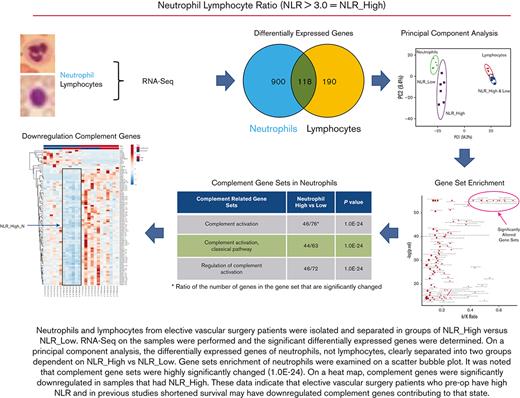
Key Points
Changes in neutrophils is the dynamic factor altering neutrophil-to-lymphocyte ratios in patients who undergo elective vascular surgery.
Patients with elevated neutrophil-to-lymphocyte ratios are associated with reduced expression of complement genes in neutrophils.
Abstract
Elevated neutrophil-to-lymphocyte ratio (NLR) in patients who undergo elective vascular surgery (EVS) have increased mortality independent of perioperative surgical outcome. To understand why high NLR is associated with higher mortality, we investigated neutrophil and lymphocyte transcriptome expression in patients undergoing EVS. Blood samples were collected from patients undergoing EVS and healthy donors for NLR calculation. RNA samples were isolated from patients' neutrophils and lymphocytes and divided into NLR_Low (<3) and NLR_High (≥3) groups (n = 6 each). Paired samples with the highest RNA integrity number (mean = 9.8 ± 0.4) were sequenced and analyzed for differential expression. Normalized data were inputted for downstream analysis using iPathwayGuide (AdvaitaBio) and gene set enrichment analysis using GenePattern and MSigDB (Broad Institute). There was no clinical difference between the patient groups with regard to clinical diagnosis, age, sex, history of hypertension, lipid abnormalities, diabetes mellitus, smoking, or statin use. The mean NLR was 4.37 ± 0.27 SEM in the NLR_High and 1.88 ± 0.16 for the NLR_Low groups. Significantly differentially expressed gene sets identified in the RNA sequence data were enriched highly (P = 1E-24) in the humoral immunity and complement systems. Neutrophils from NLR_High patients downregulated complement genes (C1QA, C1QB, C1QC, C1S, C2, CR2, C3AR1, C3, C8G, and C9 and complement regulatory genes CD59, SERPING1, C4BPA, CFH, and CFI). Downregulation of gene expressions of humoral immunity and complement within the neutrophils are associated with elevated NLR. It remains to be determined whether and how these changes contribute to increased late mortality previously observed in patients undergoing EVS.
Introduction
Cardiovascular disease (CVD) is a leading cause of death worldwide. It is due, in large part, to atherogenesis and atherosclerosis that, in addition to individual genetics and diet, is mediated by inflammatory and immune mechanisms.1-3 Recently, the differential leukocyte count (neutrophils and lymphocytes) has come into focus to predict cardiovascular (CV) outcomes. The ratio of the absolute neutrophil-to-lymphocyte count (NLR) has been recognized as a predictor of total mortality in multiple small studies of acute coronary interventions,4-6 hypertension,7,8 and heart failure.9
Previous investigations have also shown that NLR is a predictor of long-term outcomes in patients with peripheral vascular disease.10-13 Our retrospective studies of vascular surgery patient populations support this conclusion. In 1 study of 108 patients with elective endovascular abdominal aortic aneurysm repair, there was no difference in the 30-day postoperative mortality between an NLR of <4 and an NLR of >4 (P = .507). However, the 1-year, 3-year, and 5-year mortality between the groups were 4.2% vs 28.1%, 15.1% vs 64.9%, and 24% vs 90%, respectively.14 In a second study of 290 asymptomatic patients undergoing prophylactic carotid endarterectomy, the risk for stroke was noted to be significantly higher (P < .0001) in patients with an NLR of >3 (n = 116 patients, 42.6% stroke risk) than those with an NLR of <3 (n = 174 patients, 9.3% stroke risk).15 In a third study with 488 patients who underwent percutaneous interventions of femoropopliteal arteries, the 30-day mortality rates increased significantly (P = .005) with increasing NLR (1.4%, 4.3%, and 7.0% for low [<3], mid [3-4], and high [>4] NLR groups, respectively).16 Patients with a lower preoperative NLR achieved significantly greater amputation-free survival at the 4-year follow-up (low NLR 65.5%, mid NLR, 37.5%, and high NLR, 17.6% [P < .0001]).16
In this study, we performed RNA sequencing (RNA-seq) of neutrophils and lymphocytes from patients who qualified for elective vascular surgery (EVS) repair to determine potential mechanisms for elevated NLR. The studies examined whether there is a molecular basis for elevated NLR. Our investigations reveal that patients with elevated NLR who had EVS repair are distinguished by highly significant downregulation of immunoglobulin and complement genes in neutrophils.
Methods
Patients
Blood samples were collected from candidate patients for EVS procedures and healthy volunteers for NLR calculation and RNA sequence studies of neutrophils and lymphocytes. In screening patients for participation, any individual with an active local or systemic infection or obvious inflammatory disease were excluded from participation. Peripheral blood collection was approved by the institutional review board of University Hospitals Cleveland Medical Center (#01-06-02). Informed consent was obtained from each patient and healthy volunteer. All patients for EVS seen in the outpatient department were eligible. Patients in the study were characterized for the type of vascular surgery, age, sex, hypertension (blood pressure >130/80 mm Hg), diabetes mellitus (fasting blood glucose >100 mg/dL), smoking status, statin treatment, and a medical history of chronic obstructive pulmonary disease, cancer, chronic renal disease, coronary artery disease (CAD), peripheral arterial disease, cerebrovascular disease, and chronic heart failure. All patients had a complete blood count with differential white blood cell count. Patients were divided into low NLR (NLR_Low [<3]) and high NLR (NLR_High [≥3]) groups (n = 6 each) and healthy volunteers aged 18 to 71 years (n = 6). Eleven of 12 patients’ blood was drawn before surgery. One patient had blood drawn 4 months after surgery.
