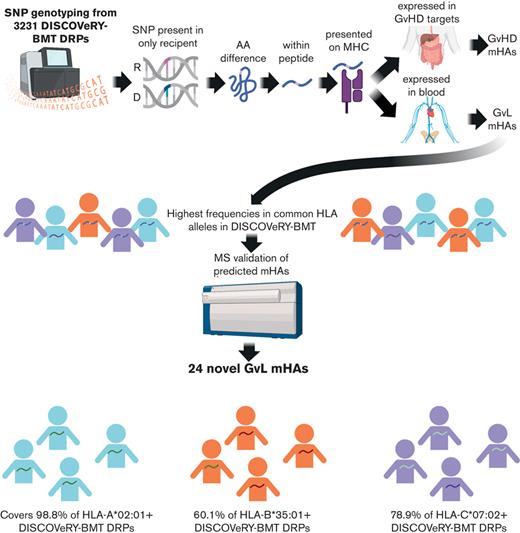
Key Points
We discovered that many predicted GVL mHAss are highly shared among DRPs in DISCOVeRY-BMT.
We validated 24 of our predicted novel GVL mHAss that are shared among patients in the DISCOVeRY-BMT data set.
Abstract
T-cell responses to minor histocompatibility antigens (mHAs) mediate graft-versus-leukemia (GVL) effects and graft-versus-host disease (GVHD) in allogeneic hematopoietic cell transplantation. Therapies that boost T-cell responses improve allogeneic hematopoietic cell transplant (alloHCT) efficacy but are limited by concurrent increases in the incidence and severity of GVHD. mHAs with expression restricted to hematopoietic tissue (GVL mHAs) are attractive targets for driving GVL without causing GVHD. Prior work to identify mHAs has focused on a small set of mHAs or population-level single-nucleotide polymorphism–association studies. We report the discovery of a large set of novel GVL mHAs based on predicted immunogenicity, tissue expression, and degree of sharing among donor-recipient pairs (DRPs) in the DISCOVeRY-BMT data set of 3231 alloHCT DRPs. The total number of predicted mHAs varied by HLA allele, and the total number and number of each class of mHA significantly differed by recipient genomic ancestry group. From the pool of predicted mHAs, we identified the smallest sets of GVL mHAs needed to cover 100% of DRPs with a given HLA allele. We used mass spectrometry to search for high-population frequency mHAs for 3 common HLA alleles. We validated 24 predicted novel GVL mHAs that are found cumulatively within 98.8%, 60.7%, and 78.9% of DRPs within DISCOVeRY-BMT that express HLA-A∗02:01, HLA-B∗35:01, and HLA-C∗07:02, respectively. We confirmed the immunogenicity of an example novel mHA via T-cell coculture with peptide-pulsed dendritic cells. This work demonstrates that the identification of shared mHAs is a feasible and promising technique for expanding mHA-targeting immunotherapeutics.
Introduction
Minor histocompatibility antigens (mHAs) are peptides derived from single-nucleotide polymorphisms (SNPs) that differ between a tissue transplant recipient and donor, such that the mHA allele is expressed by the recipient only and presented on the recipient’s major histocompatibility complex molecules.1-4 T cells that target these antigens are important mediators of the beneficial graft-versus-leukemia (GVL) effect and harmful graft-versus-host disease (GVHD) after allogeneic hematopoietic cell transplantation.5-7 Allogeneic hematopoietic cell transplant (alloHCT) is a standard therapy for eligible patients with high-risk acute myeloid leukemia (AML), the deadliest form of leukemia in the United States.8,9 It is a highly effective treatment for AML in first complete remission and reduces relapse risk by >60% versus chemotherapy alone.8,10-12 However, the prognosis is poor for patients who relapse after alloHCT. Since the development of alloHCT, transplant clinicians have experienced the “transplanter’s dilemma,” that is, interventions that boost antileukemia T-cell responses also increase GVHD incidence, and interventions preventing GVHD increase relapse rates.5,13-24 Separating GVL from GVHD is a foundational problem in transplant immunology. One approach is to use GVL mHA-directed immunotherapies.25 GVL mHAs are defined as mHAs that are only expressed in hematopoietic tissue, so T-cell responses against them can generate GVL effects without GVHD. Approximately 55 mHAs have been reported to date, including 12 validated class I GVL mHAs, and clinical trials of mHA-targeted immunotherapies have been performed as well.25-32
The majority of mHA discovery focuses on identifying personalized mHAs for individual transplant donor-recipient pairs (DRPs).9,33-37 This approach identifies mHAs that may be only applicable to a small number of patients via personalized immunotherapies. We instead seek to identify mHAs that are shared across many DRPs, allowing for an off-the-shelf approach to therapeutic mHA-targeting. We report here an innovative approach to mHA identification that allowed us to discover 24 novel GVL mHAs that are shared across many DRPs, increasing the number of known class I GVL mHAs by 200%.
Methods
Computational methods
Study population
DRP sequencing and clinical data were derived from the DISCOVeRY-BMT (Determining the Influence of Susceptibility Conveying Variants Related to One-Year mortality after BMT) study, reported to the Center for International Blood and Marrow Transplant Research from 151 transplant centers within the United States.38-42 Patients included in this study were treated for AML, acute lymphocytic leukemia (ALL), and myelodysplastic syndrome (MDS) with alloHCT. Cohort 1 consisted of 2609 10/10 HLA-matched unrelated DRPs treated from 2000 to 2008, whereas cohort 2 consisted of 572 10/10 HLA-matched unrelated DRPs treated from 2009 to 2011 and 351 8/8 (but <10/10) HLA-matched unrelated DRPs treated from 2000 to 2011.41 DRPs were excluded if the grafts were cord blood grafts or T-cell depleted or if SNP data were not available. For antigen prediction, all patients were combined.
All patients included in the DISCOVeRY-BMT study provided informed consent to be included in the Center for International Blood and Marrow Transplant Research registry. Genotyping was performed as previously described using the Illumina HumanOmni Express chip.41-43 SNP quality control was performed and the variants with minor allele frequency <0.005 were removed, leaving 637 655 and 632 823 measured SNPs for cohort 1 and cohort 2, respectively.39 We calculated the genetic distance between each DRP based on the SNP array data.44 Genomic ancestry was calculated via principal component analysis. Principal components were constructed using a set of independent SNPs in all patients self-declaring White, European, or Caucasian race and non-Hispanic ethnicity. Mean values for the first 3 eigenvectors were determined and individuals with any of the first 3 eigenvectors >2 standard deviations from each mean value were excluded. This was repeated for individuals self-declaring Black or African race and non-Hispanic ethnicity and individuals declaring Hispanic ethnicity.42,45 For this work, 3 genomic ancestry groups were assessed, including European American, Hispanic, and African American. Patients who self-reported as Asian American and Native American were included in mHA prediction work, but their genomic ancestry was not calculated, and they were excluded from ethnicity analyses owing to the small patient numbers for these groups. Student t tests and χ2 tests were performed to assess differences in the number of predicted mHAs between groups.
mHA prediction
Minor mismatches were defined as SNP loci where the recipient and donor alleles differed, and mHAs were the predicted peptides from the recipient allele of the minor mismatch.1 Each SNP allele was considered independently, such that predicted mHAs are allele-specific. All possible peptides of lengths between 8 and 11 amino acids resulting from SNP mismatches within every DRP were screened for predicted binding affinity against the recipient HLA class I alleles and expression of the source gene. We filtered for peptides with a peptide/HLA dissociation constant <500 nM using NetMHCpan.46 Peptides were called mHAs if they fit these criteria. Where multiple-length peptides derived from the same SNP met the filter, these cases were reduced to the shortest version of the peptide. This allowed us to avoid counting peptides with identical core sequences as separate entities because patients containing a specific SNP will likely have each length of that SNP-derived peptide. mHAs were then subcategorized based on source protein messenger RNA expression in AML RNA sequencing (RNA-seq) data obtained from The Cancer Genome Atlas and normal tissue protein expression data from the Genotype-Tissue Expression (GTEx) Project. Peptides were labeled “GVL” if they showed expression levels of >50 transcripts per million (TPM) in AML and <50 TPM in GVHD target organs including skin, liver, and colon. The “GVH” label indicates levels of <50 TPM in AML and >50 TPM in GVHD target organs. “Both” denotes >50 TPM in both AML and GVHD target organs. Peptides with a “GVL” label were considered for further analysis, whereas peptides with tags of “both” and “GVH” were excluded. This resulted in 1,867,836 predicted GVL mHAs across 3231 DRPs. Some of these predicted mHAs may be less applicable to patients with ALL than patients with AML/MDS in our data set, as GVL mHAs were denoted based on The Cancer Genome Atlas data for AML.
