Key Points
HDAC1 inhibition dampens IRF4 transcription through histone hyperacetylation, thereby reducing the expression of the survival mediator PIM2.
Simultaneous targeting of the intrinsic HDAC1-IRF4 axis plus externally activated PIM2 represents an efficient therapeutic option for MM.
Abstract
Multiple myeloma (MM) preferentially expands and acquires drug resistance in the bone marrow (BM). We herein examined the role of histone deacetylase 1 (HDAC1) in the constitutive activation of the master transcription factor IRF4 and the prosurvival mediator PIM2 kinase in MM cells. The knockdown or inhibition of HDAC1 by the class I HDAC inhibitor MS-275 reduced the basal expression of IRF4 and PIM2 in MM cells. Mechanistically, the inhibition of HDAC1 decreased IRF4 transcription through histone hyperacetylation and inhibiting the recruitment of RNA polymerase II at the IRF4 locus, thereby reducing IRF4-targeting genes, including PIM2. In addition to the transcriptional regulation of PIM2 by the HDAC1-IRF4 axis, PIM2 was markedly upregulated by external stimuli from BM stromal cells and interleukin-6 (IL-6). Upregulated PIM2 contributed to the attenuation of the cytotoxic effects of MS-275. Class I HDAC and PIM kinase inhibitors cooperatively suppressed MM cell growth in the presence of IL-6 and in vivo. Therefore, the present results demonstrate the potential of the simultaneous targeting of the intrinsic HDAC1-IRF4 axis plus externally activated PIM2 as an efficient therapeutic option for MM fostered in the BM.
Introduction
Epigenetic modifications contribute to oncogenesis and disease progression; therefore, various types of epigenetic modifiers have been attracting attention as therapeutic targets in the treatment of malignancies.1-4 Histone deacetylases (HDACs) are epigenetic modifiers that deacetylate lysine residues in histone tails and nonhistone proteins, thereby altering gene expression as well as protein stability and activity.5,6 Mammalian HDACs are grouped into 4 classes based on their homology with yeast enzymes, and the majority of HDAC inhibitors predominantly target class I HDACs (HDAC1, 2, 3) and a class IIa HDAC (HDAC6).7,8 Several HDAC inhibitors have already been introduced as therapeutic drugs for hematological malignancies.
Multiple myeloma (MM) is derived from long-lived plasma cells in bone marrow (BM) and remains an incurable disease, even in the era of novel treatment strategies, including proteasome inhibitors and immunomodulatory drugs,9,10 which resulted in the US Food and Drug Administration (FDA) approval of the nonselective HDAC inhibitor panobinostat based on the favorable results of the preclinical and clinical studies.11,12 The role of each HDAC isoform, including HDAC1, in MM cells has recently been deciphered. Class I HDACs are overexpressed in MM cells, and higher HDAC1 expression are associated with poor prognosis.13 Genetic ablation of HDAC1 or HDAC3 induces growth arrest and apoptosis in MM cells.14 HDAC3 promotes MM cell survival by stabilizing c-MYC and DNMT1 proteins,15 and activating STAT3.14 However, the molecular mechanisms whereby HDAC1 regulates MM cell growth and survival have largely been unknown.
Although HDACs are considered to be negative regulators of gene expression, previous studies demonstrated that they also contribute to the transcriptional upregulation of some genes.16-18 MM cells alter the activation of transcription factors (TFs), such as IRF4, c-MYC, PRDM1, and XBP1, to acquire their malignant nature.19-21 Among TFs, IRF4, the expression of which is up-regulated along with the differentiation of B cells toward plasma cells, is a master regulator of the MM phenotype.22-25 HDAC inhibitors, including panobinostat, have been shown to downregulate IRF4 expression in MM cells.26,27 However, it currently remains unclear whether HDACs directly regulate the transcription of IRF4 in MM cells.
One of the representative MM phenotypes is the high expression of the serine/threonine kinase PIM2, which is a critical anti-apoptotic mediator in MM cells.28,29 It is also a pivotal regulator of osteoblasts and osteoclastogenesis through the direct and/or indirect interaction of MM cells with BM stromal cells (BMSCs) or osteoclasts.28,30-33 In addition, IRF4 has been shown to transcriptionally regulate the expression of PIM2 in MM cells.23 Tumor microenvironment also induces PIM2 expression.30 However, it remains elusive whether PIM2 is involved in drug resistance in MM.
We herein investigated the molecular mechanisms of how HDAC1 mediates MM cell growth and survival and demonstrated the HDAC1-IRF4-PIM2 axis in MM cells.16,17 HDAC1 directly mediated the upregulation of IRF4 via histone deacetylation, whereas IRF4 upregulated PIM2. We also showed that the expression of PIM2 is sustained by BM microenvironment signaling independent of the HDAC1-IRF4-PIM2 axis. This mechanism contributes to drug resistance to HDAC inhibitors, and the combined effects of class I HDAC and PIM inhibitors overcome the protective effects of BM.