Blood separation
Whole blood (20 mL per patient) was collected in 10 mL vacutainer tubes anticoagulated with EDTA, and the blood was separated within an hour of collection. Neutrophil and lymphocyte cell populations were each isolated from 8 mL of blood using EasySep direct human cell isolation kits (STEMCELL Technologies, Vancouver, Canada) and the Big Easy EasySep magnet (STEMCELL Technologies, Vancouver, Canada) according to the manufacturer’s protocols. The magnet was chilled on ice before, and the blood cells and reagents were gently mixed by inverting the tube as opposed to pipetting. The reagent ratios were also adjusted in the following manner to optimize recovery: 400 mL of antibody mixture, 400 mL of magnetic beads, and 4 mL of phosphate-buffered saline were added to the blood before the first separation, followed by 200 mL of antibody mixture and 400 mL of beads before the second separation and 200 mL of beads before the third and final separation. A small aliquot of 200 mL was taken from the final recovery volumes for cell counting and slide preparation. The remainder of the recovery volume was centrifuged at 1000 revolutions per minute for 7 minutes to pellet the cells.
Cell counting and slide preparation
Cells resuspended in phosphate-buffered saline were further aliquoted, mixed with trypan blue, and manually counted using a hemocytometer. Typically, 9 × 106 to 15 × 106 neutrophils and 3 × 106 to 10 × 106 lymphocytes were isolated per sample. Slides of the isolated cells were prepared using a cytocentrifuge, spun at 600 revolutions per minute for 3 minutes, and then stained with Wright-Giemsa stain. The purity of each cell population was determined by manual counting, in which at least 1000 cells were distinguished per slide. The average purities of the neutrophil and lymphocyte separations by cell counting were 98.3% and 96.2%, respectively. Flow cytometry was performed on 1 sample pair of neutrophils and lymphocytes and indicated 90.6% and 92.2% purity, respectively. The other cells in the neutrophil preparation were broken down to lymphocytes (4%), monocytes (2%), eosinophils (2.3%), and basophils (0.3%). In the lymphocyte preparation, the other cells were neutrophils (1.2%), monocytes (1.4%), eosinophils (1.2%), and basophils (1.8%). Bulk RNA-seq data from all individual samples were also used to determine the extent of monocyte contamination in the cell preparations by determining whether neutrophils or lymphocytes from NLR_High or NLR_Low samples had significantly increased CD14, CD68, CD83, or CD163. Independently, the RNA-seq data were entered into estimating the proportion of immune and cancer cells (EPIC), a program designed to estimate the proportion of immune and cancer cells in bulk gene expression data.17 The reference profile used was provided by EPIC for blood circulating immune cells and contains the profile for B cells, CD4 and CD8 T cells, monocytes, neutrophils, and natural killer cells. The output reported is given as cell fractions per sample.
RNA-seq and statistical analysis
Detailed methods for RNA extraction and sequencing are presented in the supplemental Methods. Sequencing reads were assessed for quality, and the adapter was trimmed using TrimGalore! (Babraham Institute), a wrapper script for FastQC and CutAdapt. Reads passing quality control were aligned to the human reference genome GRCh38 using the STAR aligner software. The aligned reads were processed using Cufflinks version 2.2.1 for differential expression analysis using the GENCODE gene annotation for GRCh38 and gene-level expression data were reported in fragments per kilobase transcript per million reads mapped.18 Significantly differentially expressed genes were identified using a false discovery rate cutoff P value of <.05, after the Benjamini-Hochberg correction for multiple testing. Normalized fold-change and P values for all expressed genes were then used as input for downstream analysis using iPathwayGuide (AdvaitaBio) and gene set enrichment analysis (GSEA) using GenePattern and MSigDB (Broad Institute). Additional figures including heat maps and scatterplots were generated in R and ClustVis.
Results
Patient characteristics
Six pairs of patients’ samples were chosen out of 7 NLR_High and 9 NLR_Low based on the highest quality of the RNA in the pair as assessed by the RNA integrity number (RIN) (Table 1). Six controls from 8 healthy donors were also examined. In the patients included in the study, the mean ± standard deviation NLR for NLR_High was 4.37 ± 0.66 vs for NLR_Low, 1.87 ± 0.39 (P < .0001). The mean ± standard deviation of age of the patients was 61.0 ± 17.0 years for NLR_High and 64.5 ± 7.5 years for NLR_Low (P = .68). All but 1 patient had aortic aneurysmal disease, 9 were thoracic/thoracoabdominal, and 2 were abdominal. Both patients with abdominal aortic aneurysm were in the NLR_Low group. Eleven out of the 12 patients had a history of hypertension. Four out of 6 patients in both the NLR_High or NLR_Low groups had a medical history of lipid abnormalities. Four patients in the NLR_High and 3 patients in the NLR_Low groups were former smokers. One patient in the NLR_Low group was an active smoker. Five patients in the NLR_High and 4 in the NLR_Low groups were on statins. Only 3 patients in both groups were diabetic. One patient in each group had a history of CAD, and 2 patients in the NLR_Low group had peripheral artery disease.