Minimal set calculation
We developed a greedy algorithm to resolve the maximum set coverage problem and generate ranked lists of the most commonly shared mHAs for DRPs with a given HLA allele. This algorithm generates a list of the minimal set of peptides such that every DRP with a given HLA allele in the data set contains at least 1 of these mHAs. In short, the algorithm ranks every peptide within a given HLA by the study population frequency in descending order. The peptide with the highest frequency is selected and added to the mHA set, then the population frequency of every peptide is recalculated using only DRPs that do not contain an mHA in the set and, lastly the new highest frequency peptide is selected. This process is repeated until 100% of DRPs are represented by an mHA in the set. For mass spectrometric (MS) validation, we selected the peptides from the set that have nonzero RNA-seq coverage of the source gene in the cell line being used for validation. We added additional peptides for analysis by filtering the peptides not selected by the greedy algorithm for nonzero expression of the source gene, ranking in descending order of noncumulative population coverage, then selecting the necessary number of peptides to bring the total list for MS validation to 40 peptides as this was a feasible search size for MS.
Three HLA alleles were selected for mHA MS validation based on high frequency in the US ethnic groups and for including representative alleles for HLA-A, HLA-B, and HLA-C. HLA-A∗02:01 is the most common HLA-A allele among Caucasians, African Americans, and Hispanics within the United States, third most common among Asians and Pacific Islanders, and is found within 28.4% of the total population of the United States.47 HLA-B∗35:01 is the most common HLA-B allele among Asians and Pacific Islanders, is third most common among African Americans, and is fifth most common among Caucasians and Hispanics. It is found within 6.7% of the population of the United States.47 HLA-C∗07:02 is the most common HLA-C allele among Hispanics within the United States, is second most common among Caucasians and Asians and Pacific Islanders, and is seventh most common among African Americans. It is found within 15.4% of the total population of the United States.47 Two lists of 40 peptides each were searched for HLA allele B∗35:01 and HLA-C∗07:02, respectively. Two samples were sent for MS for HLA-A∗02:01. A 40-peptide search list was updated for the second sample to use the updated cell line RNA-seq data. In total, 67 peptides were searched for HLA-A∗02:01.
Experimental methods
Cell lines
The AML cell lines used for MS were U937A2, the U937 cell line stably transfected to express HLA-A∗02:01, NB4, which endogenously expresses HLA-B∗35:01, and MONOMAC1, which endogenously expresses HLA-C∗07:02.48 Cell line HLA expression data were downloaded from the TRON cell lines portal and validated by the Clinical HLA Typing Laboratory at the University of North Carolina Hospitals, with differences as reported (supplemental Figure 1).48,49 Cases where discrepancies were found between the HLA haplotype on TRON and clinical typing, the clinical typing result was used. Cell lines were maintained in culture with RPMI 1640, 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% L-glutamine.50
Immunoprecipitation and mass spectrometry
Cell lines were expanded to 1 × 108 to 5 × 108 per sample. Cells were centrifuged and washed with phosphate-buffered saline, followed by treatment with 1× cOmplete Mini EDTA-free Protease Inhibitor Cocktail tables prepared in phosphate-buffered saline (11836170001, Roche). Cells were centrifuged and the supernatant removed and cell pellets snap frozen in liquid nitrogen and placed at −80 °C. Frozen pellets were sent to Complete Omics Inc for immunoprecipitation and antigen validation and quantification by mass spectrometry through the Valid-NEO platform.51 Pellets were processed into single-cell frozen powder and then lysed. Peptide-HLA complexes were immunoprecipitated using the Valid-NEO neoantigen enrichment column preloaded with antihuman HLA-A, HLA-B, and HLA-C antibody clone W6/32 (BioXCell). After elution, dissociation, filtration, and clean-up, peptides were lyophilized before further analysis. Transition parameters for each epitope peptide were examined and curated through Valid-NEO method builder, an artificial intelligence–based biostatistical pipeline. Ions with excessive noise owing to coelution with impurities were further optimized or removed. To boost detectability, a series of computational recursive optimizations of significant ions was conducted. Each mHA sequence was individually detected and quantified in a high-throughput manner through a Valid-NEO–modified mass spectrometer.
mHA immunogenicity assessment
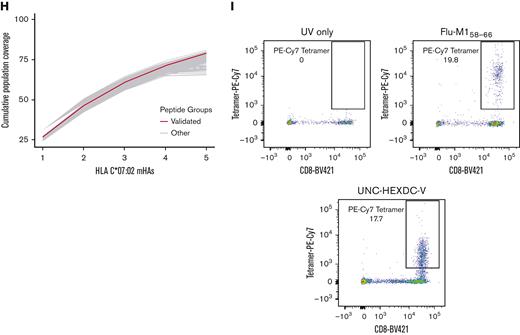
Human donor leukopaks were obtained (Gulf Coast Regional Blood Center) and genotyped for HLA∗A:02 via flow cytometry with purified antihuman HLA-A2 antibody clone BB7.2 (BioLegend). HLA-A∗02–positive samples were selected and dendritic cells (DCs) were generated via plate adherence and pulsed with mHAs not endogenous to the sample (Peptide 2.0 Inc). Naïve CD8 T cells were isolated and cocultures were initiated with mHA-pulsed DCs and naïve CD8 T cells at a 1:4 ratio and maintained for 2 weeks in culture in RPMI, 10% human serum, 1% penicillin-streptomycin, and 1% L-glutamine. The presence of mHA-specific T cells was assessed via flow cytometry with mHA tetramer staining. Tetramers were generated with Flex-T HLA-A∗02:01 Monomer UVX (280004, BioLegend) and fluorophore-conjugated streptavidin. Cells were also stained with the following: FVS700 live/dead (BD Biosciences) and CD8-BV421 (Clone: SK1, BioLegend). Cells cultured with immunodominant influenza A virus M158-66 HLA-A∗02:01–binding influenza peptide and stained with Flu-M158-66 tetramer (designated as Flu) were used as a positive control, and cells stained with tetramer exposed to UV light with no peptide (UV only) were used as negative control.52 Gating strategy is shown in supplemental Figure 4.
Results
Patient characteristics and mHA predictions
Characteristics of patients in DISCOVeRY-BMT are shown in supplemental Table 1.40 Of the total DISCOVeRY-BMT patients, 60% had a diagnosis of AML, whereas the remainder had diagnoses of ALL or MDS. Age distribution in the DISCOVeRY-BMT cohort reflects the age distribution of AML, with 60% of alloHCT recipients older than 40 years of age. Most transplant recipients in the data set received bone marrow–derived grafts. Using the SNP typing data from these patients, we predicted a total of 9,241,788 mHAs in the DISCOVeRY-BMT data set. The prediction pathway was as follows: (1) identification of SNPs is present only in the recipient of DRP from SNP typing data, (2) which generates an amino acid difference, (3) within a predicted peptide, and (4) that binds major histocompatibility complex allele(s) expressed by the DRP. These predicted mHAs were then (1) categorized as GVL or GVH based on tissue expression and (2) lists of the most highly shared GVL mHAs were generated and (3) validated via MS.