Materials and methods
For a more detailed description of the materials and methods used in the present study, see supplemental information.
Cell lines, primary samples, and plasmids
The human MM cell lines, RPMI 8226, MM.1S, U266, and NCI-H929 were purchased from the American Type Culture Collection (ATCC) (Manassas, VA). KMS-11 and INA-6 cells were kindly provided by Takemi Otsuki (Kawasaki Medical University, Okayama, Japan, JCRB1179) and Renate Burger (University of Kiel, Kiel, Germany), respectively. RPMI 8226 cells expressing luciferase (RPMI 8226-Luc) were generated by retrovirally transducing the MSCV-Luc vector into RPMI 8226 cells. The 293T cell line was also obtained from ATCC. The MM cell lines were cultured in RPMI 1640 (Sigma-Aldrich, St. Louis, MO) and supplemented with 10% heat-inactivated fetal bovine serum (iFBS), 100 U/mL penicillin (Sigma), and 100 μg/mL streptomycin (Sigma). 293T cells were cultured in DMEM (Sigma) with 10% iFBS. Cell lines were checked for contamination with mycoplasma using the MycoAlert mycoplasma detection kit (Lonza, Basel, Switzerland).
Primary samples were obtained from patients who were diagnosed with MM at the Department of Hematology in Tokushima University Hospital or Tokushima Prefectural Central Hospital, with written informed consent according to the Declaration of Helsinki under the protocols of the Institutional Review Board (3842-1 in Tokushima University Hospital, 16-8 in Tokushima Prefectural Central Hospital). Mononuclear cells were separated from peripheral blood or BM aspirates using Ficoll-Paque PLUS (GE Healthcare Bio-Sciences AB, Uppsala, Sweden), and primary CD138-positive cells were then purified using anti-CD138 magnetic activated cell-separation microbeads (Miltenyi Biotec, San Diego, CA). Patient-derived BMSCs were cultured as adherent cells from BM mononuclear cells. BMSC conditioning media (BMSC-CM) were harvested after a 48-hour culture of BMSCs under semiconfluent conditions, and 20% of BMSC-CM was used in subsequent experiments.
HDAC1, HDAC3, IRF4, PIM2 and STAT3 pLKO.1 short hairpin RNA (shRNA) vectors were purchased from Sigma. A luciferase pLKO.1 shRNA vector (shLuc) was used as a negative control of transfection. The RNAi Consortium clone ID and target sequence of each vector are listed in supplemental Table 2. Human HDAC1 complementary DNA (cDNA), which was obtained from the plasmid (kindly provided by Eric Verdin, Addgene plasmid #13820), was amplified with FLAG-tag using polymerase chain reaction (PCR) and then ligated into the EcoRI and HpaI sites of the pMSCV-neo retroviral expression vector (Takara Bio USA, San Jose, CA). MSCV-IRF4 was established in a previous study.24 MSCV-Luc was kindly provided by Scott W. Lowe, Addgene plasmid #18782.
Transduction
Lentiviral and retroviral production was performed using 293T cells as previously described.15,24 In brief, pLKO-based plasmids were transfected into 293T cells in combination with pCMV-dvpr and VSV-G for lentiviral packaging, and the pMSCV plasmid with pMD-MLV and VSV-G for retroviral packaging, using TransIT-LT1 Transfection Reagent (Mirus Bio, Madison, WI). Virus-containing media were then harvested according to previous methods. MM cells were cultured with virus-containing media in the presence of polybrene (Santa-Cruz) for 5 hours. After 24 hours, shRNA- or cDNA-induced MM cells were selected using 1 μg/mL puromycin (Sigma-Aldrich) for 48 hours or 400 μg/mL G418 (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) for at least 7 days, respectively. Selected cells were subjected to the following experiments.
Cell proliferation assay
Cells were seeded at a density of 3 × 104 cells per well and cultured for 48 to 72 hours. Cell viability was assessed using Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Kumamoto, Japan).
RNA-sequencing (RNA-seq) analysis
Total RNA was extracted from RPMI 8226 cells with shHDAC1 or control shRNA after 2 days of transduction in biological triplicates. RNAs were then treated with the TURBO DNA-free Kit (Invitrogen, Waltham, MA) to remove contaminating DNAs. The libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA), and subjected to 75-bp single read sequencing on an Illumina HiSeq 2000. Sequencing reads were aligned against the hg19 genome (GRCh37), and alignments were performed with the STAR aligner. A differential gene expression analysis with read normalization was performed using DESeq. Differentially expressed transcripts were selected based on >20.5-fold changes with adjusted P < .05. RNA-seq raw data are deposited in the GEO database under the accession code GSE193298.
