package wateRmelon.31 Probes that were not detected (detection P value > .0001) in >10% of the samples and those hybridizing to multiple locations in the genome (cross-reactive) were removed.32-34 Raw β values were normalized using the dasen approach,31 and adjustments for both chip and chip positions were accomplished using ComBat35 in the R package sva.36
Five individuals each had 2 technical replicates, which were correlated with each other. One replicate for each sample was randomly chosen for inclusion in statistical analysis (5 replicates were excluded). One additional sample was excluded based on the presence of a sex mismatch between the reported sex and the DNAm-predicted sex. DNAm-predicted sex was generated using the R package minfi, which estimates the X and Y chromosome copy numbers from total methylation signals.29 No other samples were excluded based on the remaining QC metrics: average fluorescence signal intensity below 2000 arbitrary units, <50% of the mean intensity of all samples, or >10% of probes were not detectable (detection P value >.0001). Ninety samples were included in subsequent statistical analyses.
Statistical analysis
Epigenetic age calculations for the Horvath, Hannum, PhenoAge, and GrimAge clocks were performed using the DNA Methylation Age Calculator.10,37 Methylation data containing β-values for CpG probe identifiers and phenotype data containing tissue type (blood), reported chronological age, and reported sex were uploaded to the calculator. Three values were obtained from the calculator: epigenetic age estimates, age acceleration (chronological age subtracted from epigenetic age), and epigenetic age residuals (residuals from regressing epigenetic age on chronological age). DunedinPACE rates were calculated with the β values for CpG probe identifiers using the R package DunedinPACE.15 Epigenetic age residuals were not relevant for DunedinPACE because these values were standardized to 1 and controlled for chronological age. Upon assessment of epigenetic age values, 1 sample was a distinct outlier in multiple clocks relative to the other samples and was excluded from further analysis, leaving 89 samples for subsequent analysis.
Genetic ancestry estimates were calculated using the program ADMIXTURE.38 The WGS data for each of the 91 samples were obtained using the NHLBI TOPMed program. These data were filtered for biallelic single nucleotide polymorphisms (SNPs) with ≥ 1% minor allele frequency and pruned for variants under linkage disequilibrium using the PLINK program (window size 50, step size 10, pairwise r2 threshold 0.1 [--indep-pairwise 50 10 0.1]).39 The remaining variants were run using ADMIXTURE to calculate the estimated percentages of ancestry. Two ancestral populations were assumed, corresponding to the African and European components. The larger “P1” estimate (presumably African) was used as the “genetic ancestry” value in this study.
Associations between epigenetic age and various demographical and clinical phenotypes were calculated in R. Welch t tests were performed on biological sex and SCD genotype. The biological sex was treated as a dichotomous categorical phenotype. SCD genotype was dichotomized into “severe” (SS and SB0thalassemia) and “less severe” (SC and SB+thalassemia) categories based on prior assessments of SCD severity based on genotype.8 SCD severity (measured with the ASCQ-Me checklist) was treated as a polytomous ordinal phenotype (4 categories, scores of 0 through 3); epigenetic age and age residuals were regressed on this phenotype using linear models. Associations between chronological age and the proportion of African genetic ancestry were calculated using the Pearson correlation coefficient.
Two known modifiers of SCD progression, BCL11A polymorphism and β-globin SCD haplotypes, were also generated. Both modifiers were obtained from the TOPMed sequencing data, as described above. Genotypes for the BCL11A polymorphism rs11886868, known to be associated with fetal hemoglobin levels,40 were transformed into an ordinal “dosage” metric, counting the number of alternate alleles “T” present. Epigenetic age and age residuals were regressed on this metric using linear models. β-globin haplotypes were generated for the individuals in this study using a previously described method.41 Genome sequences for chromosome 11 (the location of the β-globin locus) were phased using the program SHAPEIT542 and run through the haplotype classifier script,41 generating β-globin haplotypes. This script was edited to update the genomic coordinates to the GRCh38 format used by the TOPMed sequence data, convert the script to Python version 3, and fix the issues by parsing the input. The haplotypes were dichotomized into 2 groups, 1 for homozygous Benin calls and the other for all other calls, owing to the high frequency of the Benin haplotype in SCDIC. Associations with this phenotype were calculated using Welch t tests.
Results
Table 1 provides a summary of the sociodemographic and clinical characteristics of the 89 individuals included in this study. Most were female, never married, had at least a college education, and were not currently used. The mean chronological age was 30.6 years. Most individuals had the HbSS SCD genotype and low SCD severity as measured by the ASCQ-Me checklist.