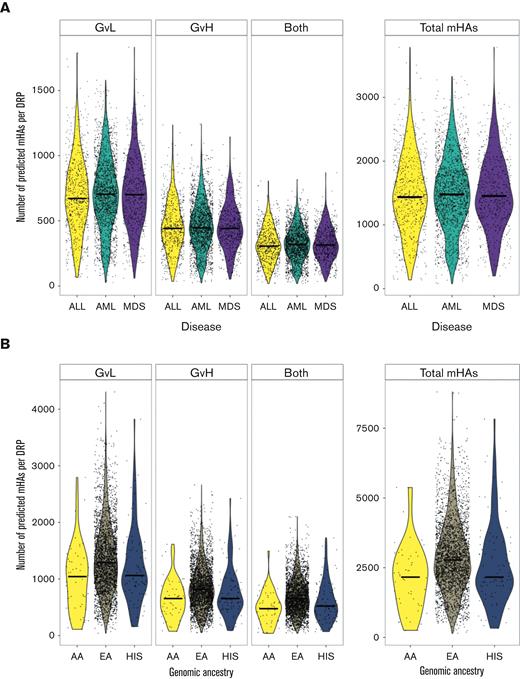
The number of predicted mHAs did not vary by disease type (Figure 1A). The self-reported ethnicity and genomic ancestry of alloHCT recipients in this data set mirror the general distribution of alloHCT recipients in the United States, with a predominance of patients with European American ancestry.53
Predicted mHAs by disease type and genomic ancestry. (A) Shows the number of each category of predicted mHA per DRP by disease type. “GVL” denotes expression in leukemia cells, “GVH” denotes expression in GVH target organs, and “both” denotes expression in both. (B) Shows the number of each category of predicted mHA per DRP by genomic ancestry, including patients identifying as European American (EA), African American (AA), or Hispanic (HIS).
Predicted mHAs by disease type and genomic ancestry. (A) Shows the number of each category of predicted mHA per DRP by disease type. “GVL” denotes expression in leukemia cells, “GVH” denotes expression in GVH target organs, and “both” denotes expression in both. (B) Shows the number of each category of predicted mHA per DRP by genomic ancestry, including patients identifying as European American (EA), African American (AA), or Hispanic (HIS).
A large number of mHAs were predicted for each genomic ancestry group assessed in this study, with 75 918 total predicted mHAs for European American, 27 557 mHAs for African American, and 39 272 mHAs for Hispanic (Figure 1B). mHAs were then assigned tags based on the expression of the source gene in AML and GVHD target tissues. The mean total predicted mHAs per DRP across all ethnicities was 1476, with a mean of 704 predicted GVL mHAs. The number of predicted mHAs differed significantly by genomic ancestry group, with European American >Hispanic >African American for the number of mHAs labeled as GVL, GVH, and both as well as total mHAs per DRP.
Predicted GVL mHAs were identified for 56 HLA alleles found in DISCOVeRY-BMT alloHCT recipients
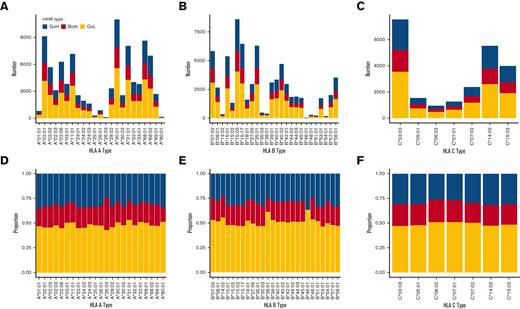
A total of 23 HLA-A alleles, 26 HLA-B alleles, and 7 HLA-C alleles were represented in DISCOVeRY-BMT. The total number of predicted mHAs that bind each allele varied widely, from 82 to 11 017 for HLA-A alleles, 19 to 8585 for HLA-B alleles, and 946 to 7537 for HLA-C alleles (Figure 2A-C). However, our method predicted GVL mHAs for every HLA allele represented within DISCOVeRY-BMT. Next, we looked at the proportion of mHAs classified as GVL, GVH, or both for each HLA allele. GVL mHA comprised approximately half of all predicted mHAs for each HLA allele (Figure 2D-F).
Number and proportion of predicted mHAs by HLA allele within the study population. mHAs classified as “GVL” broadly represent mHAs that are desirable to target for antileukemia effects with minimal GVHD. mHAs classified as “GVH” represent mHAs that are undesirable to target as they are predicted to correspond to GVHD and have no GVL effects. The “both” category represents peptides that are predicted to lead to both GVL and GVH effects. (A) Shows counts of each predicted class of mHA for HLA-A alleles represented in the patient data set. (B) Shows counts for HLA-B alleles represented in the patient data set. (C) Shows counts for HLA-C alleles represented in the patient data set. (D) Shows the proportion of predicted mHAs corresponding to each mHA class for HLA-A alleles. (E) Shows the proportion for HLA-B alleles. (F) Shows the proportion of HLA-C alleles.
Number and proportion of predicted mHAs by HLA allele within the study population. mHAs classified as “GVL” broadly represent mHAs that are desirable to target for antileukemia effects with minimal GVHD. mHAs classified as “GVH” represent mHAs that are undesirable to target as they are predicted to correspond to GVHD and have no GVL effects. The “both” category represents peptides that are predicted to lead to both GVL and GVH effects. (A) Shows counts of each predicted class of mHA for HLA-A alleles represented in the patient data set. (B) Shows counts for HLA-B alleles represented in the patient data set. (C) Shows counts for HLA-C alleles represented in the patient data set. (D) Shows the proportion of predicted mHAs corresponding to each mHA class for HLA-A alleles. (E) Shows the proportion for HLA-B alleles. (F) Shows the proportion of HLA-C alleles.
Genetic distance between the donor and recipient does not correlate with the number of predicted GVL mHAs
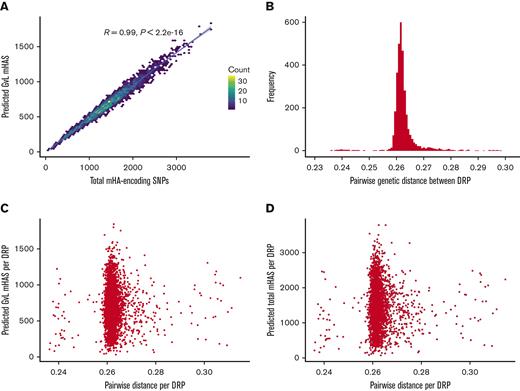
Next, we assessed whether the overall genetic distance between the donor and recipient correlated with predicted total mHAs or GVL mHAs. We saw a strong positive correlation between the total number of mHA-encoding SNPs and the number of predicted GVL mHAs (Figure 3A). We observed a narrow range of pairwise genetic distance across all DRPs within DISCOVeRY-BMT (Figure 3B), likely because a large number of rare SNPs were genotyped leading to high denominators of total SNPs and low numerators of SNPs that differ in genetic distance calculations. Still, distance values were consistent with previously reported data for healthy pairs.44 We found no correlation between genetic distance and predicted GVL mHAs (Figure 3C) or total mHAs (Figure 3D).