Clinical characteristics of elective vascular surgery patients examined by RNA-seq
| Patient ID . | NLR ratio . | Diagnosis . | Age . | Sex . | Hypertension . | High lipids . | Smoking . | Statins . |
|---|---|---|---|---|---|---|---|---|
| H1 | 3.71 | TAAA | 75 | M | X | X | Former | X |
| H2 | 4.13 | TAAA | 81∗ | F | X | X | Former | X |
| H3 | 4.71 | DTA | 65† | F | X | — | — | X |
| H4 | 5.09 | DTA | 51 | M | X | X | Former | |
| H5 | 3.58 | TAAA | 34 | M | X | — | — | X |
| H6 | 5.02 | TAAA | 62 | M | X | X | Former | X |
| L1 | 1.27 | TAAA | 68†,‡ | M | X | X | Former | X |
| L2 | 2.28 | TAAA | 53 | M | X | X | — | — |
| L3 | 2.06 | Perivisceral aortic occlusion | 60∗,‡ | F | X | X | Former | X |
| L4 | 1.76 | AAA | 67‡ | M | X | — | — | — |
| L5 | 1.65 | TAAA | 75∗,† | F | X | X | Former | X |
| L6 | 2.25 | AAA | 64†,‡ | M | — | — | X | X |
| Patient ID . | NLR ratio . | Diagnosis . | Age . | Sex . | Hypertension . | High lipids . | Smoking . | Statins . |
|---|---|---|---|---|---|---|---|---|
| H1 | 3.71 | TAAA | 75 | M | X | X | Former | X |
| H2 | 4.13 | TAAA | 81∗ | F | X | X | Former | X |
| H3 | 4.71 | DTA | 65† | F | X | — | — | X |
| H4 | 5.09 | DTA | 51 | M | X | X | Former | |
| H5 | 3.58 | TAAA | 34 | M | X | — | — | X |
| H6 | 5.02 | TAAA | 62 | M | X | X | Former | X |
| L1 | 1.27 | TAAA | 68†,‡ | M | X | X | Former | X |
| L2 | 2.28 | TAAA | 53 | M | X | X | — | — |
| L3 | 2.06 | Perivisceral aortic occlusion | 60∗,‡ | F | X | X | Former | X |
| L4 | 1.76 | AAA | 67‡ | M | X | — | — | — |
| L5 | 1.65 | TAAA | 75∗,† | F | X | X | Former | X |
| L6 | 2.25 | AAA | 64†,‡ | M | — | — | X | X |
AAA, abdominal aortic aneurysm; DTA, descending thoracic aortic aneurysm; F, female; M, male; TAAA, thoracoabdominal aortic aneurysm.
Diabetes mellitus.
CAD.
Peripheral arterial disease.
Whole transcriptome
We observed global differences in the whole transcriptome when comparing gene expression profiles between neutrophils and lymphocytes (Figure 1). This finding was not surprising as cell-type–specific gene expression has been previously characterized as different.19 In the neutrophil section, across the patients in the NLR_High, NLR_Low, and NLR_Control groups, there were many genes upregulated that were clearly not upregulated in the lymphocyte section and vice versa. At the whole transcriptome level, the cell-specific differences between neutrophils and lymphocytes were distinct and dominant regardless of the NLR or whether a patient or control. However, on closer examination, differences began to appear. A principal component analysis plot showed that NLR_Control for either neutrophils or lymphocytes overlapped with NLR_High and NLR_Low in both cells (supplemental Figure 1).
Whole transcriptome of RNA from neutrophils and lymphocytes collected from patients with NLR, high (NLR_High), low (NLR_Low), or control (NLR_Control). The “N” or “L” letter code in the sample name means that either neutrophils or lymphocytes were the source of the RNA. There were 6 patients within each category. Blue=reduced level of expression; red=high level of expression.
Whole transcriptome of RNA from neutrophils and lymphocytes collected from patients with NLR, high (NLR_High), low (NLR_Low), or control (NLR_Control). The “N” or “L” letter code in the sample name means that either neutrophils or lymphocytes were the source of the RNA. There were 6 patients within each category. Blue=reduced level of expression; red=high level of expression.
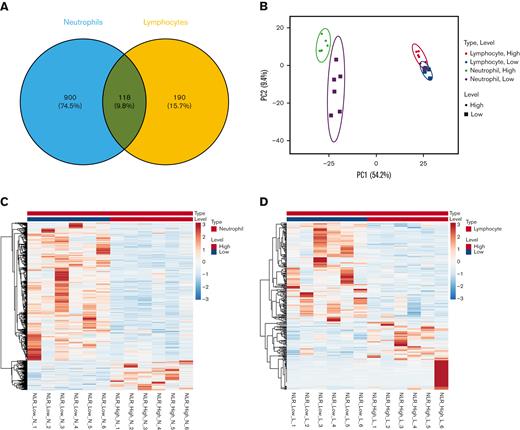
Investigations next sought to find additional differences between NLR_High and NLR_Low groups in the significantly differentially expressed genes. It was observed that 900 (74.5%) significantly differentially expressed genes were observed in neutrophils in NLR_High vs NLR_Low groups (Figure 2A). In comparison, lymphocytes had only 190 (15.7%) significantly differentially expressed genes. A principal component analysis plot of NLR_High genes vs NLR_Low genes in neutrophils and lymphocytes revealed that the group of NLR_High vs NLR_Low genes in the neutrophils segregated into 2 distinct areas, whereas the NLR_High and NLR_Low lymphocytes genes overlapped (Figure 2B). Again, as with the whole transcriptome data, NLR_Control overlapped with NRL_High and NLR_Low significantly differentially expressed genes in both neutrophils and lymphocytes (supplemental Figure 2). Most of the significantly differentially expressed genes in NLR_High of neutrophils were downregulated from NLR_Low (Figure 2C). A similar but much lower number of genes was downregulated in NLR_High of lymphocytes than that of neutrophils (Figure 2D). These data suggested that NLR levels were mostly associated with altered gene expression in neutrophils. Furthermore, it was needed to be compared with patients from its disease group NLR_Low, not NLR-Control, because the former group clearly separated from NLR_High but not NLR_Control (Figure 2B; supplemental Figure 2).
Significantly differentially expressed genes from neutrophils and lymphocytes. (A) The number of significantly differentially expressed genes in neutrophils (blue) and lymphocytes (orange). (B) The principal component analysis plot of significantly differentially expressed genes in neutrophils (green and purple) and lymphocytes (red and blue) in NLR_High (circles) and NLR_Low (squares). (C) A heat map of significantly differentially expressed genes from NLR_High vs NLR_Low neutrophil RNA. (D) A heat map of significantly differentially expressed genes from NLR_High vs NLR_Low lymphocyte RNA.
Significantly differentially expressed genes from neutrophils and lymphocytes. (A) The number of significantly differentially expressed genes in neutrophils (blue) and lymphocytes (orange). (B) The principal component analysis plot of significantly differentially expressed genes in neutrophils (green and purple) and lymphocytes (red and blue) in NLR_High (circles) and NLR_Low (squares). (C) A heat map of significantly differentially expressed genes from NLR_High vs NLR_Low neutrophil RNA. (D) A heat map of significantly differentially expressed genes from NLR_High vs NLR_Low lymphocyte RNA.