Demographic and clinical characteristics of the 89 individuals with SCDIC
| Characteristic . | . |
|---|---|
| Sex: female | 54 (60.67%) |
| Age | 30.64 (7.97) |
| Marital status | |
| Married | 10 (11.24%) |
| Never married | 68 (76.40%) |
| Other | 11 (12.36%) |
| Education | |
| Less than high school | 10 (11.24%) |
| High school | 16 (17.98%) |
| Some college | 36 (40.45%) |
| College | 17 (19.10%) |
| Graduate school | 10 (11.24%) |
| Employment | |
| Working now | 38 (42.70%) |
| Disabled | 24 (26.97%) |
| Student | 14 (15.73%) |
| Other | 13 (14.61%) |
| Genotype | |
| Hb SS or sickle cell anemia | 61 (68.54%) |
| Hb SC disease | 20 (22.47%) |
| Hb S β0 thalassemia | 5 (5.62%) |
| Hb S β+ thalassemia | 3 (3.37%) |
| Sickle cell attacks in last 12 months | |
| 0 | 10 (11.63%) |
| 1 | 8 (9.30%) |
| 2 | 12 (13.95%) |
| 3 | 10 (11.63%) |
| 4 or more | 46 (53.49%) |
| Sickle cell disease severity | 1.03 (0.91) |
| Current hydroxyurea use | 52 (60.47%) |
| Characteristic . | . |
|---|---|
| Sex: female | 54 (60.67%) |
| Age | 30.64 (7.97) |
| Marital status | |
| Married | 10 (11.24%) |
| Never married | 68 (76.40%) |
| Other | 11 (12.36%) |
| Education | |
| Less than high school | 10 (11.24%) |
| High school | 16 (17.98%) |
| Some college | 36 (40.45%) |
| College | 17 (19.10%) |
| Graduate school | 10 (11.24%) |
| Employment | |
| Working now | 38 (42.70%) |
| Disabled | 24 (26.97%) |
| Student | 14 (15.73%) |
| Other | 13 (14.61%) |
| Genotype | |
| Hb SS or sickle cell anemia | 61 (68.54%) |
| Hb SC disease | 20 (22.47%) |
| Hb S β0 thalassemia | 5 (5.62%) |
| Hb S β+ thalassemia | 3 (3.37%) |
| Sickle cell attacks in last 12 months | |
| 0 | 10 (11.63%) |
| 1 | 8 (9.30%) |
| 2 | 12 (13.95%) |
| 3 | 10 (11.63%) |
| 4 or more | 46 (53.49%) |
| Sickle cell disease severity | 1.03 (0.91) |
| Current hydroxyurea use | 52 (60.47%) |
The n-sizes and percentages are presented for categorical characteristics. Means and standard deviations are presented for continuous characteristics.
Epigenetic age values and age acceleration/deceleration
Epigenetic age values were compared with the chronological age (Table 2). Scatterplots of these comparisons are shown in supplemental Figure 1. Epigenetic age was highly correlated with chronological age across 4 of the clocks evaluated (Horvath: r = 0.88, P < 1e-5; Hannum: r = 0.89, P < 1e-5; PhenoAge: r = 0.85, P < 1e-5; GrimAge: r = 0.88, P < 1e-5). DunedinPACE showed a weak correlation with chronological age (r = 0.34, P = .001).