Chromatin immunoprecipitation (ChIP)-seq analysis
Publicly available ChIP-seq data (HDAC1 [GSM2302869], H3K27Ac [GSM894083], RNA polymerase II (RNA Pol II) [GSM1070127], and IRF4 [GSM1195560] in MM.1S cells and IRF4 [GSM2481669] in KK1 cells) processed in ChIP-Atlas34 were downloaded and visualized using the Integrative Genomics Viewer (Broad Institute). Computational processing in ChIP-Atlas is available (https://github.com/inutano/chip-atlas/wiki#experimentList_schema). Briefly, Fastq files were aligned to the reference human genome (hg19) with Bowtie 2. SAM-formatted files were binarized into the BAM format using SAMtools and then sorted before removing PCR duplicates. BedGraph-formatted coverage scores were calculated with bedtools, and BedGraph files were binarized into the BigWig format using UCSC bedGraphToBigWig tool. Peak-call was performed using MACS2. HDAC1 ChIP enrichment values around H3K27Ac or RNA Pol II ChIP peaks were calculated using the computeMatrix tool and visualized using the plotHeatmap tool on the Galaxy platform (https://usegalaxy.org).
Murine xenograft models
All animal studies were performed under a protocol approved by the Animal Ethics Committee of Tokushima University (T30-1). Five-week-old male C.B-17/Icr-scid/scidJcl (SCID) mice were purchased from CLEA Japan (Tokyo, Japan). Mice were subcutaneously transplanted with 5 × 106 RPMI 8226-Luc cells in the right flank 1 day after an intraperitoneal injection of 100 μg of a rabbit antiacialo-GM1 antibody (FUJIFILM Wako Pure Chemical Corporation). After the confirmation of a tumor volume of ≥50 mm3, mice were randomly grouped into 4 groups and then treated with an intraperitoneal injection of PBS as the vehicle control, per os of MS-275 (3.5 mg/kg) 3 days a week, an intraperitoneal injection of SMI-16a (20 mg/kg) 5 days a week, or MS-275 (3.5 mg/kg) in combination with SMI-16a (20 mg/kg). MS-275 was dissolved in DMSO/30% PEG300/ddH2O, and SMI-16a was dissolved in DMSO/50% PEG400/PBS. Tumor sizes and body weights were measured once every 3 days. Tumor volumes were calculated with the formula: 0.5(a × b2), where “a” is the long diameter and “b” is the short diameter of the tumor. Tumors were also visualized using the IVIS Imaging System on days 1, 14, and 28. Mice were sacrificed when the tumor reached 2 cm in length or 2 cm3 in volume or if mice appeared moribund to prevent unnecessary morbidity.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism (GraphPad Software, version 7) or Statcel 3 (OMS Publisher, Tokyo, Japan). The Student t test was performed to compare 2 groups and the Tukey-Kramer multiple comparison test for pairwise comparisons among multiple groups. Differences in survival were evaluated by the Log-rank test. P values < .05 were considered to be significant.
Results
The genetic knockdown or pharmacological inhibition of HDAC1 downregulates IRF4 and PIM2 expression in MM cells
Because the biological significance of HDAC1 in MM cells remains unclear, we initially examined its expression in primary tumor cells from patients with MM. Among class I HDACs, HDAC1 and HDAC3 were expressed at significantly higher levels in MM cells than in normal plasma cells (supplemental Figure 1A). Moreover, HDAC1 expression was positively associated with disease progression from monoclonal gammopathy of undetermined significance to plasma cell leukemia (supplemental Figure 1B). Because HDAC1 expression was elevated in MM cells and increased in association with disease progression, we examined the effects of the depletion of HDAC1 in MM cells. The knockdown of HDAC1 induced apoptosis in 3 MM cell lines using the shRNA lentiviral system (supplemental Figure 1C). These results suggest that HDAC1 is indispensable for MM cell survival.