Degree of genetic distance versus the number of predicted GVL mHAs by DRP in DISCOVeRY-BMT data set. (A) Shows the number of total SNPs that differ and are predicted to lead to an mHA versus the number of predicted GVL mHAs per patient. (B) Shows the distribution of pairwise distance values for every DRP in the DISCOVeRY-BMT data set. Pairwise genetic distance value is calculated as the mean of (1-.5(number of shared alleles at SNP locus)) for every genotyped SNP locus for a DRP. (C) Shows pairwise genetic distance versus the number of predicted total mHAs per DRP. (D) Shows pairwise genetic distance versus the number of predicted GVL mHAs per DRP.
Degree of genetic distance versus the number of predicted GVL mHAs by DRP in DISCOVeRY-BMT data set. (A) Shows the number of total SNPs that differ and are predicted to lead to an mHA versus the number of predicted GVL mHAs per patient. (B) Shows the distribution of pairwise distance values for every DRP in the DISCOVeRY-BMT data set. Pairwise genetic distance value is calculated as the mean of (1-.5(number of shared alleles at SNP locus)) for every genotyped SNP locus for a DRP. (C) Shows pairwise genetic distance versus the number of predicted total mHAs per DRP. (D) Shows pairwise genetic distance versus the number of predicted GVL mHAs per DRP.
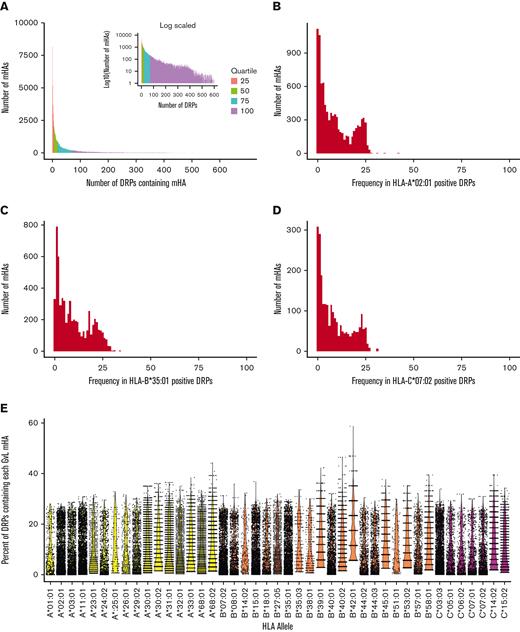
Most predicted mHAs are private, but a small number are widely shared among patients with any given HLA allele
We evaluated sharing of predicted mHAs within the DISCOVeRY-BMT cohort. Of our predicted mHAs, the majority were found within <10 DRPs. However, 38.7% of our predicted mHAs were shared by 1% or more of the study population and 4% were shared by 10% or more of the study population (Figure 4A). Next, we assessed sharing of mHAs within individual HLA alleles. For the 3 HLA alleles focused on in this work, the population frequency of predicted mHAs shows a bimodal distribution. Most mHAs are unshared, but a group of mHAs covers ∼20% to 30% of patients (Figure 4B,D). Finally, we assessed predicted mHA frequency across all HLA alleles represented by >0.5% of DISCOVeRY-BMT patients. The same bimodal distribution of mHA population frequency was observed across most HLA alleles (Figure 4E).
Degree of sharing of predicted mHAs across the study population. (A) Shows the distribution of predicted mHAs by the number of patients in the DISCOVeRY-BMT cohorts that possess them. Most mHAs are shared by ≤10 patients. Inlaid are the same data with y transformed y-axis to highlight the tail of the distribution. Data are colored by quartile of number of patients for each mHA. (B) Shows the distribution of predicted HLA-A∗02:01 mHAs by population frequency in DRPs with HLA-A∗02:01. (C) Shows the distribution of predicted HLA-B∗35:01 mHAs by population frequency in DRPs with HLA-B∗35:01. (D) Shows the distribution of predicted HLA-C∗07:02 mHAs by population frequency in DRPs with HLA-C∗07:02. (E) Shows the percentage of DISCOVeRY-BMT cohort with each HLA allele covered by each predicted GVL mHA that binds that HLA allele, for all HLA alleles representing >0.5% of DISCOVeRY-BMT patients.
Degree of sharing of predicted mHAs across the study population. (A) Shows the distribution of predicted mHAs by the number of patients in the DISCOVeRY-BMT cohorts that possess them. Most mHAs are shared by ≤10 patients. Inlaid are the same data with y transformed y-axis to highlight the tail of the distribution. Data are colored by quartile of number of patients for each mHA. (B) Shows the distribution of predicted HLA-A∗02:01 mHAs by population frequency in DRPs with HLA-A∗02:01. (C) Shows the distribution of predicted HLA-B∗35:01 mHAs by population frequency in DRPs with HLA-B∗35:01. (D) Shows the distribution of predicted HLA-C∗07:02 mHAs by population frequency in DRPs with HLA-C∗07:02. (E) Shows the percentage of DISCOVeRY-BMT cohort with each HLA allele covered by each predicted GVL mHA that binds that HLA allele, for all HLA alleles representing >0.5% of DISCOVeRY-BMT patients.
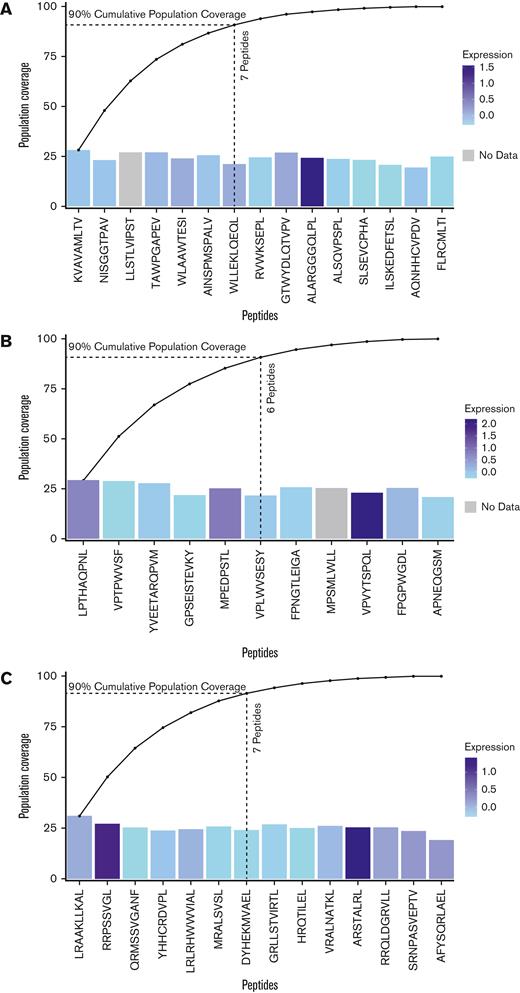
For 3 HLA alleles common in prevalent USt ethnic groups, 11 to 15 GVL mHA peptides cover 100% of patients in DISCOVeRY-BMT that express the given allele
We selected 3 HLA alleles to generate minimal mHA sets with our greedy algorithm. Together, HLA-A∗02:01, HLA-B∗35:01, and HLA-C∗07:02 represent a set of common alleles within the US population and within the major ethnic groups found in the DISCOVeRY-BMT population. For the most common HLA allele in the United States, HLA∗A02:01, a set of 15 GVL mHAs is needed to ensure that every DRP with this HLA allele has at least 1 of the 15 (Figure 5A). Only 7 peptides are needed to reach 90% coverage. The noncumulative population frequencies for each of these top 15 peptides range from 19.4% to 28.3%. We obtained similar results with HLA-B∗35:01, 11 peptides were needed to reach 100% population coverage and 6 peptides were needed to reach 90%, with noncumulative population frequencies between 20.9% and 29.3% (Figure 5B). HLA∗C07:02 also showed similar results, with 14 peptides needed to reach 100% population coverage and 7 peptides needed to reach 90%. Noncumulative frequencies ranged from 19.3% to 31.1% (Figure 5C). A total of 40 peptides gives 100% population coverage of 3 HLA alleles that are among the most common in major ethnic groups in the United States.