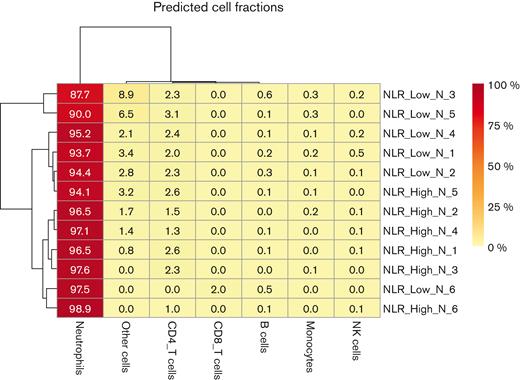
Before investigating differential expression of genes and gene set enrichment, additional studies characterized the degree of monocyte contamination in the neutrophil preparation. Using the RNA-seq data, we observed that the expression of the monocyte markers CD14, CD68, CD83, and CD163 in NLR_High vs NLR_Low neutrophils or lymphocytes preparations were not significantly different. The EPIC analysis of the neutrophil RNA-seq data showed that nonimmune cells (“other cells”) and CD4 T cells were the major contaminants in the neutrophil preparations (Figure 3). Except NLR_Low CD4 T cells and B cells, none of the other contaminating cells were significantly different in the NLR_Low neutrophils vs the NLR_High neutrophils. In the neutrophil preparation, monocyte contamination by EPIC analysis was only 0.17% ± 0.05% for the NLR_Low vs 0.1% ± 0.033% for the NLR_High preparations (P = .12). This value indicates that monocyte contamination was an order of magnitude lower when determined by RNA-seq data analysis vs flow cytometry.
The RNA-seq data in this investigation were used as an independent means to determine other cell contamination in the neutrophil and lymphocyte preparations. Only data from the neutrophils are shown. An EPIC analysis was performed to estimate the proportion of immune and cancer cells in bulk gene expression data. This analysis yielded the percentage of neutrophils, monocytes, B cells, CD4 T cells, CD8 T cells, natural killer (NK) cells, and other cells in the neutrophil preparation.
The RNA-seq data in this investigation were used as an independent means to determine other cell contamination in the neutrophil and lymphocyte preparations. Only data from the neutrophils are shown. An EPIC analysis was performed to estimate the proportion of immune and cancer cells in bulk gene expression data. This analysis yielded the percentage of neutrophils, monocytes, B cells, CD4 T cells, CD8 T cells, natural killer (NK) cells, and other cells in the neutrophil preparation.
To examine the nature of the differentially expressed genes, we performed a GSEA with Gene Ontology (GO) categories for biological processes using the significantly differentially expressed gene sets between NLR_High and NLR_Low. The results are shown in the scatter bubble plots (Figure 4). Segregation of gene sets into upper right of each of the plots shows (1) a high percentage of the genes in the gene sets that are significantly changed and (2) a high degree of significance for enrichment of the gene set. In our sample, the scatter bubble plots showed the enrichment of both complement and humoral response GO categories independently for both neutrophils and the lymphocytes. In neutrophils, the gene sets for classical pathway of complement activation, regulation of protein activation cascade, and humoral immune response mediated by circulating immunoglobulin were the most significant with the highest percentage of gene set members significantly changed (Figure 4A). Independently, the identical gene sets in lymphocytes also were observed to have a high ratio of significantly changed genes and a degree of significance as in the neutrophil studies (Figure 4B).
Scatter bubble plot of significantly expressed GO gene set categories for biologic processes between NLR_High and NLR_Low. The ratio of k:K on the abscissa is the number of genes that are significantly differentially expressed (k) compared with the total number of genes in the category (K). The ordinate is the significance of the enrichment expressed as the log (P value). (A) GO gene sets derived from neutrophil RNA-seq comparing NLR_High to NLR_Low. (B) GO gene sets derived from lymphocyte RNA-seq. Gene sets in the lower left corner of both plots are the least significant. Gene sets in the upper right corner of both plots are the most significant.
Scatter bubble plot of significantly expressed GO gene set categories for biologic processes between NLR_High and NLR_Low. The ratio of k:K on the abscissa is the number of genes that are significantly differentially expressed (k) compared with the total number of genes in the category (K). The ordinate is the significance of the enrichment expressed as the log (P value). (A) GO gene sets derived from neutrophil RNA-seq comparing NLR_High to NLR_Low. (B) GO gene sets derived from lymphocyte RNA-seq. Gene sets in the lower left corner of both plots are the least significant. Gene sets in the upper right corner of both plots are the most significant.
Table 2 lists the most significantly changed immunology gene sets in neutrophils (Table 2, Panel A) and lymphocytes (Table 2, Panel B). This study compared gene expression in neutrophils or lymphocytes between NLR_High samples and NLR_Low samples. Healthy NLR_Control samples were not compared as their expression levels overlapped with NLR_High and NLR_Low patient samples in both the whole transcriptome and significantly differentially expressed gene sets. In addition, the healthy NLR_Control samples were not matched in age, disease, and surgical condition (supplemental Figures 1 and 2). In both neutrophils (Table 2) and lymphocytes (Table 2), the data indicated that the expression of gene sets related to the immune system were highly significantly different when NLR_High is compared with NLR_Low. On RNA-seq, each cell type has the same findings as the other cell type. In both panels, significance levels of the major gene sets maximized at P < 1E-24. These data were highly significant regardless of whether the neutrophil or lymphocyte was examined.