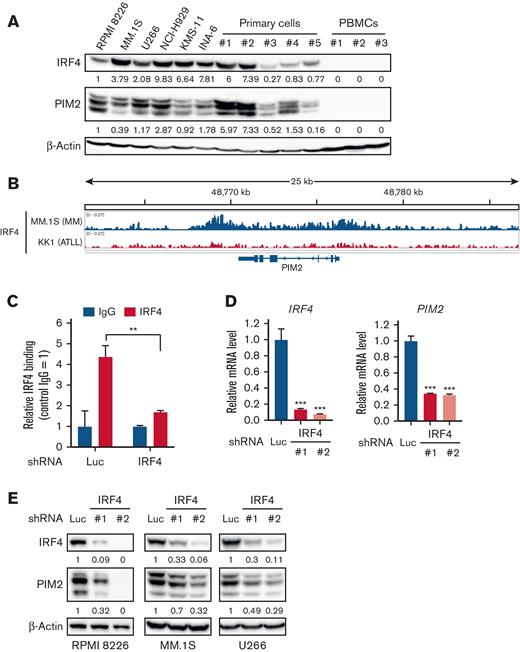
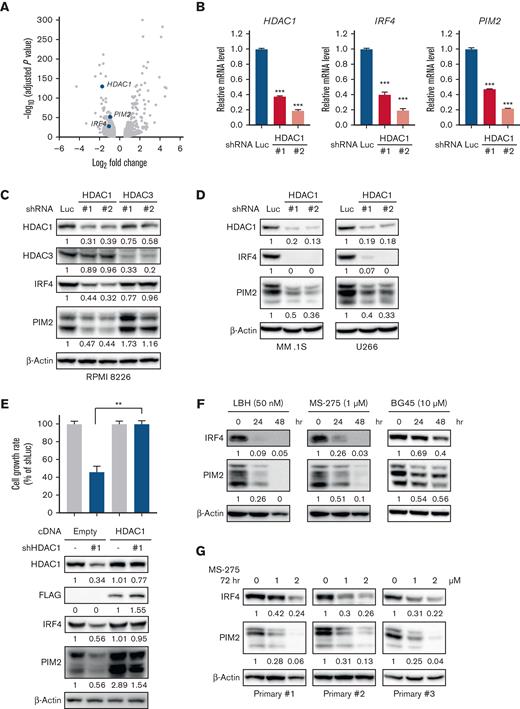
To delineate the functional roles of HDAC1 in MM cell survival, we next performed RNA-seq in RPMI 8226 cells transduced with HDAC1 shRNA (Figure 1A). The knockdown of HDAC1 increased or decreased the expression of 1008 or 795 genes (LogFC > 0.5, adjP < .05), respectively (supplemental Table 1). Because IRF4 and PIM2 have both been identified as therapeutic targets in MM cells,23 we were interested in the downregulation of these genes. We confirmed the downregulation of IRF4 and PIM2 at both mRNA and protein levels following the knockdown of HDAC1 in 3 MM cell lines (Figure 1B-D). Because we previously reported that HDAC3 is also a therapeutic target in MM cells,14,15 we compared the knockdown of HDAC1 vs HDAC3 in MM cells. The knockdown of HDAC1 downregulated IRF4 and PIM2, whereas that of HDAC3 did not affect the expression of IRF4 or PIM2, suggesting that IRF4 and PIM2 expression is specifically regulated by HDAC1 (Figure 1C). Enforced expression of HDAC1 cDNA rescued the downregulation of IRF4 and PIM2 and growth inhibition mediated by HDAC1 knockdown in RPMI 8226 cells (Figure 1E), confirming the observed phenotype is derived from the on-target effect of HDAC1 knockdown.
The knockdown or inhibition of HDAC1 downregulates IRF4 and PIM2 expression in MM cells. (A-D) RPMI 8226, MM.1S, U266 cells were transduced with shLuc (control shRNA targeting luciferase), shHDAC1 (#1, #2), or HDAC3 (#1, #2). Total RNA or whole cell lysates were extracted from transduced cells, followed by each assay. (A) RNA-seq was performed using the RNAs extracted from HDAC1-knockdown (shHDAC1 #1) or control RPMI 8226 cells. RNA-seq expression data shows as a volcano plot selected based on >20.5-fold changes (x-axis) with adjusted P < .05 (y-axis). (B) Total RNA extracted from HDAC1-knockdown RPMI 8226 cells was subjected to Q-PCR. GAPDH served as an internal control. Values represent the amount of mRNA relative to shLuc control, defined as 1. Error bars show the standard deviation (SD) of triplicates. ∗∗∗P < .001 from the control; the Tukey-Kramer multiple comparison test. (C-D) The whole cell lysates extracted were subjected to immunoblotting using the indicated antibodies. β-Actin served as a loading control. (E) RPMI 8226 cells were transduced with either HDAC1-FLAG cDNA or Empty as a control by a retrovirus. Cells were further transduced with either shHDAC1 #1 (targeting 3′ UTR of HDAC1) or shLuc. After puromycin selection, cell viability for 48 hours was assessed by the CCK-8 assay. The cell growth rate in RPMI 8226 cells induced Empty or HDAC1-FLAG cDNA with the shLuc set as 100% for control. ∗∗P < .01 significantly different from the transduced cells with Empty cDNA with shHDAC1; the Tukey-Kramer multiple comparison test. The whole cell lysates extracted were subjected to immunoblotting using indicated antibodies. β-Actin served as a loading control. (F-G) RPMI 8226 cells (F) and primary CD138-positive cells (G) were treated with LBH589, MS-275, or BG45 at the indicated concentration and time course, and whole cell lysates were then extracted from treated cells and subjected to immunoblotting using the indicated antibodies. β-Actin served as a loading control. Relative expression levels of each target, which are normalized to its loading control, are shown below for each immunoblotting image.
