TO THE EDITOR:
Although acute myeloid leukemia (AML) is caused by somatically acquired gene mutations, germline variants can also modulate disease characteristics such as sensitivity to treatment and are therefore associated with patient outcomes. Several single-nucleotide polymorphisms (SNPs) have been identified in the past decade to be significantly associated with patient characteristics such as survival, therapy response, and other outcome variables in AML.1-3 Furthermore, susceptibility loci are increasingly recognized as being associated with the risk to develop leukemia.4-6 Although these led to some intriguing hypotheses, SNPs are currently not included in routine risk assessment.7 Guidelines provided by the European LeukaemiaNet (ELN) in 2017 and 2022 improved risk stratification of patients with AML; however, recent publications suggest that further improvement beyond ELN2017 is possible.7,8 In this context, SNPs present attractive candidates because they can easily be implemented in current routine panel sequencing analysis.
Multiple studies have reported associations between germline polymorphisms and disease characteristics in AML.1,9-14 For example, SNP rs12036333 (G>A, LINC01139;CHRM3) has been associated with inferior overall survival (OS) and relapse-free survival (RFS).9,10 Furthermore, many studies reported associations of SNPs with clinical characteristics (supplemental Table 1). However, these associations are often derived from small cohorts and have not been systematically validated so far. In addition, to our knowledge, there has not yet been a comprehensive analysis of the prognostic/clinical relevance of common SNPs in genes linked to AML pathogenesis. The aim of our study, therefore, was to validate previously identified SNPs that were associated with AML outcomes in prior publications in a large and uniformly treated patient cohort. As a secondary goal, we investigated the potential prognostic role of SNPs in genes commonly affected by somatic mutations in AML, which are included in most routinely used diagnostic panels, because such polymorphisms might allow any easily implementable refinement of risk classifications. Local ethic committees of participating institutions approved all protocols, and patients were treated according to the Declaration of Helsinki.
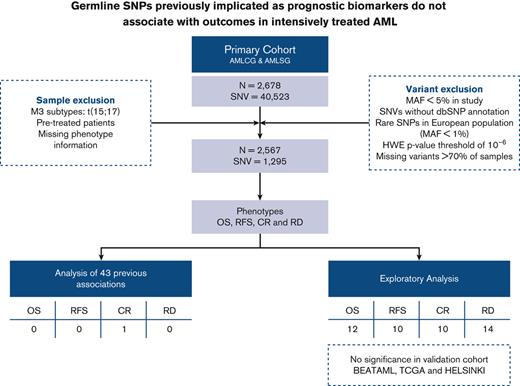
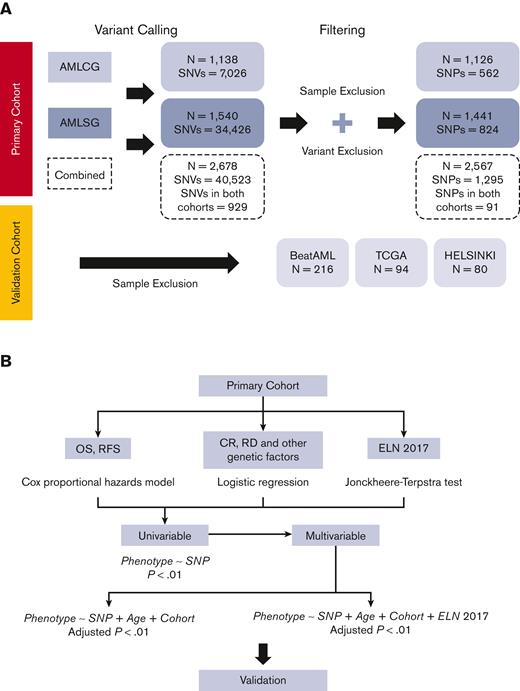
We studied associations between germline variants and treatment response as well as survival in the context of age and the ELN2017 risk classification. Our primary cohort included 2678 intensively treated patients with AML from the German AML Co-operative Group (AMLCG, n = 1138) and the German-Austrian AML Study Group (AMLSG, n = 1540). Patient characteristics of the primary cohorts are shown in supplemental Table 2. Details regarding treatment protocols and inclusion criteria are published elsewhere.15,16 Only participants matching the filtering criteria (shown in Figure 1A) were included in the analyses. Significant results from these analyses were validated in intensively treated patients from the BEAT AML (n = 216), TCGA (n = 94), and HELSINKI (n = 80) cohorts.20-22 Information regarding sequence processing, variant calling, and filtering is provided in the supplemental Information. Although our primary cohorts resulted in 40 523 called SNVs, the application of filtering criteria lead to 2567 samples and 1295 SNPs (variants present in at least 1% of the European population, detailed in Figure 1A), which were further used in the downstream analyses.