Patient population cumulative coverage by shared GVL mHAs. (A) Coverage of DISCOVeRY-BMT patients with HLA-A∗02:01 allele with predicted GVL mHAs. Noncumulative independent population frequencies of each of the top 15 peptides within the HLA-A∗02:01 population range from 19.4% to 28.3%, shown as bar heights. The colors of the bars show z-scores of expression for the genes that contain each peptide from The Cancer Genome Atlas AML sample expression data (TCGA_AML). Cumulative population coverage by the 15 predicted GVL mHAs needed to reach 100% corresponding coverage is shown as an overlaid line graph. Dotted lines indicate 7 peptides needed to reach 90% population coverage. (B) Shows coverage of DISCOVeRY-BMT patients with HLA-B∗35:01 allele with predicted GVL mHAs. Eleven predicted GVL mHAs correspond to 100% cumulative population coverage and 6 correspond to 90% coverage for this HLA allele. Noncumulative coverage by the top 11 peptides for this HLA allele range from 20.9% to 29.3%. (C) Shows coverage of DISCOVeRY-BMT patients with HLA-C∗07:02. Fourteen predicted GVL mHAs correspond to 100% cumulative population coverage and 7 correspond to 90% for this allele. Noncumulative coverage for these mHAs ranges from 19.3% to 31.1%.
Patient population cumulative coverage by shared GVL mHAs. (A) Coverage of DISCOVeRY-BMT patients with HLA-A∗02:01 allele with predicted GVL mHAs. Noncumulative independent population frequencies of each of the top 15 peptides within the HLA-A∗02:01 population range from 19.4% to 28.3%, shown as bar heights. The colors of the bars show z-scores of expression for the genes that contain each peptide from The Cancer Genome Atlas AML sample expression data (TCGA_AML). Cumulative population coverage by the 15 predicted GVL mHAs needed to reach 100% corresponding coverage is shown as an overlaid line graph. Dotted lines indicate 7 peptides needed to reach 90% population coverage. (B) Shows coverage of DISCOVeRY-BMT patients with HLA-B∗35:01 allele with predicted GVL mHAs. Eleven predicted GVL mHAs correspond to 100% cumulative population coverage and 6 correspond to 90% coverage for this HLA allele. Noncumulative coverage by the top 11 peptides for this HLA allele range from 20.9% to 29.3%. (C) Shows coverage of DISCOVeRY-BMT patients with HLA-C∗07:02. Fourteen predicted GVL mHAs correspond to 100% cumulative population coverage and 7 correspond to 90% for this allele. Noncumulative coverage for these mHAs ranges from 19.3% to 31.1%.
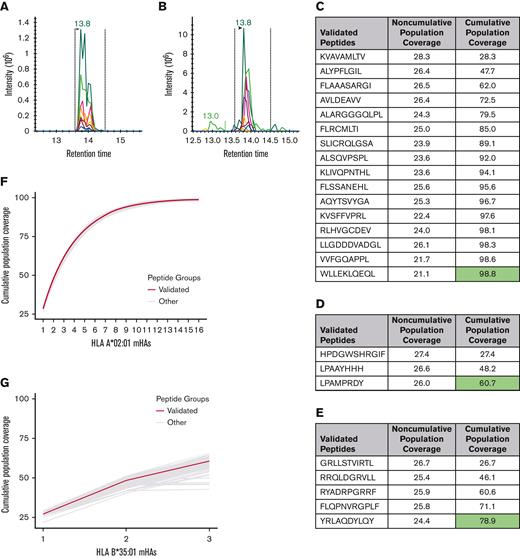
24 novel GVL mHAs were validated using mass spectrometry
We employed mass spectrometry to validate the HLA presentation of predicted GVL mHAs. Of the 67 peptides searched for HLA-A∗02:01 across 2 U937A2 cell line samples, we positively identified 17 peptides. Of the 40 searched for HLA-B∗35:01 using an NB4 cell line, we identified 3 peptides, and of the 40 searched for C∗07:02 using a MONOMAC1 cell line, we identified 5 peptides. Representative spectra are shown for a heavy-labeled peptide standard and endogenous identified peptide from an immunoprecipitated NB4 cell sample (Figure 6A-B). From the list of 17 validated peptides for HLA-A∗02:01, peptide VLDIEQFSV is also known as UNC-GRK4-V and was previously identified by our group as a GVL mHA using the U937A2 cell line.1,29 Mass spectrometry analysis was blinded to the peptide’s status as previously identified. As this peptide is previously known, a total of 16 novel HLA-A∗02:01 binding mHAs were discovered. These 16 novel HLA-A∗02:01–binding mHAs cumulatively cover 98.8% of patients with positive HLA-A∗02:01 in the DISCOVeRY-BMT data set, with individual peptide population frequencies between 21.1% and 28.3% (Figure 6C). The 3 novel HLA-B∗35:01 binding mHAs cover 60.7% of the HLA-B∗35:01–positive DISCOVeRY-BMT population, with population frequencies of 26.0% to 27.6% (Figure 6D). The 5 novel HLA-C∗07:02–binding mHAs give cumulative HLA-C∗07:02–positive DISCOVeRY-BMT patient coverage of 78.9%, with independent frequencies of 24.4% to 26.7% (Figure 6E). The characteristics of all novel mHAs are shown in Table 1.54,55 One novel mHA, UNC-BCL2A1-Y, is derived from the same SNP as the previously identified mHA ACC1Y.56 These 2 mHAs overlap by 5 amino acids, and ACC1Y binds HLA-A∗24:02, while our novel UNC-BCL2A1-Y binds HLA-C∗07:02. We also demonstrated immunogenicity of 1 example novel mHA, UNC-HEXDC-V, via tetramer staining of CD8 T cells cocultured with mHA-pulsed DCs (Figure 6I).
Mass spectrometry validation of predicted GVL mHAs for HLA-A∗02:01, B∗35:01, and C∗07:02. (A) Shows representative spectra for heavy-labeled peptide standard for HLA-C∗07:02–binding mHA LPAAYHHH. (B) Shows endogenous LPAAYHHH peptide identified from immunoprecipitated peptide sample from cell line MONOMAC1. (C) Shows all novel identified peptides from cell line U937A2 sample. “Noncumulative population coverage” shows the percentage of DRPs expressing HLA-A∗02:01 within the DISCOVeRY-BMT data set where the recipient expresses the mHA allele and the donor does not. “Cumulative population coverage” shows the output from the greedy algorithm calculating total population coverage by each peptide and the ones preceding it, with a total of 98.8% population coverage by the 10 peptides. (D) Shows all identified peptides from cell line NB4 sample, with a 60.7% cumulative coverage of DRPs expressing HLA-B∗35:01 within the data set by the 3 peptides. (E) Shows all identified peptides from cell line MONOMAC1, with a 78.9% cumulative coverage of HLA-C∗07:02–expressing DRPs within the data set. (F) Shows cumulative coverage by the 16 novel confirmed HLA-A∗02:01–binding mHAs and 1000 simulated sets of 16 peptides from the set of mHAs searched by mass spectrometry. Cumulative coverage by confirmed peptides is shown in blue, whereas each simulated run is shown as an individual gray line. (G) Shows cumulative coverage for the 3 confirmed HLA-B∗35:01–binding mHAs and 1000 simulated sets of 3 peptides. (H) Shows cumulative coverage for the 5 confirmed HLA-C∗07:02–binding mHAs and 1000 simulated sets of 5 peptides. (I) Shows flow cytometry staining from the mHA immunogenicity experiment. “UV only” shows negative control stained with tetramer exposed to UV light with no peptide. “Flu-M158-66” shows CD8 T cells cocultured with M158-66 pulsed DCs, stained with M158-66 tetramer. “UNC-HEXDC-V” shows CD8 T cells cocultured with novel mHA UNC-HEXDC-V stained with UNC-HEXDC-V tetramer.