The knockdown or inhibition of HDAC1 downregulates IRF4 and PIM2 expression in MM cells. (A-D) RPMI 8226, MM.1S, U266 cells were transduced with shLuc (control shRNA targeting luciferase), shHDAC1 (#1, #2), or HDAC3 (#1, #2). Total RNA or whole cell lysates were extracted from transduced cells, followed by each assay. (A) RNA-seq was performed using the RNAs extracted from HDAC1-knockdown (shHDAC1 #1) or control RPMI 8226 cells. RNA-seq expression data shows as a volcano plot selected based on >20.5-fold changes (x-axis) with adjusted P < .05 (y-axis). (B) Total RNA extracted from HDAC1-knockdown RPMI 8226 cells was subjected to Q-PCR. GAPDH served as an internal control. Values represent the amount of mRNA relative to shLuc control, defined as 1. Error bars show the standard deviation (SD) of triplicates. ∗∗∗P < .001 from the control; the Tukey-Kramer multiple comparison test. (C-D) The whole cell lysates extracted were subjected to immunoblotting using the indicated antibodies. β-Actin served as a loading control. (E) RPMI 8226 cells were transduced with either HDAC1-FLAG cDNA or Empty as a control by a retrovirus. Cells were further transduced with either shHDAC1 #1 (targeting 3′ UTR of HDAC1) or shLuc. After puromycin selection, cell viability for 48 hours was assessed by the CCK-8 assay. The cell growth rate in RPMI 8226 cells induced Empty or HDAC1-FLAG cDNA with the shLuc set as 100% for control. ∗∗P < .01 significantly different from the transduced cells with Empty cDNA with shHDAC1; the Tukey-Kramer multiple comparison test. The whole cell lysates extracted were subjected to immunoblotting using indicated antibodies. β-Actin served as a loading control. (F-G) RPMI 8226 cells (F) and primary CD138-positive cells (G) were treated with LBH589, MS-275, or BG45 at the indicated concentration and time course, and whole cell lysates were then extracted from treated cells and subjected to immunoblotting using the indicated antibodies. β-Actin served as a loading control. Relative expression levels of each target, which are normalized to its loading control, are shown below for each immunoblotting image.
Because we confirmed the downregulation of IRF4 and PIM2 by the knockdown of HDAC1, we examined the impact of the pharmacological inhibition of HDAC1 on IRF4 and PIM2 using non-selective (panobinostat, LBH589) and class I selective (entinostat, MS-275) HDAC inhibitors. LBH589 and MS-275 both down-regulated IRF4 and PIM2 expression in 3 MM cell lines, whereas the HDAC3 selective inhibitor (BG45) did not (Figure 1F and supplemental Figure 2A-B). MS-275 also downregulated IRF4 and PIM2 expression in CD138-positive primary tumor cells from patients with MM (Figure 1G). Collectively, these results indicate that HDAC1 regulates IRF4 and PIM2 expression in MM cells.
IRF4 is a crucial downstream target of HDAC1 in MM cells
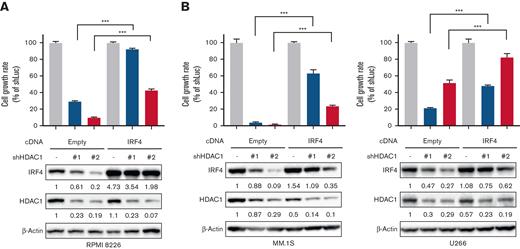
IRF4 plays pivotal roles in MM cell survival.23 Therefore, we transduced IRF4 cDNA in MM cell lines using a retroviral system to investigate whether the downregulation of IRF4 is involved in the HDAC1 knockdown-induced inhibition of MM cell growth. The overexpression of IRF4 partially protected MM cells from the growth inhibitory effects of the knockdown of HDAC1 (Figure 2A-B), suggesting that the downregulation of IRF4 mediates the inhibition of MM cell growth by the suppression of HDAC1 in MM cells.
IRF4 is a crucial target of HDAC1 in MM cells. (A-B) RPMI 8226, MM.1S, or U266 cells were transduced with IRF4 cDNA or Empty as a control by a retrovirus. Cells were further knocked down with HDAC1 or Luc shRNA. After puromycin selection, cell viability for 48 hours was assessed by the CCK-8 assay, and the whole cell lysates extracted were subjected to immunoblotting using the indicated antibodies. β-Actin served as a loading control. Relative expression levels of each target, which are normalized to its loading control, are shown below for each immunoblotting image. Error bars show the SD of triplicates. The cell growth rate in each cell line induced Empty or IRF4 cDNA with the shLuc set as 100% for control. ∗∗∗P < .001 significantly different from each cell induced with Empty cDNA with shHDAC1; the Tukey-Kramer multiple comparison test.