Flow diagram of cohorts and statistical analysis. (A) Flow diagram of cohorts under study. The difference in the number of variants called is due to the difference in sequencing types and the region of coverage. Sample exclusion comprise those (1) with missing phenotype information, (2) with M3 sub-types: t(15;17), and (3) exclusion of pretreated patients limiting the analysis to patients receiving AML-directed induction treatment. Variant exclusion consist of those (1) that are not annotated in the dbSNP database; (2) with major allele frequency (MAF) < 1% in European population based on 1000 Genomes project (excluding rare SNPs in European population); (3) with MAF <5% in our primary cohort (excluding rare SNPs in our cohort); (4) which failed with Hardy-Weinberg equilibrium P-value threshold of 10−6; and (5) which are missing in more than 70% of samples.17 (B) Statistical analyses applied. Cox proportional hazards models were applied for detecting differences in OS and RFS, whereas logistic models were used for CR, RD, and other genetic factors. In the case of ELN2017 association with SNPs, we used the Jonckheere-Terpstra test to identify ordered differences among different groups.18 SNPs significant in the univariate analysis (unadjusted P < .01) were included in a multivariable model and stratified for age and the ELN2017 classification. We also added cohort as covariate for those SNPs that were present in both AMLCG and AMLSG data sets. For comparison, we also performed multivariable models excluding the stratification for ELN2017. The P values from the multivariable models were corrected for multiple testing using the Benjamini-Hochberg method.19 CR, complete remission; RD, refractory disease; SNV, single nucleotide variant; TGCA, The Cancer Genome Atlas.
Flow diagram of cohorts and statistical analysis. (A) Flow diagram of cohorts under study. The difference in the number of variants called is due to the difference in sequencing types and the region of coverage. Sample exclusion comprise those (1) with missing phenotype information, (2) with M3 sub-types: t(15;17), and (3) exclusion of pretreated patients limiting the analysis to patients receiving AML-directed induction treatment. Variant exclusion consist of those (1) that are not annotated in the dbSNP database; (2) with major allele frequency (MAF) < 1% in European population based on 1000 Genomes project (excluding rare SNPs in European population); (3) with MAF <5% in our primary cohort (excluding rare SNPs in our cohort); (4) which failed with Hardy-Weinberg equilibrium P-value threshold of 10−6; and (5) which are missing in more than 70% of samples.17 (B) Statistical analyses applied. Cox proportional hazards models were applied for detecting differences in OS and RFS, whereas logistic models were used for CR, RD, and other genetic factors. In the case of ELN2017 association with SNPs, we used the Jonckheere-Terpstra test to identify ordered differences among different groups.18 SNPs significant in the univariate analysis (unadjusted P < .01) were included in a multivariable model and stratified for age and the ELN2017 classification. We also added cohort as covariate for those SNPs that were present in both AMLCG and AMLSG data sets. For comparison, we also performed multivariable models excluding the stratification for ELN2017. The P values from the multivariable models were corrected for multiple testing using the Benjamini-Hochberg method.19 CR, complete remission; RD, refractory disease; SNV, single nucleotide variant; TGCA, The Cancer Genome Atlas.
A flow diagram of the applied statistical analyses is shown in Figure 1B. Significant results from the primary analyses were validated in the combined validation cohorts.
First, we tested outcome associations of 43 SNPs (previously reported to be prognostic) for OS, RFS, CR, and RD (supplemental Table 1). Second, we evaluated 12 SNPs reported to be prognostic in defined patient subgroups within these subsets. P < .01 was considered significant for the previously published SNPs.
Among the 43 prognostically associated SNPs from previous publications, only 1 SNP, rs1130609 (T>G, RRM2), correlated significantly with improved CR (odds ratio = 1.84, P = .0013) and reduced RD (odds ratio = 0.56, P = .0079), after adjustment for age, cohort, and the ELN2017 classification (supplemental Table 3). In the subgroup analyses, none of them showed any signal in their respective subgroups even before adjustment for age and the ELN2017 classification (supplemental Table 4 and supplemental Figure 1).
In a second step, we performed an explorative analysis including all filtered SNPs in the recurrently mutated AML driver genes covered by next-generation sequencing analyses of our cohorts and sought to identify novel associations with outcomes (OS, RFS, CR, and RD). The definitions of the outcomes of interests were defined elsewhere.23
The univariate analysis of common SNPs on postinduction therapy response and survival revealed 43 unique SNPs (3.5%) to be significantly associated (P < .01) with the respective phenotypes (supplemental Figure 2). Among those significant associations, 33 SNPs and 46 associations remained significant after adjustment for age and cohort. Only 16 associations (34.8%) persisted after accounting for the ELN2017 risk classification (Figure 2). None of the SNPs retained its significance in the validation analyses (Figure 2 and supplemental Figure 3).