Mass spectrometry validation of predicted GVL mHAs for HLA-A∗02:01, B∗35:01, and C∗07:02. (A) Shows representative spectra for heavy-labeled peptide standard for HLA-C∗07:02–binding mHA LPAAYHHH. (B) Shows endogenous LPAAYHHH peptide identified from immunoprecipitated peptide sample from cell line MONOMAC1. (C) Shows all novel identified peptides from cell line U937A2 sample. “Noncumulative population coverage” shows the percentage of DRPs expressing HLA-A∗02:01 within the DISCOVeRY-BMT data set where the recipient expresses the mHA allele and the donor does not. “Cumulative population coverage” shows the output from the greedy algorithm calculating total population coverage by each peptide and the ones preceding it, with a total of 98.8% population coverage by the 10 peptides. (D) Shows all identified peptides from cell line NB4 sample, with a 60.7% cumulative coverage of DRPs expressing HLA-B∗35:01 within the data set by the 3 peptides. (E) Shows all identified peptides from cell line MONOMAC1, with a 78.9% cumulative coverage of HLA-C∗07:02–expressing DRPs within the data set. (F) Shows cumulative coverage by the 16 novel confirmed HLA-A∗02:01–binding mHAs and 1000 simulated sets of 16 peptides from the set of mHAs searched by mass spectrometry. Cumulative coverage by confirmed peptides is shown in blue, whereas each simulated run is shown as an individual gray line. (G) Shows cumulative coverage for the 3 confirmed HLA-B∗35:01–binding mHAs and 1000 simulated sets of 3 peptides. (H) Shows cumulative coverage for the 5 confirmed HLA-C∗07:02–binding mHAs and 1000 simulated sets of 5 peptides. (I) Shows flow cytometry staining from the mHA immunogenicity experiment. “UV only” shows negative control stained with tetramer exposed to UV light with no peptide. “Flu-M158-66” shows CD8 T cells cocultured with M158-66 pulsed DCs, stained with M158-66 tetramer. “UNC-HEXDC-V” shows CD8 T cells cocultured with novel mHA UNC-HEXDC-V stained with UNC-HEXDC-V tetramer.
Novel mHA characteristics
| mHA name . | mHA . | HLA allele . | Gene . | Chromosome . | rsID of SNP . | Donor amino acid . | Recipient amino acid . | Major allele . | Minor allele . | MAF in TOPMED . | MAF in ALFA . | MAF in 1000 genomes . | Peptide length . | Binding affinity . | Frequency in DISCOVeRY-BMT patients with corresponding HLA . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UNC-IQCE-V | AVLDEAVV | A∗02:01 | IQCE | 11 | rs2293404 | A | V | C | T | 0.35 | 0.29 | 0.41 | 8 | 412.7421 | 26.4 |
| UNC-GLRX3-S | FLSSANEHL | A∗02:01 | GLRX3 | 10 | rs2274217 | P | S | C | T | 0.21 | 0.25 | 0.19 | 9 | 14.9364 | 25.6 |
| UNC-SLC25A37-V | AQYTSVYGA | A∗02:01 | SLC25A37 | 8 | rs2942194 | I | V | A | G | 0.21 | 0.26 | 0.17 | 9 | 45.2204 | 25.3 |
| UNC-ARHGEF18-Q | SLICRQLGSA | A∗02:01 | ARHGEF18 | 19 | rs2287918 | R | Q | G | A | 0.19 | 0.24 | 0.17 | 10 | 367.3796 | 23.9 |
| UNC-DPP3-H | KLIVQPNTHL | A∗02:01 | DPP3 | 11 | rs2305535 | R | H | G | A | 0.19 | 0.22 | 0.21 | 10 | 218.2962 | 23.6 |
| UNC-HEXDC-V | RLHVGCDEV | A∗02:01 | HEXDC | 17 | rs4789773 | I | V | A | G | 0.45 | 0.37 | 0.56 | 9 | 357.9789 | 24 |
| UNC-TOP1MT-W | WLLEKLQEQL | A∗02:01 | TOP1MT | 8 | rs2293925 | R | W | C | T | 0.36 | 0.43 | 0.46 | 10 | 8.8962 | 21.1 |
| UNC-USP4-V | KVSFFVPRL | A∗02:01 | USP4 | 3 | rs35446411 | L | V | T | G | 0.13 | 0.16 | 0.09 | 9 | 445.1234 | 22.4 |
| UNC-AHRR-A | VVFGQAPPL | A∗02:01 | AHRR | 5 | rs2292596 | P | A | C | G | 0.30 | 0.35 | 0.38 | 9 | 311.398 | 21.7 |
| UNC-FPR1-K | KVAVAMLTV | A∗02:01 | FPR1 | 19 | rs1042229 | N | K | T | G | — | 0.45 | 0.37 | 9 | 178.6526 | 28.3 |
| UNC-FLT3-G | ALARGGGQLPL | A∗02:01 | FLT3 | 13 | rs12872889 | D | G | A | G | 0.35 | 0.31 | 0.37 | 11 | 257.8616 | 24.3 |
| UNC-GDPD5-A | ALSQVPSPL | A∗02:01 | GDPD5 | 11 | rs571353 | T | A | A | G | 0.34 | 0.28 | 0.33 | 9 | 94.9686 | 23.6 |
| UNC-SLC26A8-M | FLRCMLTI | A∗02:01 | SLC26A8 | 6 | rs743923 | V | M | G | A | 0.30 | 0.25 | 0.26 | 8 | 100.0366 | 25 |
| UNC-FPGS-I | FLAAASARGI | A∗02:01 | FPGS | 9 | rs10760502 | V | I | C | T | 0.28 | 0.35 | 0.22 | 10 | 23.9163 | 26.5 |
| UNC-NDUFAF1-L | ALYPFLGIL | A∗02:01 | NDUFAF1 | 15 | rs3204853 | R | L | C | A | 0.17 | 0.24 | 0.12 | 9 | 164.2741 | 26.4 |
| UNC-WDR62-L | LLGDDDVADGL | A∗02:01 | WDR62 | 19 | rs2285745 | S | L | C | T | 0.32 | 0.35 | 0.35 | 11 | 182.9111 | 26.1 |
| UNC-POLL-W | HPDGWSHRGIF | B∗35:01 | POLL | 10 | rs3730477 | R | W | C | T | 0.16 | 0.21 | 0.10 | 11 | 31.3362 | 27.4 |
| UNC-HLX-P | LPAAYHHH | B∗35:01 | HLX | 1 | rs12141189 | S | P | T | C | 0.23 | 0.23 | 0.21 | 8 | 285.0737 | 26.6 |
| UNC-NEK4-A | LPAMPRDY | B∗35:01 | NEK4 | 3 | rs1029871 | P | A | G | C | 0.33 | 0.39 | 0.31 | 8 | 39.4681 | 26 |
| UNC-MARCH2-T | GRLLSTVIRTL | C∗07:02 | MARCH2 | 19 | rs1133893 | A | T | C | T | 0.24 | 0.32 | 0.20 | 11 | 175.8924 | 26.7 |
| UNC-GAA-R | RRQLDGRVLL | C∗07:02 | GAA | 17 | rs1042395 | H | R | A | G | 0.36 | 0.28 | 0.40 | 10 | 375.2756 | 25.4 |