Immunology gene sets in neutrophils and lymphocytes
| Neutrophils . | . | . |
|---|---|---|
| GO immune–related gene sets . | Neutrophil high vs low . | P value . |
| Immune response | 231/1506∗ | 1.00E-24† |
| Immune system process | 291/2090 | 1.00E-24 |
| Humoral immune response mediated by circulating immunoglobin | 46/72 | 1.00E-24 |
| Humoral immune response | 74/155 | 1.00E-24 |
| Adaptive immune response | 94/347 | 1.00E-24 |
| Regulation of humoral immune response | 48/81 | 1.00E-24 |
| Immunoglobulin-mediated immune response | 50/128 | 3.40E-24 |
| B-cell–mediated immunity | 50/130 | 7.80E-24 |
| Regulation of immune system process | 172/1110 | 9.80E-23 |
| Lymphocyte-mediated immunity | 62/221 | 6.90E-21 |
| Neutrophils . | . | . |
|---|---|---|
| GO immune–related gene sets . | Neutrophil high vs low . | P value . |
| Immune response | 231/1506∗ | 1.00E-24† |
| Immune system process | 291/2090 | 1.00E-24 |
| Humoral immune response mediated by circulating immunoglobin | 46/72 | 1.00E-24 |
| Humoral immune response | 74/155 | 1.00E-24 |
| Adaptive immune response | 94/347 | 1.00E-24 |
| Regulation of humoral immune response | 48/81 | 1.00E-24 |
| Immunoglobulin-mediated immune response | 50/128 | 3.40E-24 |
| B-cell–mediated immunity | 50/130 | 7.80E-24 |
| Regulation of immune system process | 172/1110 | 9.80E-23 |
| Lymphocyte-mediated immunity | 62/221 | 6.90E-21 |
| Lymphocytes . | . | . |
|---|---|---|
| GO immune–related gene sets . | Lymphocyte high vs low . | P value . |
| Immune response–regulating cell surface receptor signaling pathway | 31/122∗ | 1.00E-24 |
| Immunoglobulin-mediated immune response | 32/132 | 1.00E-24 |
| Humoral immune response mediated by circulating immunoglobulin | 32/72 | 1.00E-24 |
| Humoral immune response | 37/152 | 1.00E-24 |
| B-cell–mediated immunity | 32/134 | 1.00E-24 |
| Regulation of humoral immune response | 32/79 | 1.00E-24 |
| Immune system process | 144/2230 | 4.80E-21 |
| Lymphocyte-mediated immunity | 34/229 | 8.30E-21 |
| Adaptive immune response | 45/448 | 2.50E-20 |
| Adaptive immune response based on somatic recombination | 33/234 | 1.70E-19 |
| Lymphocytes . | . | . |
|---|---|---|
| GO immune–related gene sets . | Lymphocyte high vs low . | P value . |
| Immune response–regulating cell surface receptor signaling pathway | 31/122∗ | 1.00E-24 |
| Immunoglobulin-mediated immune response | 32/132 | 1.00E-24 |
| Humoral immune response mediated by circulating immunoglobulin | 32/72 | 1.00E-24 |
| Humoral immune response | 37/152 | 1.00E-24 |
| B-cell–mediated immunity | 32/134 | 1.00E-24 |
| Regulation of humoral immune response | 32/79 | 1.00E-24 |
| Immune system process | 144/2230 | 4.80E-21 |
| Lymphocyte-mediated immunity | 34/229 | 8.30E-21 |
| Adaptive immune response | 45/448 | 2.50E-20 |
| Adaptive immune response based on somatic recombination | 33/234 | 1.70E-19 |
Ratio indicates the number of genes in the gene set that are significantly changed.
The P value represents the degree of change in RNA expression in the members of the gene set when comparing the messenger RNA changes from lymphocyte samples with NLR_High to NLR_Low.
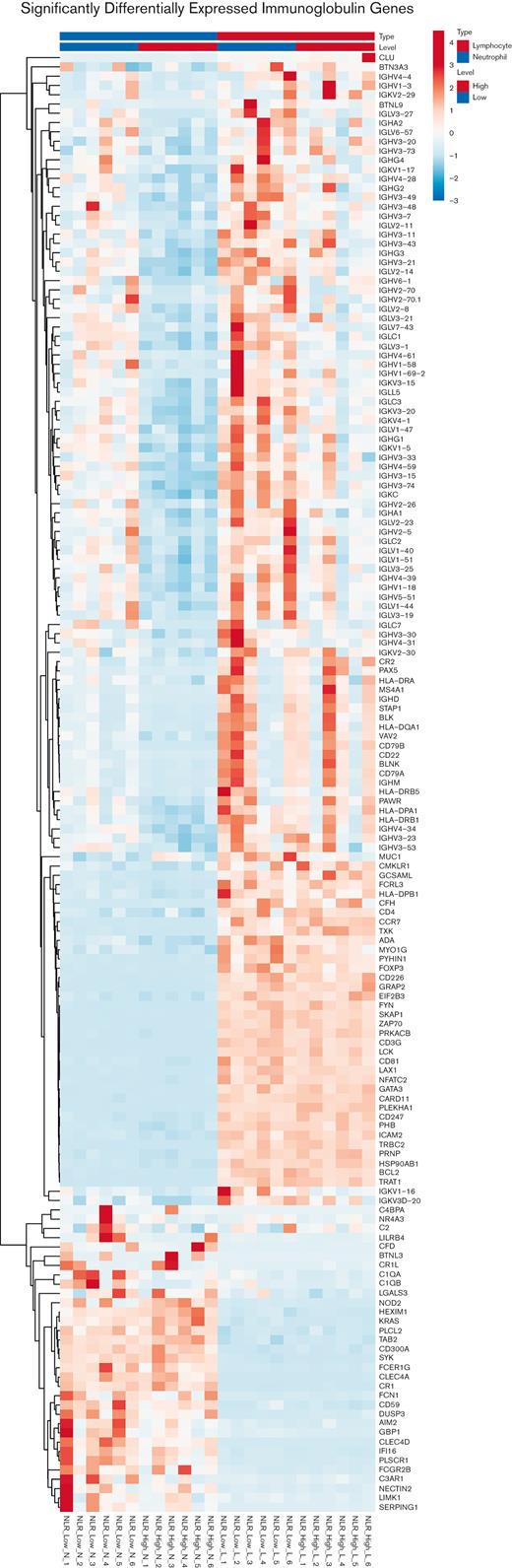
To focus on the highly significant content area of this RNA-seq investigation, significantly differentially expressed immunoglobulin gene expression was examined. First, a principal component analysis plot of the neutrophil genes showed that for NLR_High vs NLR_Low, the significantly differentially expressed immunoglobulin genes segregated into 2 groups (supplemental Figure 3A). Alternatively, the lymphocytes genes of NLR_High vs NLR_Low overlapped (supplemental Figure 3A). Next, a combined heat map of immune-related gene expression between NLR_High with NLR_Low was compared in neutrophils and lymphocytes (Figure 5). Overall, when the entire heat map is examined, there is obvious downregulation of the entire gene family of immunoglobulin genes in the neutrophil NLR_High group compared with that of NLR_Low. Most genes (69/156, 44%) were immunoglobulin related. Only 12 of 156 (7.7%) genes were complement related: C1QA, C1QB, Serping1, C2, C2R, C4BPA, C3AR1, CR1, CR1L, CFH, CFD, and CD59. In the lymphocytes group, there was also downregulation of mostly immunoglobulin genes in the NLR_High category.