Epigenetic clock values, residuals, and associations with various clinical characteristics
| . | Median (SD) . | Welch t test–P value . | Linear regressions–P value . | Pearson’s correlation coefficient - r (P value) . | ||||
|---|---|---|---|---|---|---|---|---|
| Biological sex . | SCD genotype . | β-globin haplotype . | SCD disease severity . | BCL11A SNP . | Chronological age . | Genetic ancestry . | ||
| DNAmAge (Horvath) age | 0.40 (4.48) | .89 | .61 | .28 | .93 | .99 | .88 (<1e-5)∗∗ | .212 (.057) |
| DNAmAge (Hannum) age | −3.98 (3.86) | .62 | .11 | .89 | .92 | .71 | .89 (<1e-5)∗∗ | .173 (.120) |
| PhenoAge age | −8.65 (6.00) | .23 | .02∗ | .34 | .90 | .26 | .85 (<1e-5)∗∗ | .092 (.411) |
| GrimAge age | 11.90 (4.02) | .96 | .19 | .52 | .67 | .69 | .88 (<1e-5∗)∗∗ | .176 (.114) |
| DunedinPACE rate | 1.14 (0.12) | .07 | <.001∗∗ | .41 | .56 | .79 | .34 (.001)∗∗ | .041 (.722) |
| DNAmAge (Horvath) residuals | 0.07 (4.50) | .25 | .61 | .29 | .83 | .54 | NA | .181 (.104) |
| DNAmAge (Hannum) residuals | 0.24 (3.78) | .96 | .07 | .30 | .54 | .87 | NA | −.001 (.994) |
| PhenoAge residuals | 0.08 (5.89) | .10 | .03∗ | .39 | .59 | .11 | NA | −.135 (.226) |
| GrimAge residuals | −0.30 (3.94) | .25 | .10 | .96 | .95 | .81 | NA | .015 (.894) |
| . | Median (SD) . | Welch t test–P value . | Linear regressions–P value . | Pearson’s correlation coefficient - r (P value) . | ||||
|---|---|---|---|---|---|---|---|---|
| Biological sex . | SCD genotype . | β-globin haplotype . | SCD disease severity . | BCL11A SNP . | Chronological age . | Genetic ancestry . | ||
| DNAmAge (Horvath) age | 0.40 (4.48) | .89 | .61 | .28 | .93 | .99 | .88 (<1e-5)∗∗ | .212 (.057) |
| DNAmAge (Hannum) age | −3.98 (3.86) | .62 | .11 | .89 | .92 | .71 | .89 (<1e-5)∗∗ | .173 (.120) |
| PhenoAge age | −8.65 (6.00) | .23 | .02∗ | .34 | .90 | .26 | .85 (<1e-5)∗∗ | .092 (.411) |
| GrimAge age | 11.90 (4.02) | .96 | .19 | .52 | .67 | .69 | .88 (<1e-5∗)∗∗ | .176 (.114) |
| DunedinPACE rate | 1.14 (0.12) | .07 | <.001∗∗ | .41 | .56 | .79 | .34 (.001)∗∗ | .041 (.722) |
| DNAmAge (Horvath) residuals | 0.07 (4.50) | .25 | .61 | .29 | .83 | .54 | NA | .181 (.104) |
| DNAmAge (Hannum) residuals | 0.24 (3.78) | .96 | .07 | .30 | .54 | .87 | NA | −.001 (.994) |
| PhenoAge residuals | 0.08 (5.89) | .10 | .03∗ | .39 | .59 | .11 | NA | −.135 (.226) |
| GrimAge residuals | −0.30 (3.94) | .25 | .10 | .96 | .95 | .81 | NA | .015 (.894) |
Associations between sex and sickle cell genotype (dichotomous continuous variables) were calculated using the Welch t tests. Associations with SCD severity, β-globin haplotype, and BCL11A SNP (polytomous ordinal variables) were calculated using linear regression. Associations between chronological age and genetic ancestry (continuous variables) were calculated using Pearson correlation coefficient. ∗P < 0.05; ∗∗P < 0.001.
The average age acceleration/deceleration values varied widely between the clocks. Regressing age acceleration/deceleration on chronological age yielded the following results. Three of the 5 epigenetic clocks deviated significantly from chronological age: Hannum (−3.98 years; P = 1.6e-17) and PhenoAge (−8.65 years; P = 2.3e-23) showed large deceleration, and GrimAge (11.90 years; P = 7.3e-49) showed large acceleration. Horvath epigenetic aging was not significant (0.40 years; P = .62). An increased pace of aging was observed using the DunedinPACE clock (1.14 biological year per chronological year; P = .001).
Epigenetic age residuals
The summary statistics of epigenetic age residuals are shown in Table 2. The PhenoAge residuals were significantly associated with the SCD genotype (P = .03), none of the other residuals were associated with the other phenotypes. Plots of the residuals compared with chronological age are shown in supplemental Figure 2. The residuals were randomly scattered around zero, showing that the assumption of homoscedasticity was preserved.