| UNC-RNASE3-R | RYADRPGRRF | C∗07:02 | RNASE3 | 14 | rs2073342 | T | R | C | G | 0.36 | 0.29 | 0.36 | 10 | 147.8419 | 25.9 |
| UNC-SNX19-V | FLQPNVRGPLF | C∗07:02 | SNX19 | 11 | rs3751037 | L | V | G | C | 0.30 | 0.29 | 0.27 | 11 | 161.7018 | 25.8 |
| UNC-BCL2A1-Y | YRLAQDYLQY | C∗07:02 | BCL2A1 | 15 | rs1138357 | C | Y | G | A | 0.28 | 0.26 | 0.35 | 10 | 188.9083 | 24.4 |
| mHA name . | mHA . | HLA allele . | Gene . | Chromosome . | rsID of SNP . | Donor amino acid . | Recipient amino acid . | Major allele . | Minor allele . | MAF in TOPMED . | MAF in ALFA . | MAF in 1000 genomes . | Peptide length . | Binding affinity . | Frequency in DISCOVeRY-BMT patients with corresponding HLA . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UNC-IQCE-V | AVLDEAVV | A∗02:01 | IQCE | 11 | rs2293404 | A | V | C | T | 0.35 | 0.29 | 0.41 | 8 | 412.7421 | 26.4 |
| UNC-GLRX3-S | FLSSANEHL | A∗02:01 | GLRX3 | 10 | rs2274217 | P | S | C | T | 0.21 | 0.25 | 0.19 | 9 | 14.9364 | 25.6 |
| UNC-SLC25A37-V | AQYTSVYGA | A∗02:01 | SLC25A37 | 8 | rs2942194 | I | V | A | G | 0.21 | 0.26 | 0.17 | 9 | 45.2204 | 25.3 |
| UNC-ARHGEF18-Q | SLICRQLGSA | A∗02:01 | ARHGEF18 | 19 | rs2287918 | R | Q | G | A | 0.19 | 0.24 | 0.17 | 10 | 367.3796 | 23.9 |
| UNC-DPP3-H | KLIVQPNTHL | A∗02:01 | DPP3 | 11 | rs2305535 | R | H | G | A | 0.19 | 0.22 | 0.21 | 10 | 218.2962 | 23.6 |
| UNC-HEXDC-V | RLHVGCDEV | A∗02:01 | HEXDC | 17 | rs4789773 | I | V | A | G | 0.45 | 0.37 | 0.56 | 9 | 357.9789 | 24 |
| UNC-TOP1MT-W | WLLEKLQEQL | A∗02:01 | TOP1MT | 8 | rs2293925 | R | W | C | T | 0.36 | 0.43 | 0.46 | 10 | 8.8962 | 21.1 |
| UNC-USP4-V | KVSFFVPRL | A∗02:01 | USP4 | 3 | rs35446411 | L | V | T | G | 0.13 | 0.16 | 0.09 | 9 | 445.1234 | 22.4 |
| UNC-AHRR-A | VVFGQAPPL | A∗02:01 | AHRR | 5 | rs2292596 | P | A | C | G | 0.30 | 0.35 | 0.38 | 9 | 311.398 | 21.7 |
| UNC-FPR1-K | KVAVAMLTV | A∗02:01 | FPR1 | 19 | rs1042229 | N | K | T | G | — | 0.45 | 0.37 | 9 | 178.6526 | 28.3 |
| UNC-FLT3-G | ALARGGGQLPL | A∗02:01 | FLT3 | 13 | rs12872889 | D | G | A | G | 0.35 | 0.31 | 0.37 | 11 | 257.8616 | 24.3 |
| UNC-GDPD5-A | ALSQVPSPL | A∗02:01 | GDPD5 | 11 | rs571353 | T | A | A | G | 0.34 | 0.28 | 0.33 | 9 | 94.9686 | 23.6 |
| UNC-SLC26A8-M | FLRCMLTI | A∗02:01 | SLC26A8 | 6 | rs743923 | V | M | G | A | 0.30 | 0.25 | 0.26 | 8 | 100.0366 | 25 |
| UNC-FPGS-I | FLAAASARGI | A∗02:01 | FPGS | 9 | rs10760502 | V | I | C | T | 0.28 | 0.35 | 0.22 | 10 | 23.9163 | 26.5 |
| UNC-NDUFAF1-L | ALYPFLGIL | A∗02:01 | NDUFAF1 | 15 | rs3204853 | R | L | C | A | 0.17 | 0.24 | 0.12 | 9 | 164.2741 | 26.4 |
| UNC-WDR62-L | LLGDDDVADGL | A∗02:01 | WDR62 | 19 | rs2285745 | S | L | C | T | 0.32 | 0.35 | 0.35 | 11 | 182.9111 | 26.1 |
| UNC-POLL-W | HPDGWSHRGIF | B∗35:01 | POLL | 10 | rs3730477 | R | W | C | T | 0.16 | 0.21 | 0.10 | 11 | 31.3362 | 27.4 |
| UNC-HLX-P | LPAAYHHH | B∗35:01 | HLX | 1 | rs12141189 | S | P | T | C | 0.23 | 0.23 | 0.21 | 8 | 285.0737 | 26.6 |
| UNC-NEK4-A | LPAMPRDY | B∗35:01 | NEK4 | 3 | rs1029871 | P | A | G | C | 0.33 | 0.39 | 0.31 | 8 | 39.4681 | 26 |
| UNC-MARCH2-T | GRLLSTVIRTL | C∗07:02 | MARCH2 | 19 | rs1133893 | A | T | C | T | 0.24 | 0.32 | 0.20 | 11 | 175.8924 | 26.7 |
| UNC-GAA-R | RRQLDGRVLL | C∗07:02 | GAA | 17 | rs1042395 | H | R | A | G | 0.36 | 0.28 | 0.40 | 10 | 375.2756 | 25.4 |
| UNC-RNASE3-R | RYADRPGRRF | C∗07:02 | RNASE3 | 14 | rs2073342 | T | R | C | G | 0.36 | 0.29 | 0.36 | 10 | 147.8419 | 25.9 |
| UNC-SNX19-V | FLQPNVRGPLF | C∗07:02 | SNX19 | 11 | rs3751037 | L | V | G | C | 0.30 | 0.29 | 0.27 | 11 | 161.7018 | 25.8 |
| UNC-BCL2A1-Y | YRLAQDYLQY | C∗07:02 | BCL2A1 | 15 | rs1138357 | C | Y | G | A | 0.28 | 0.26 | 0.35 | 10 | 188.9083 | 24.4 |
16 novel GVL mHAs that bind HLA-A∗02:01, 3 that bind B∗35:01, and 5 that bind C∗07:02 and were validated by mass spectrometry are shown.
MAF, minor allele frequency.
To evaluate the generalizability of our discovery process, we calculated the range of cumulative coverage that would be obtained with a subset of the number of peptides that we validated from the searched lists. For each HLA allele, 1000 random sets of peptides were selected from the searched peptide list and cumulative coverage by each set was calculated. The range of cumulative coverage by the 1000 random sets of 16 HLA-A∗02:01 peptides was from 97.4% to 99.7%, by the 1000 random sets of 3 HLA-B∗35:01 peptides was from 42.8% to 66.4%, and by the 1000 random sets of 5 HLA-C∗07:02 peptides was from 65.4% to 80.7% (Figure 6F-H).
Discussion
Discovery and characterization of novel mHAs is crucial for enhancing immune monitoring in alloHCT, predicting clinical outcomes based on donor and recipient genetics, and improving outcomes by optimizing donor selection and/or specifically targeting GVL mHAs. We built upon previous work to perform the first population-level survey of mHA peptides, taking a new approach by predicting mHAs common among recipients with diverse HLA alleles. This ensures that therapeutics targeting our newly identified mHAs would apply to as broad of a recipient population as possible.