Heat map of significantly differentially expressed genes in the immunoglobulin gene set in neutrophils (left) and lymphocytes (right). The genes listed on the right of each heat map are the members of the immunoglobulin gene set. The RNA is from neutrophils or lymphocytes collected from patients with NLR that were high (NLR_High) or low (NLR_Low). There were 6 patients within each category. Six individual samples from NLR_High are compared with 6 individual samples from NLR_Low from both neutrophils and lymphocytes. Please notice the "blue" column in the upper half on the left for genes expressed in neutrophils with "high" NLR. Blue=reduced level of expression; red=high level of expression.
Heat map of significantly differentially expressed genes in the immunoglobulin gene set in neutrophils (left) and lymphocytes (right). The genes listed on the right of each heat map are the members of the immunoglobulin gene set. The RNA is from neutrophils or lymphocytes collected from patients with NLR that were high (NLR_High) or low (NLR_Low). There were 6 patients within each category. Six individual samples from NLR_High are compared with 6 individual samples from NLR_Low from both neutrophils and lymphocytes. Please notice the "blue" column in the upper half on the left for genes expressed in neutrophils with "high" NLR. Blue=reduced level of expression; red=high level of expression.
Complement gene sets in neutrophil and lymphocyte are detailed in Table 3. In both neutrophil (Table 3) and lymphocyte (Table 3), expression of genes in 3 gene sets related to the complement system were highly significantly different (P < 1E-24) when NLR_High samples were compared with NLR_Low samples. This observation was the same whether the comparison was between RNA from neutrophils or lymphocytes. The 3 gene sets include complement activation, complement activation classical pathway, and regulation of complement activation. A principal component analysis plot of the neutrophil genes again showed that for NLR_High vs NLR_Low, the significantly differentially expressed complement genes segregated into 2 groups (supplemental Figure 3B). Alternatively, the lymphocyte genes of NLR_High vs NLR_Low overlap (supplemental Figure 3B).
Complement gene sets in neutrophils and lymphocytes
| Neutrophils . | . | . |
|---|---|---|
| GO complement–related gene sets . | Neutrophil high vs low . | P value . |
| Complement activation | 46/76∗ | 1.00E-24† |
| Complement activation, classical pathway | 44/63 | 1.00E-24 |
| Complement receptor–mediated signaling pathway | 2/5 | .0475 |
| Regulation of complement activation | 46/72 | 1.00E-24 |
| Regulation of complement activation, lectin pathway | 1/1 | .0744 |
| Regulation of complement-dependent cytotoxicity | 1/4 | .266 |
| Negative regulation of complement activation | 2/5 | .0475 |
| Negative regulation of complement activation, lectin pathway | 1/1 | .0744 |
| Neutrophils . | . | . |
|---|---|---|
| GO complement–related gene sets . | Neutrophil high vs low . | P value . |
| Complement activation | 46/76∗ | 1.00E-24† |
| Complement activation, classical pathway | 44/63 | 1.00E-24 |
| Complement receptor–mediated signaling pathway | 2/5 | .0475 |
| Regulation of complement activation | 46/72 | 1.00E-24 |
| Regulation of complement activation, lectin pathway | 1/1 | .0744 |
| Regulation of complement-dependent cytotoxicity | 1/4 | .266 |
| Negative regulation of complement activation | 2/5 | .0475 |
| Negative regulation of complement activation, lectin pathway | 1/1 | .0744 |
| Lymphocytes . | . | . |
|---|---|---|
| GO complement–related gene sets . | Neutrophil high vs low . | P value . |
| Complement activation | 32/76∗ | 1.00E-24† |
| Complement activation, classical pathway | 32/62 | 1.00E-24 |
| Regulation of complement activation | 32/71 | 1.00E-24 |
| Lymphocytes . | . | . |
|---|---|---|
| GO complement–related gene sets . | Neutrophil high vs low . | P value . |
| Complement activation | 32/76∗ | 1.00E-24† |
| Complement activation, classical pathway | 32/62 | 1.00E-24 |
| Regulation of complement activation | 32/71 | 1.00E-24 |
Ratio indicates the number of genes in the gene set that are significantly changed.
The P value represents the degree of change in RNA expression in the members of the gene set when comparing the messenger RNA changes from lymphocyte samples with NLR_High to NLR_Low.
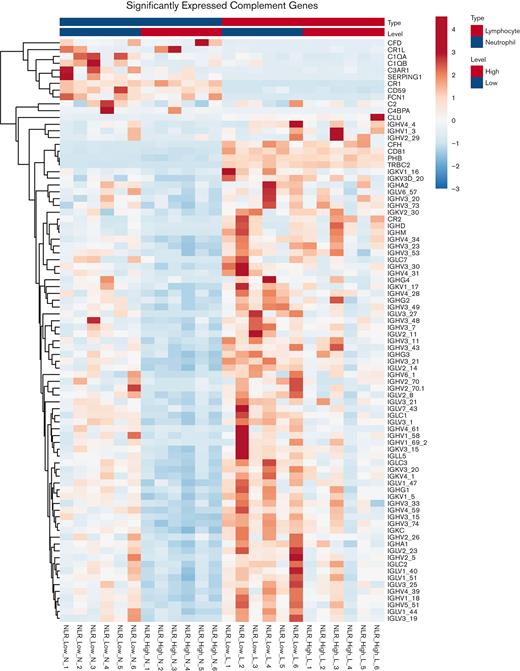
A heat map of neutrophils and lymphocytes expression of a complement-related gene set comparing NLR_High with NLR_Low is shown in Figure 6. In the neutrophils, an enormous number of genes were downregulated in the NLR_High population. In this gene set, similar to the immunoglobulin gene sets, most genes (69/86, 80%) were immunoglobulin related. Only 12 of 86 (14%) were complement component and regulatory genes (C1QA, C1QB, SERPING1, C2, CR2, C4BPA, C3AR1, CR1, CR1L, CD59, CFH, and CFD). In the lymphocyte group overall, there were fewer expressions of complement genes, and immunoglobulin genes were slightly more downregulated in the NLR_High group.