IRF4 is a crucial target of HDAC1 in MM cells. (A-B) RPMI 8226, MM.1S, or U266 cells were transduced with IRF4 cDNA or Empty as a control by a retrovirus. Cells were further knocked down with HDAC1 or Luc shRNA. After puromycin selection, cell viability for 48 hours was assessed by the CCK-8 assay, and the whole cell lysates extracted were subjected to immunoblotting using the indicated antibodies. β-Actin served as a loading control. Relative expression levels of each target, which are normalized to its loading control, are shown below for each immunoblotting image. Error bars show the SD of triplicates. The cell growth rate in each cell line induced Empty or IRF4 cDNA with the shLuc set as 100% for control. ∗∗∗P < .001 significantly different from each cell induced with Empty cDNA with shHDAC1; the Tukey-Kramer multiple comparison test.
HDAC1 regulates IRF4 expression by fine-tuning the histone acetyl status in MM cells
The inhibition of HDAC is generally considered to induce the transcriptional upregulation of genes through histone acetylation. However, in this study, the inhibition or knockdown of HDAC1 reduced IRF4 and PIM2 expression. MS-275 reduced IRF4 and PIM2 expression in association with increases in the acetylation of histone H3 in MM cell lines (supplemental Figure 3A-B). To clarify the molecular mechanisms by which the inhibition/knockdown of HDAC1 triggers the transcriptional downregulation of these genes, we analyzed publicly available ChIP-seq data. The enrichment analysis revealed that HDAC1 bound around H3K27Ac- and RNA Pol II-enriched regions in MM.1S cells (Figure 3A), suggesting that HDAC1 is recruited to transcriptionally active genes. To further clarify the relationship between HDAC1 and the positive/negative transcription of genes, we combined HDAC1 knockdown RNA-seq data with HDAC1 ChIP-seq data (Figure 3B). As expected, HDAC1-bound genes were not only upregulated but also downregulated in MM cells. HDAC1 bound to the promotor and enhancer regions of IRF4, which were marked with H3K27Ac (Figure 3C). Therefore, we evaluated the H3K27Ac status after the inhibition of HDAC and showed that MS-275 increased H3K27Ac levels around the IRF4 promotor and enhancer regions in MM.1S and RPMI 8226 cells (Figure 3D). Similar to previous findings showing that histone hyperacetylation removed RNA pol II from core regulatory binding sites in rhabdomyosarcoma cells,35 we observed a decrease in RNA pol II binding around IRF4 promotor regions in MS-275-treated MM cells (Figure 3E). Consistent with the results, HDAC1 knockdown elevated the acetylation levels of H3K27 and reduced RNA pol II binding around the IRF4 promotor (supplemental Figure 3C-D), suggesting that the hyperacetylation of H3K27 by the inhibition of HDAC1 prevents RNA pol II binding to the IRF4 promotor in MM cells. Collectively, these results suggest that HDAC1 induces appropriate histone acetylation at the promotor and enhancer regions of the transcriptionally active IRF4 gene to mediate IRF4 transcription.
HDAC1 epigenetically regulates IRF4 expression by fine-tuning histone acetylation in MM cells. (A) HDAC1 enrichment around H3K27Ac sites (left) and RNA Pol II binding sites (right) for all genes was analyzed using publicly available ChIP-seq data (GSM2302869 [HDAC1], GSM894083 [H3K27Ac], and GSM1070127 [RNA Pol II]). Heatmaps of HDAC1 levels at H3K27Ac sites or RNA Pol II-binding sites in MM.1S cells are shown (bottom). Each row indicated ±5 kb centered on the H3K27Ac or RNA Pol II sites. The mean signal in the same intervals is plotted (top). (B) RNA-seq data of HDAC1-knockdown RPMI 8226 cells (supplemental Table 1) allocated HDAC1-related (violet dots) and -nonrelated (gray dots) genes based on ChIP-seq data (GSM2302869). Genes shown as a volcano plot selected based on fold changes (x-axis) with adjusted P (y-axis). (C) The distribution of HDAC1, H3K27Ac, and RNA Pol II binding at the IRF4 locus in MM.1S cells was analyzed (GSM2302869 [HDAC1], GSM894083 [H3K27Ac], and GSM1070127 [RNA Pol II]). The x-axis shows the genomic position. (D-E) MM.1S and RPMI 8226 cells were treated with 1 μM of MS-275 for 24 hours and were then subjected to ChIP-Q-PCR for (D) H3K27Ac levels around the IRF4 gene or (E) RNA Pol II binding around the TSS of the IRF4 gene. Results were normalized to control immunoglobulin G (IgG) in each gene position. Error bars show the SD of triplicates. ∗∗P < .01, ∗∗∗P < .001 significantly different from the condition without MS-275 at each gene position; the Student t test. Ns, not significant.