We evaluated mHAs for a total of 56 HLA-A, HLA-B, and HLA-C alleles called in 3231 DISCOVeRY-BMT recipients. Despite large differences in the total number of mHAs per HLA allele, ∼50% of predicted mHAs for each HLA allele are GVL. Therefore, we expect that every HLA allele will present a set of GVL mHAs. The majority of GVL mHAs are shared among <10 patients in the data set, highlighting the largely private nature of the mHA landscape. That said, for each HLA allele, we predicted a small number of highly shared mHA expressed by 20% to 25% of the recipient population. For all HLA alleles studied, 6 to 8 mHA peptides would cover >80% of recipients that express that allele, and 11 to 15 mHA would cover 100% of recipients. Conceptually, targeting a small number of shared GVL mHAs could treat a majority of alloHCT recipients regardless of race or ethnicity.
Using mass spectrometry, we validated a total of 24 novel GVL mHAs, an increase from the 12 class I GVL mHAs that have been discovered since Goulmy et al reported the first discovered GVL mHA, HA-1, in 1983.1,25,28,30-32 The 16 novel GVL mHAs found for HLA-A∗02:01 together cover 98.8% of patients with positive HLA-A∗02:01 in the DISCOVeRY-BMT data set, the 3 for HLA-B∗35:01 cover 60.7% of HLA-B∗35-:01–positive DISCOVeRY-BMT patients, and the 5 for HLA-C∗07:02 cover 78.9% of HLA-C∗07:02–positive DISCOVeRY-BMT patients. Furthermore, we confirmed the immunogenicity of 1 predicted novel mHA, UNC-HEXDC-V, via tetramer staining of T cells cocultured with mHA-pulsed DCs. We expect that these novel mHAs will serve as future targets for antigen-directed therapeutics.
We genotyped 7 DRPs from the Lineberger Cancer Center Tissue Procurement Facility, University of North Carolina expressing HLA-A∗02:01, 1 expressing HLA-B∗35:01 and 5 expressing HLA-C∗07:02 for the majority of the novel mHAs for the corresponding HLA alleles (supplemental Figure 3). We found appropriate minor antigen mismatches for a potential use of these mHAs in 58% of the genotyped DRPs, highlighting their utility for future work. This does not align perfectly with the predicted coverage of DISCOVeRY-BMT patients with these mHAs, but is likely explained by the small patient count and different patient populations. However, most of the patients genotyped could use treatments targeting these mHAs. We also genotyped the 7 HLA-A∗02:01–positive DRPs for the previously known GVL mHAs HA-1 and UTA2-1 and discovered they were targetable in 0% of these DRPs. We also assessed allele frequencies in the DISCOVeRY-BMT population and found that most of the 11 previously known class I GVL mHAs are not targetable for any patients in this data set (supplemental Table 2). This emphasizes the expanded utility of finding shared mHAs over traditional methods.
Our study is limited in important ways. We biologically validated predicted GVL mHAs for 3 HLA alleles that were selected based on their high frequency of expression within diverse ethnic groups. In the future, mHAs for additional HLA alleles should be validated. Furthermore, we validated GVL mHAs in a single AML cell line for each HLA allele. This is sufficient to establish that the mHAs are capable of being presented; however, antigen expression, HLA expression, and antigen presentation efficiency will be heterogeneous across patient samples. Further studies of primary AML samples will be required to estimate the frequency of expression of each GVL mHA in AML. We validated more mHAs for HLA-A∗02:01 than the other HLA alleles, which is likely not only due to running 2 samples for this allele but also because the cell line U937A2 is engineered to express HLA-A∗02:01 and presents larger quantities of it on its cell surface than endogenously expressed HLA alleles. NB4 endogenously expresses HLA-B∗35:01 and MONOMAC1 endogenously expresses HLA-C∗07:02. In addition, though this study includes in vivo validation of immunogenicity of 1 of our novel mHAs with a healthy donor sample, future work will identify mHA-specific T cells for all mHAs validated in this work. We will also assess T-cell responses to the novel GVL mHAs in alloHCT recipients to better understand determinants of GVL mHA immunogenicity.
This work increases the number of known validated class I GVL mHAs by 200%, and these mHAs are unique in being specifically identified for their high population prevalence in the corresponding HLA-expressing DRPs. Targeting these newly discovered mHAs could greatly expand the capacity for the treatment of patients with AML with GVL mHA-targeting immunotherapies.
Acknowledgments
The authors acknowledge the participation of all the patients and donors who consented to the biorepository and research database, as well as all transplant centers which participated in the Center for International Blood and Marrow Transplant Research database and biorepository studies.
The authors acknowledge the Genotype-Tissue Expression (GTEx) Project, which is supported by the Common Fund of the Office of the Director of the National Institutes of Health (NIH) and by National Cancer Institute (NCI), National Human Genome Research Institute, National Heart, Lung, and Blood Institute (NHLBI), National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke.
This work was supported by the University of North Carolina University Cancer Research Fund (B.V.); the NIH (1F30CA268748) (K.S.O.), (5R37CA247676-03 formerly 1R01CA247676) (B.V. and P.A.), and (NHLBI, R01 HL102278 and NCI, R03 CA188733) (L.S.-C. and T.H.), and the DISCOVeRY-BMT (NIH R01 HL102278).
The Center for International Blood and Marrow Transplant Research is supported primarily by Public Health Service U24CA076518 NCI, the NHLBI, and the National Institute of Allergy and Infectious Diseases (NIAID); NHLBI and NCI (U24HL138660 and U24HL157560); NCI (U24CA233032); NHLBI (OT3HL147741 and U01HL128568); Health Resources and Services Administration (HRSA) (HHSH250201700005C, HHSH250201700006C, and HHSH250201700007C); and the Office of Naval Research (N00014-20-1-2832 and N00014-21-1-2954).
The views expressed in this article do not reflect the official policy or position of the NIH, the Department of the Navy, the Department of Defense, Health Resources and Services Administration, or any other agency of the US Government.
Some figures were created with BioRender.com.
Authorship
Contribution: B.V., P.A., and K.S.O. conceived the project, designed experiments, and interpreted experimental results; K.S.O. prepared the manuscript, generated figures, performed experiments, survival analyses, and computational minor histocompatibility antigen prioritization; O.J., S.D., D.B., and S.P.V.II performed minor histocompatibility antigen prediction and assisted with computational algorithm generation; S.B. performed coculture experiment; H.T. assisted with ethnicity analyses; M.D. and D.D. assisted with experiments; T.S. assisted with HLA typing comparisons; Q.Z. and A.W. performed data quality control; Y.W. analyzed the data; C.A.H., L.P., and X.S. performed genotyping interpretation; M.C.P. and S.R.S. acquired data and interpreted analyses; P.L.M. interpreted data analyses; E.W. performed HLA typing of cell lines; T.H. conceived and designed the DISCOVeRY-BMT study and acquired and interpreted data; L.S.-C. provided data, assisted with study conception and design, and assisted with data analyses; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Benjamin Vincent, The University of North Carolina at Chapel Hill, 5230E Marsico Hall, 125 Mason Farm Road, Chapel Hill, NC 27599; e-mail: benjamin_vincent@med.unc.edu.
References
Author notes
∗P.A. and B.V. are joint last authors.
The RNA sequencing data reported in this article have been deposited to the Gene Expression Omnibus database (accession number GSE212013).
DISCOVeRY-BMT data are available via request to Center for International Blood and Marrow Transplant Research.
Data are available on request from the corresponding author, Benjamin Vincent (benjamin_vincent@med.unc.edu).
The full-text version of this article contains a data supplement.