Heat map of significantly differentially expressed genes in the complement gene set in neutrophils (left) and lymphocytes (right). The genes listed on the right of each heat map are the members of the complement gene set. The RNA is from neutrophils or lymphocytes collected from patients with high (NLR_High) or low (NLR_Low) NLR. Six individual samples from NLR_High are compared with 6 individual samples from NLR_Low from both neutrophils and lymphocytes.
Heat map of significantly differentially expressed genes in the complement gene set in neutrophils (left) and lymphocytes (right). The genes listed on the right of each heat map are the members of the complement gene set. The RNA is from neutrophils or lymphocytes collected from patients with high (NLR_High) or low (NLR_Low) NLR. Six individual samples from NLR_High are compared with 6 individual samples from NLR_Low from both neutrophils and lymphocytes.
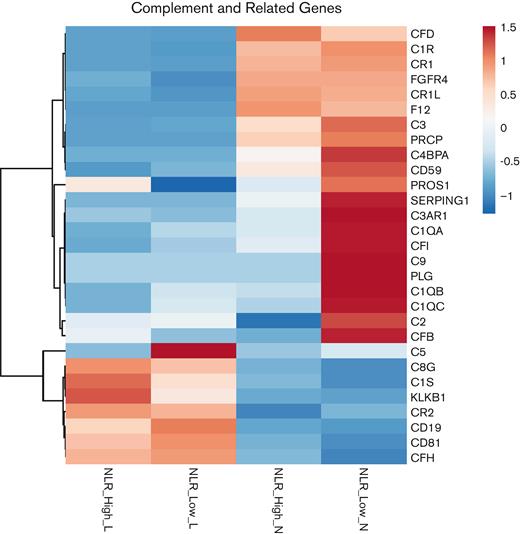
Because there is a greater understanding of the genes of the complement system compared with the genes of the immunoglobulin systems, attention was focused on the complement system as a potential candidate for target genes related to high and low NLR. A composite heat map comparing NLR_High with NLR_Low was prepared showing the expression of complement system and related genes in both neutrophil and lymphocyte samples (Figure 7). For the most part, the lymphocyte is not a reservoir cell system for the complement system. Most complement genes are not upregulated in lymphocytes. Only C1S, CR2, C5, C8G, C5, and CFB were present with a difference in expression between NLR_High and NLR_Low lymphocytes (Figure 7). CD81 in lymphocytes is a B-cell tetraspanin. C1S and C8G expressions seem to be significantly higher in lymphocyte NLR_High than those of lymphocyte NLR_Low, similar to KLKB1, the gene for plasma prekallikrein.
Composite heat map of complement and related genes from neutrophil or lymphocyte NLR_High or NLR_Low. In each category, the composite heat map was prepared from 6 individual RNA-seq samples from each cell type.
Composite heat map of complement and related genes from neutrophil or lymphocyte NLR_High or NLR_Low. In each category, the composite heat map was prepared from 6 individual RNA-seq samples from each cell type.
By contrast, in neutrophils, NLR_High showed downregulation of complement component genes C1QA, C1QB, C1QC, SERPING1, C1R, C4BPA, C3, and C3AR1, and their complement regulatory (inhibitory) genes CD59, CFB, and CFI. Only C2 and CFD genes were upregulated in NLR_High_N patients. CD59 and C9 are relatively upregulated in neutrophil NLR_Low_N compared with those of NLR_High_N. FGFR4 is a growth factor receptor. CFH was present but appeared not to be influenced by neutrophil NLR_High or NLR_Low. It is more highly expressed in lymphocytes.
Discussion
The NLR on the differential leukocyte count has become a widely used, simple marker to recognize potential deleterious outcomes in a wide range of disorders, including CVD, lymphoma, multiple solid tumors, and inflammatory disorders such as systemic lupus erythematosus and systemic sclerosis. In our own retrospective investigations on patients with atherosclerotic CVD, we observed that patients with a high NLR have a higher late mortality rate after elective aortic aneurysm repair, a higher late stroke/death rate after prophylactic carotid endarterectomy, and a decreased amputation-free survival after elective femoral-popliteal revascularization if their NLR is elevated.14-16 The present investigations were initiated to find a mechanistic basis for high NLR.
To develop an unbiased approach as to mechanism(s) of differential effects of NLR on CV outcomes, we performed RNA-seq on both neutrophils and lymphocytes from the same patients. For this study, all patients with aortic disease were prospectively collected and grouped into NLR categories by their results. The final 6 samples in each of the 2 cell types were selected by the highest level of their RIN in the prepared RNA of the paired neutrophil and lymphocyte samples without regard to sex, age, or previous medical conditions. As can be seen in Table 1, the patients in the NLR_High and NLR_Low groups were quite similar and only distinguished by their NLR.
The quality of the RNA used in this study is shown by the RIN and the complete segregation of the pattern of gene expression between neutrophils and lymphocytes in all patient groups collected (high NLR, low NLR, and controls) (Figure 1). A principal component analysis plot shows that NLR_Control data were irrelevant in this investigation because the patients’ NLR overlapped with the NLR_High and NLR_Low in patients’ neutrophil and lymphocyte populations. The GSEA analysis provided great insight as to which gene sets were enriched in the GO gene sets of biologic processes, and the scatter bubble plot clearly demonstrated this by plotting the log significance vs the ratio of the number of gene altered over the total number of genes in the gene sets. When reviewed, the gene sets that separate themselves apart from the entire list were those in humoral (immunoglobulin) immunity and complement system (Tables 2 and 3) based on ratio and statistical significance in an unbiased discovery approach.