Associations with demographic and clinical phenotypes
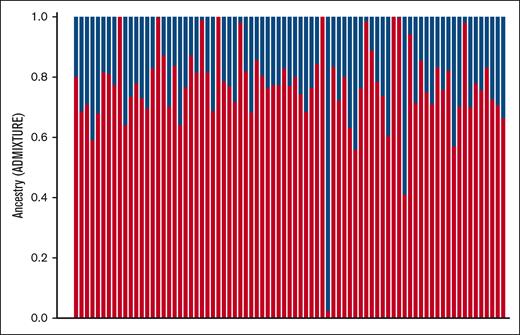
An ancestry plot of the SCDIC individuals generated in ADMIXTURE is shown in Figure 1. On average, the SCDIC cohort had 77% African ancestry. The majority component, estimated to be the “African” component, falls within the previously published estimates for African Americans.27,43
Estimated ancestry components of SCDIC samples using ADMIXTURE. The P1 component (red bars) is hypothesized to be the African component (average 77%), and the P2 component (blue bars) is hypothesized to be the European component (average 23%).
Estimated ancestry components of SCDIC samples using ADMIXTURE. The P1 component (red bars) is hypothesized to be the African component (average 77%), and the P2 component (blue bars) is hypothesized to be the European component (average 23%).
Associations of epigenetic age and epigenetic age residuals with biological sex, SCD genotype, SCD disease severity, African genetic ancestry, BCL11A rs11886868 genotype, and β-globin haplotype are shown in Table 2. The SCD genotype was associated with PhenoAge (P = .02) and DunedinPACE (P < .001), but not with any other clock. Biological sex, SCD disease severity, genetic ancestry, BCL11A genotype, and β-globin haplotype (49.4% homozygous Benin, 50.6% not homozygous Benin) were not associated with any clock age or age residual.
Discussion
In this study, we report our findings on epigenetic aging in a cohort of adult individuals with SCD. Given that this is the first study on epigenetic aging in an SCD context to our knowledge, we not only report the epigenetic aging estimates of the Duke SCDIC Research Registry cohort and their associations with demographic and clinical variables, but also assess the utility of the 5 epigenetic clocks used to calculate these estimates in an African American and SCD context. Among the 5 clocks examined, Horvath estimated minimal age acceleration, Hannum and PhenoAge estimated age deceleration, and GrimAge and DunedinPACE estimated age acceleration. Consequently, the GrimAge and DunedinPACE clocks lend support to our hypothesis of epigenetic age acceleration in people with SCD.
First-generation Horvath and Hannum clocks produced estimates of age acceleration in healthy individuals.10,12 The overall lack of significance of first-generation clocks may be due to their design for predicting aging in relatively healthy individuals. The second-generation PhenoAge and GrimAge clocks estimate “composite” age, which combines chronological age and biological markers informative for cellular aging and epigenetic regulation processes.13,14 This may explain the very limited association observed between SCD genotype and these clocks and not with any of the other phenotypes. These observations align with previous assessments of these clocks.23 Notably, the third-generation DunedinPACE clock uses longitudinal measurements of methylation probes, as opposed to the single-timepoint measurements used by the other 4 clocks. Longitudinal measurements can more accurately convey age-dependent methylation changes by accounting for chronological age. This is particularly beneficial when characterizing SCD, given that SCD pathogenesis occurs in an age-dependent manner (eg, renal function and early mortality).44-46 Future development of epigenetic clocks may benefit from longitudinal methylation data, which may better model the effects of natural aging (as opposed to accelerated cellular aging caused by chronic diseases). This may allow signals of accelerated aging to be more readily identified and unconfounded with the natural chronological age.
The variability in the age acceleration measures is also reflective of differences in the clocks. A comparison of shared CpG probes is presented in supplemental Table 1. The vast majority of probes were unique to each clock, with only a few common across multiple clocks and none common to 4 or all 5 clocks. The difference in the used probes likely led to inconsistencies in the per-individual epigenetic age estimates across the 5 clocks. The clocks also differed based on the biomarkers measured, affecting which probes were selected. For example, GrimAge selected probes that corresponded with their 12 surrogate biomarkers of plasma protein levels in addition to smoking pack-years, whereas Horvath and Hannum selected probes agnostic of any covariates besides chronological age. As epigenetic clocks are continually being refined, both in the CpG probes used and the outcomes measured, there is an opportunity for SCD-specific biomarkers and outcomes to be used to develop an SCD epigenetic clock in future studies.47,48
Moreover, the clocks were trained mostly on healthy tissues of European descent. Their applicability to other chronic diseases, such as SCD and other populations with differing genetic ancestries, remains to be investigated. Previous studies have discovered differential epigenetic aging when analyzed against the genetic background,49,50 motivating our own analysis of ancestry in individuals with SCD.