HDAC1 epigenetically regulates IRF4 expression by fine-tuning histone acetylation in MM cells. (A) HDAC1 enrichment around H3K27Ac sites (left) and RNA Pol II binding sites (right) for all genes was analyzed using publicly available ChIP-seq data (GSM2302869 [HDAC1], GSM894083 [H3K27Ac], and GSM1070127 [RNA Pol II]). Heatmaps of HDAC1 levels at H3K27Ac sites or RNA Pol II-binding sites in MM.1S cells are shown (bottom). Each row indicated ±5 kb centered on the H3K27Ac or RNA Pol II sites. The mean signal in the same intervals is plotted (top). (B) RNA-seq data of HDAC1-knockdown RPMI 8226 cells (supplemental Table 1) allocated HDAC1-related (violet dots) and -nonrelated (gray dots) genes based on ChIP-seq data (GSM2302869). Genes shown as a volcano plot selected based on fold changes (x-axis) with adjusted P (y-axis). (C) The distribution of HDAC1, H3K27Ac, and RNA Pol II binding at the IRF4 locus in MM.1S cells was analyzed (GSM2302869 [HDAC1], GSM894083 [H3K27Ac], and GSM1070127 [RNA Pol II]). The x-axis shows the genomic position. (D-E) MM.1S and RPMI 8226 cells were treated with 1 μM of MS-275 for 24 hours and were then subjected to ChIP-Q-PCR for (D) H3K27Ac levels around the IRF4 gene or (E) RNA Pol II binding around the TSS of the IRF4 gene. Results were normalized to control immunoglobulin G (IgG) in each gene position. Error bars show the SD of triplicates. ∗∗P < .01, ∗∗∗P < .001 significantly different from the condition without MS-275 at each gene position; the Student t test. Ns, not significant.
IRF4 transcriptionally regulates PIM2 expression in MM cells
The expression of IRF4 and PIM2 was higher in MM cell lines and primary tumor cells from patients with MM than in normal peripheral blood mononuclear cells or other malignant cell lines (Figure 4A and supplemental Figure 4A). Because IRF4 is one of the major TFs playing a crucial role in the pathogenesis of MM,23 we examined the relationship between IRF4 and PIM2 expression in primary MM cells (n = 559) using the publicly available dataset GSE2658 and observed a positive correlation (r = 0.169, P < 6.26237 × 10−5) between the genes (supplemental Figure 4B), suggesting that IRF4 transcriptionally regulates PIM2 expression. We demonstrated IRF4 binding in the PIM2 locus of the myeloma cell line MM.1S, but not in the adult T-cell leukemia/lymphoma cell line KK1 (Figure 4B). We also confirmed IRF4 binding on the identified site in PIM2 by the ChIP qualitative polymerase chain reaction (Q-PCR) assay in RPMI 8226, MM.1S, and U266 cells (Figure 4C and supplemental Figure 4C). The knockdown of IRF4 significantly downregulated the expression of PIM2 at the mRNA and protein levels in these MM cell lines (Figure 4D-E and supplemental Figure 4D). Collectively, these results suggest that IRF4 transcriptionally regulates PIM2.
PIM2 expression is transcriptionally regulated by IRF4 in MM cells. (A) Whole cell lysates, which were extracted from the MM cell lines, RPMI 8226, MM.1S, U266, NCI-H929, KMS-11, and INA-6, primary CD138-positive cells derived from patients with MM, and PBMCs from healthy donors were subjected to immunoblotting. β-Actin served as a loading control. (B) Publicly available ChIP-seq data (GSM1195560) were analyzed for the distribution of IRF4 binding at the PIM2 locus in MM.1S cells. The x-axis shows the genomic position. KK1, which is an adult T-cell leukemia/lymphoma cell line, was used as a negative control (GSM2481669). (C) RPMI 8226 cells were transduced with shLuc or shIRF4 (#1). Transduced cells were subjected to a ChIP-Q-PCR analysis for IRF4 occupancy on the PIM2 gene. Results were normalized to control IgG. Error bars show the SD of triplicates. ∗∗P < .01 significantly different from the shLuc condition; the Student t test. (D) Total RNA was extracted from RPMI 8226 cells transduced with shLuc or shIRF4 (#1, #2) and then subjected to Q-PCR. GAPDH served as an internal control. Values represent the amount of mRNA relative to the shLuc control. Error bars show the SD of triplicates. ∗∗∗P < .001 significantly different from the control; the Tukey-Kramer multiple comparison test. (E) RPMI 8226, MM.1S, and U266 cells were transduced with shLuc or shIRF4 (#1, #2). The whole cell lysates extracted were subjected to immunoblotting using the indicated antibodies. β-Actin served as a loading control. Relative expression levels of each target, which are normalized to its loading control, are shown below for each immunoblotting image. PBMCs, peripheral blood mononuclear cells.