Multivariable analysis of common SNPs stratified for age and cohort. The colors represent if the analysis was also accounted for the ELN2017 classification. The Cox proportional hazard model was used for both OS and RFS, whereas logistic regression was used for CR. The numbers represent odds ratio/hazard ratio in its respective analysis. The red line indicates the adjusted P value cutoff of .01 after multiple testing correction. In the case of the validation cohort, we did not adjust the P values for multiple testing correction.
Multivariable analysis of common SNPs stratified for age and cohort. The colors represent if the analysis was also accounted for the ELN2017 classification. The Cox proportional hazard model was used for both OS and RFS, whereas logistic regression was used for CR. The numbers represent odds ratio/hazard ratio in its respective analysis. The red line indicates the adjusted P value cutoff of .01 after multiple testing correction. In the case of the validation cohort, we did not adjust the P values for multiple testing correction.
A more detailed description of the results is given in the supplemental Information section.
In summary, to our knowledge, our analysis represents one of the first and largest studies of SNPs previously associated with AML outcomes and, more broadly, those identified by targeted sequencing panels, in large, homogeneously treated AML patient cohorts. For 43 SNPs previously reported as having prognostic or predictive relevance in AML, we were unable to corroborate their significance after accounting for the ELN2017 risk classification, with the exception of rs1130609 (T>G, RRM2).
Limitations of previous reports include smaller cohort sizes, a focus on individual SNPs, and lack of validation in independent cohorts.
Our exploratory investigation of potential new prognostic SNPs in AML is limited by the fact that we relied on a targeted sequencing panel focused on a small region of the genome, and therefore missed many potentially prognostic or predictive SNPs. However, we focused on the most commonly mutated genes in AML, which might improve the likelihood to detect clinically relevant SNPs, and tried to validate additional, previously published variants.
Although we identified several significant associations between SNPs and response variables in the univariate analyses, only few variants persisted after adjustment for age and ELN2017 risk. Their impacts were further analyzed in independent datasets, but the results were not validated. One reason may be the small effect size of germline variants, which may require a larger sample size than those available in our validation cohorts. Hence, we identified no novel SNPs that can improve current AML risk stratification algorithms (for more details, see supplemental Information).
Our study underscores the importance of including routinely used risk models such as the ELN2017 classification in reports of novel prognostic markers. Approximately 65% of SNPs that were significant after stratifying for age and study cohort failed to meet the significance threshold after adjusting for ELN2017. This suggests that a large portion of their predictive value is already accounted for within the ELN classification. This also accounts for the recently published ELN2022 classification that has not included SNPs as additional prognostic variables as well.24
Our analysis highlights the importance of interpreting candidate prognostic markers in large and homogeneous data sets, as well as validating them in independent cohorts. Even cohorts of several thousands of patients might be limited in identifying all possible associations. Therefore, the accumulation of genetic information in the next decades will become critical for our ability to elucidate the genetic context and background of AML to improve risk stratification and patient outcome.
Acknowledgments: The authors thank all patients and investigators who contributed data to our analysis.
This work is supported by a grant of the Wilhelm-Sander-Stiftung (no. 2013.086.2), funding from the Deutsche José Carreras Leukämie-Stiftung (DJCLS 10 R/2021), the Physician Scientists Grant (G-509200-004) from the Helmholtz Zentrum München (T.H.), the German Cancer Consortium (Deutsches Konsortium für Translationale Krebsforschung, Heidelberg, Germany), a grant from Deutsche Forschungsgemeinschaft (DFG SFB 1243, TP A06 and TP A07; K.H.M., K.S. and T.H.), and the BMBF grant 01ZZ1804B (DIFUTURE; A.M.N.B.).
Contribution: A.M.N.B., K.H.M., and T.H. conceived and designed the analysis; A.M.N.B., N.B., S.A.B., V.J., M.R.-T., H.G., A.D., S.S., K.S., K.H.M., and T.H. provided and analyzed data; A.M.N.B., V.J., and U.M. provided bioinformatics support; M.R.-T., H.G., B.K., A.D., S.S., K.H.M., and K.S. characterized patient samples; C.S., D.G., W.E.B., B.J.W., U.K., J.B., and W.H. coordinated the AMLCG clinical trials; A.M.N.B., K.H.M., and T.H. wrote the manuscript; and all authors approved the final manuscript.
Conflict-of-interest disclosure: H.G. is a current employee of Roche Pharma AG, Grenzach-Wyhlen, Germany. The remaining authors declare no competing financial interests.
Correspondence: Tobias Herold, Department of Medicine III, University Hospital, LMU Munich, Marchioninistr. 15, 81377 Munich, Germany; e-mail: tobias.herold@med.uni-muenchen.de.
References
Author notes
Although legal restrictions prohibit the authors from publicly sharing raw sequencing data, the AMLSG data set is publicly available through the European Genome-Phenome Archive (EGAS00001000275).
The full-text version of this article contains a data supplement.
K.H.M. and T.H. contributed equally to this study.