Epigenetic age and demographic and clinical phenotypes
As this is the first study on epigenetic aging in an adult population with SCD, no prior reference point exists for these associations. Under the hypothesis that SCD causes physiological stress and cellular damage that result in epigenetic modifications, more severe forms of SCD should cause increased stress and damage, resulting in larger epigenetic age differences. This is evidenced by prior studies demonstrating that SCD genotype affects clinical presentation and disease progression; individuals with SS or Sβ0-thalassemia genotypes have a higher risk of acute chest syndrome, pulmonary hypertension, and lower oxyhemoglobin than those with SC or Sβ+-thalassemia.51 The evidence here supports this expectation, in which SCD genotype is significantly associated with PhenoAge age and DunedinPACE rate (Table 2).
We did not find any association between epigenetic age and biological sex for any clocks. This differs from the Hannum study, which found an association between sex and the aging rate in 2 cohorts of Caucasian and Hispanic individuals.12 Other studies either adjusted for reported sex (PhenoAge,13 GrimAge14) when constructing epigenetic aging metrics or did not find an association with sex.52
We did not find an association between epigenetic aging and SCD severity in this study. Most of the 89 individuals had an SCD severity score of 0 or 1 and the vast majority of the “yes” responses were for daily pain medication or regular blood transfusions. Patients with more severe SCD progression typically experience complications, such as spleen removal, retinopathy, or stroke, which are also indicated on the ASCQ-Me checklist. Given that these individuals are relatively young (mean age 30.64), their overall SCD severity is lower and thus, they do not present with many of the symptoms that often emerge later in life.44
Previous studies have found associations between epigenetic age and genetic ancestry,13 which were not observed here. Importantly, these studies evaluated differences across multiple ancestral groups, whereas this study evaluated differences within a single ancestral group. Larger cohorts of individuals with African ancestry may be needed to determine whether variation within a single ancestral group influences epigenetic aging in SCD. Calculations of estimated ancestry in our cohort were consistent with those of prior studies describing African American genetic ancestry in other cohorts, thus, supporting our observed ancestry clusters.53-55
No association was found between epigenetic age and the BCL11A or β-globin haplotypes. Although there are rich data supporting these modifiers in SCD, it is possible that a larger and older sample of patients with SCD will be needed to determine any effect on epigenetic aging.
Epigenetic age and epigenetic age residuals
Given the strong correlation between epigenetic age and chronological age for 4 of the 5 clocks examined (r > 0.7; Table 2), we wanted to conceptualize the component of methylation age independent of the natural aging effects of chronologic age. After accounting for the effects of chronological age in Horvath, Hannum, PhenoAge, and GrimAge, the clock residuals showed relatively similar patterns of association with the demographic and clinical phenotypes. Specifically, PhenoAge residuals were significantly associated with the SCD genotype, but none of the other clocks were associated with any of the other phenotypes.
Horvath, Hannum, PhenoAge, and GrimAge epigenetic clocks scale the epigenetic age to units of biological years to facilitate comparisons with biological age. DunedinPACE is a rate metric of 1 biological year per chronological year; therefore, in an individual with no epigenetic age acceleration or deceleration, the pace of aging would remain the same regardless of chronological age. Any correlation between the pace of aging and chronological age indicated age-dependent acceleration/deceleration. We observed a weak but statistically significant correlation between chronological age and DunedinPACE pace of aging. This provides evidence for both age acceleration measured by DunedinPACE in our study and age acceleration, which increases as patients naturally age. Given the weak correlation, the residuals for DunedinPACE were not calculated.
Limitations
Although this study is the first to present novel data on epigenetic aging in SCD, it is an exploratory study based on a relatively small cohort of 89 samples. Future studies will benefit from using additional samples when assessing the characteristics described in this study. Regarding genetic ancestry, differences within a single population (such as African Americans) are likely to be smaller than the differences between multiple populations with larger variation in ancestry. Other studies noting the significant effects of genetic ancestry on epigenetic age have analyzed multiple cohorts from different genetic backgrounds.49,50
Furthermore, aging was assessed here in a SCD case-only cohort. To better understand the effects of background genetic ancestry, epigenetic ages could be calculated in control populations of healthy age- and sex-matched Black or African American individuals with methylation data and compared with those of individuals with SCD. This would allow comparisons between cases and controls across the clinical and genetic characteristics previously described, including direct comparisons of epigenetic ages and components of genetic ancestry. Epigenetic data generation on additional SCD-relevant cohorts would allow comparisons between multiple cohorts.