The large number of humoral immunity immunoglobulin genes involved is difficult to work with because important differences between these genes are not well known. Alternatively, much more is known about the complement system, and the focus on the classical complement system that is activated by immunoglobulin is a recognizable target for inflammation and disease related to immunoglobulins. Our data suggest that classical complement component genes C1QA, C1QB, C1R, CR1, C1S, SERPING1, C2, C4BPA, C3 C3AR1, C8G, C9, and several regulatory genes (CD59, CFB CFI, and CFH) are downregulated in neutrophils of patients with high NLR (Figure 7). These findings are striking for 3 reasons. First, the genes involved are those mostly of the classical complement pathway. Second, in high NLR patients, these complement genes are, for the most part, downregulated compared with that of low NLR patients. Last, more F12 is expressed in neutrophils from NLR_High than those from NLR_Low samples. Enzymatic forms of the protein product of F12 (factor XII [FXII]) activate the classical pathway of complement and neutrophils.20 By contrast, PLG, the gene for plasminogen and activator of FXII, is downregulated in lymphocytes and neutrophils from patients with in NLR_High, suggesting that FXII activation itself may be a regulator of complement activation.
In addition, it has been shown that the classical complement system is involved in accelerated CVD. Complement component 1q (C1q) is the initial factor of the complement pathway, which has an important role in both innate and acquired immune system.21 C1q has been shown to have dual roles, positive and negative effects on atherosclerosis.22 In the classical pathway of complement, C1q has a protective role in early-stage atherosclerosis by regulating macrophage molecular signal through a complement independent pathway, modulating the uptake of atherogenic lipoprotein, mediating apoptotic cells and cell debris elimination.23,24 A recent study by Jia et al23 indicates that reduced serum C1q is associated with CAD. Another study by Guo et al24 indicates that increased C1q activity is associated with CAD. Finally, recent studies indicate that patients with diabetes with reduced C1q have shortened survival.25 These data indicate that examination of the complement system may be important in understanding the inflammation-mediated pathogenesis of CVD.
Recently, a retrospective investigation of the data from the CANTOS, JUPITER, SPIRE-1, SPIRE-2, and CIRT trials on 60 087 patients was conducted to determine whether NLR predicts major adverse CV events and is modified by anti-inflammatory therapy.26 NLR modestly correlated (ie, r2 ≤ 0.26) with interleukin 6, C-reactive protein, and fibrinogen levels but minimally with lipids.25 In all 5 trials, NLR predicted incident CV events and death in patients with NLR >3.08.26 Although lipid-lowering had no impact on NLR, anti-inflammatory therapy with canakinumab, an interleukin 1β inhibitor lowered NLR (P < .0001).26 These data suggest that NLR may be a marker for CVD independent of lipids and the influence of statins. This ad hoc analysis of multiple prospective randomized trials suggests that NLR may be an independent risk factor for mortality in CVD.
There are some limitations in this study. Our neutrophil preparations had 4% lymphocyte and 2% monocyte contamination by flow cytometry. Because the neutrophils are not known to synthesize immunoglobulin, the neutrophil immunoglobin gene changes could be from contaminating B cells. On the EPIC analysis, there was a significant increase in the B-cell contamination of the NLR-Low samples. This observation suggests that an increase in the level of immunoglobulin genes in the NLR_Low neutrophil samples may arise from B-cell contamination. Thus, reduced immunoglobulin genes in NLR_High samples may be due to increased B cells in the NLR_Low samples and not necessarily reduced immunoglobulin gene levels in the NLR_High samples. Likewise, neutrophils are not known to be complement-producing cells. Two percentage contamination with monocytes could be the source of the complement genes. However, using the RNA-seq data, we observed that there is no significant difference in monocyte contamination of the neutrophil preparations between NLR_Low and NLR_High. Furthermore, performing an EPIC analysis of the RNA-seq data shows that monocyte contamination was <0.1% (<1:1000) of the neutrophil preparations, indicating that the RNA-seq was unlikely influenced by contaminating monocytes (Figure 3). The contaminating B cells in the NLR_Low samples do not influence the complement data observed. Finally, even now, it is not known that the changes we observed in complement and immunoglobulin gene are specific to vascular surgery patients with high NLR or older patients in general. NLR and genetic studies need to be performed on healthy age- and sex-matched patients without CVD.
In sum, the cardinal finding in this investigation is that, in high NLR states, many complement component genes and some of its regulators are downregulated in neutrophils. It is unclear as to the overall meaning and activation state of the identified downregulated complement genes. Because the complement gene sets are strikingly decreased, we speculate that individuals with NLR_High and shortened postoperative survival have loss of major CV protective mechanisms provided by the complement systems. This assessment needs to be examined prospectively in clinical investigations for the underlying mechanism(s) for this outcome.
Acknowledgments
This research was supported by the Genomics Core Facility of the Case Western Reserve University School of Medicine’s Genetics and Genome Sciences Department and National Institutes of Health grants AI130131, HL144113, HL143402, and CA223301 (A.H.S.).
Authorship
Contribution: A.H.S. and J.S.C. conceived the project; J.S.C., S.S., J.L., and A.H.S. performed patient selection and blood collections; J.L., A.M., and H.M. performed neutrophil and lymphocyte preparations and RNA sequencing; and E.R.C., F.L., J.S.C., and A.H.S. performed data analysis and manuscript preparation.
Conflict-of-interest disclosure: A.H.S. is on the Data Safety Monitoring Committee for Intellia and the Scientific Advisory Board for Rezolute, Inc. The remaining authors declare no competing financial interests.
Correspondence: Alvin H. Schmaier, Department of Medicine, Case Western Reserve University, 2103 Cornell Rd, WRB 2-130, Cleveland, OH 44106; e-mail: schmaier@case.edu.
References
Author notes
∗J.S.C. and A.H.S. are joint senior authors.
The RNA sequencing data have been uploaded as a supplement. The sequencing data have also been submitted to the Gene Expression Omnibus for data sharing (accession number GSE211190).
The full-text version of this article contains a data supplement.