Conclusion
To our knowledge, this study is the first to calculate epigenetic age values for an SCD cohort of 89 adult individuals. We found that each epigenetic clock yielded varying results: of the 5 clocks presented, GrimAge and DunedinPACE reported epigenetic age acceleration, Hannum and PhenoAge reported age deceleration, and Horvath reported neither acceleration nor deceleration. GrimAge and DunedinPACE were associated with SCD genotype, and all 5 clocks were correlated with chronological age. The GrimAge and DunedinPACE clocks may be particularly relevant to assess in future studies on epigenetic aging in SCD. As this is the first study on epigenetic aging in an SCD cohort, additional analyses are needed to further characterize and validate SCD-related epigenetic aging. The development of future clocks may better model chronic disease processes by including longitudinal methylation data as well as incorporating cohorts from diverse genetic ancestries. The results of this study allow for the investigation of the interpretability of epigenetic ages when evaluating SCD clinical phenotypes.
Acknowledgments
B.M.L. is a PhD candidate at the Duke University. This work is submitted in partial fulfillment of the requirements for his PhD. The authors also acknowledge the editorial assistance of Donnalee Frega.
This work was supported by funding from the National Institute of Nursing Research (R21NR020017). The SCDIC Research Registry data and biospecimen collection were supported by funding from the National Heart, Lung, and Blood Institute (NHLBI) (U01HL133964) and the Duke University Clinical & Translational Sciences Institute’s Special Populations Core Pilot grant funded by the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1TR002553). The content of this manuscript is solely the responsibility of the authors and does not represent the policies or views of the funding agencies.
The 91 samples from the Duke SCDIC Research Registry were included in the OMG_SCD study for sequencing through the Trans-Omics in Precision Medicine (TOPMed) program. Molecular data for the TOPMed program was supported by the NHLBI. Genome sequencing for "NHLBI TOPMed: OMG_SCD" (phs001608) was performed at the Baylor College of Medicine Human Genome Sequencing Center (HHSN268201600033I). Core support, including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering, was provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Core support, including phenotype harmonization, data management, sample identity QC, and general program coordination, was provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). The authors gratefully acknowledge the studies and participants who provided biological samples and data for the TOPMed.
Authorship
Contribution: B.M.L. processed the methylation data, calculated the epigenetic age values, analyzed and interpreted the data, and wrote the manuscript; D.H. calculated associations between sickle cell genotype and sickle cell disease severity and analyzed and interpreted the data; M.E.G. provided scripts to process the methylation data and interpreted the data; P.T. and N.S. collected phenotypic data for the Duke SCDIC registry; Q.Y., N.S., and F.S.L. designed the study and contributed to the interpretation of the data; A.E.A.-K. and M.R.K. designed the study, obtained funding for the research, analyzed and interpreted the data, and provided oversight; and all authors reviewed, edited, and approved the manuscript. The NHLBI Trans-Omics in Precision Medicine Consortium generated data and their associated Publications Committee reviewed and approved the final version of this manuscript.
Conflict-of-interest disclosure: N.S. is a consultant for Global Blood Therapeutics (GBT)/Pfizer, Forma, Agios, Vertex, and bluebird bio; is a speaker for GBT/Pfizer and Alexion; and performs research on GBT/Pfizer. P.T. is a consultant for CSL Behring. The remaining authors declare no competing financial interests.
A complete list of the members of the NHLBI Trans-Omics in Precision Medicine Consortium appears in supplemental Material.
Correspondence: Mitchell R. Knisely, Duke University School of Nursing, 307 Trent Dr, DUMC 3322, Durham, NC 27710; email: mitchell.knisely@duke.edu.
References
Author notes
Sequencing data are available in dbGaP under accession number phs001608. The authors have ethical restrictions on openly releasing the complete data set to the public. However, data set requests for clinical data can be made to the Sickle Cell Disease Implementation Consortium (SCDIC) and their data coordinating center at RTI International. Requests will be reviewed by the SCDIC Publications Committee. Clinical data set requests can be sent to SCDIC at scdic-publications-subcommittee@rtiresearch.org or +1 301-230-4674.
Data generated from the biospecimens used in this study are available on request from the corresponding author, Mitchell R. Knisely (mitchell.knisely@duke.edu).
The full-text version of this article contains a data supplement.