PIM2 expression is transcriptionally regulated by IRF4 in MM cells. (A) Whole cell lysates, which were extracted from the MM cell lines, RPMI 8226, MM.1S, U266, NCI-H929, KMS-11, and INA-6, primary CD138-positive cells derived from patients with MM, and PBMCs from healthy donors were subjected to immunoblotting. β-Actin served as a loading control. (B) Publicly available ChIP-seq data (GSM1195560) were analyzed for the distribution of IRF4 binding at the PIM2 locus in MM.1S cells. The x-axis shows the genomic position. KK1, which is an adult T-cell leukemia/lymphoma cell line, was used as a negative control (GSM2481669). (C) RPMI 8226 cells were transduced with shLuc or shIRF4 (#1). Transduced cells were subjected to a ChIP-Q-PCR analysis for IRF4 occupancy on the PIM2 gene. Results were normalized to control IgG. Error bars show the SD of triplicates. ∗∗P < .01 significantly different from the shLuc condition; the Student t test. (D) Total RNA was extracted from RPMI 8226 cells transduced with shLuc or shIRF4 (#1, #2) and then subjected to Q-PCR. GAPDH served as an internal control. Values represent the amount of mRNA relative to the shLuc control. Error bars show the SD of triplicates. ∗∗∗P < .001 significantly different from the control; the Tukey-Kramer multiple comparison test. (E) RPMI 8226, MM.1S, and U266 cells were transduced with shLuc or shIRF4 (#1, #2). The whole cell lysates extracted were subjected to immunoblotting using the indicated antibodies. β-Actin served as a loading control. Relative expression levels of each target, which are normalized to its loading control, are shown below for each immunoblotting image. PBMCs, peripheral blood mononuclear cells.
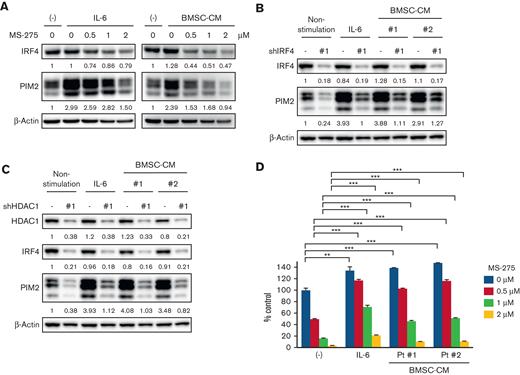
PIM2 expression is partly induced by interleukin-6 (IL-6) or BMSC independently of the HDAC1-IRF4 axis
A previous study reported that PIM2 was upregulated by MM-relevant soluble factors (ie, IL-6 and tumor necrosis factor-α) in the BM microenvironment.30 Therefore, we investigated whether IL-6 or BMSC-CM induces PIM2 independently of the HDAC1-IRF4 axis. Although IRF4 expression was not altered by IL-6 or BMSC-CM, PIM2 expression was markedly upregulated by these stimulations (Figure 5A and supplemental Figure 5A). Moreover, PIM2 was partially recovered in IRF4- or HDAC1-knockdown MM.1S and RPMI 8226 cells in the presence of IL-6 or BMSC-CM (Figure 5B-C and supplemental Figure 5B-C). These results confirmed that PIM2 is partly modulated independently of IRF4 or HDAC1 activity in the BM microenvironment. Because JAK/STAT3 signaling pathway is activated by IL-6 in MM cells, we investigated the effect of STAT3 knockdown on PIM2 expression in MM cells. STAT3 knockdown significantly reduced PIM2 expression even in the presence of IL-6 (supplemental Figure 5D), indicating the transcriptional regulation of STAT3 on PIM2 expression in MM cells under the condition of IL-6 stimulation.
MS-275 downregulated PIM2 expression and induced MM cell growth inhibition via apoptosis in a dose-dependent manner, even in the presence of IL-6 or BMSC-CM (Figure 5A,D and supplemental Figure 5A,E,F). However, the cell viability and PIM2 expression remained higher in the presence of IL-6 or BMSC-CM compared to those without these treatments, suggesting that IL-6-mediated STAT3 activation partially protects MM cells from the HDAC inhibitor-induced cytotoxicity by upregulating PIM2 expression.
PIM2 expression is regulated not only by the intrinsic axis, but also by extrinsic stimulations in MM cells. (A) MM.1S cells were treated with MS-275 at the indicated concentration in the presence of 10 ng/mL of IL-6 or patient-derived BMSC-CM (20% of total cell culture media) for 48 hours. The whole cell lysates extracted were subjected to immunoblotting with the indicated antibodies. (B-C) MM.1S cells were transduced with shLuc, shIRF4 (#1) (B), or shHDAC1 (#1) (C). Transduced cells were stimulated or cocultured with IL-6 (10 ng/mL) or BMSC-CM (20% of total cell culture media) for 48 hours, and whole cell lysates were then extracted. Lysates were subjected to immunoblotting using the indicated antibodies. β-Actin served as the loading control. Relative expression levels of each target, which are normalized to its loading control, are shown below for each immunoblotting image. (D) MM.1S cells were treated with MS-275 at the indicated concentration in the presence of IL-6 (10 ng/mL) or BMSC-CM (20% of total cell culture media) for 48 hours. Cell viability was assessed by the CCK-8 assay. Error bars show the SD of triplicates. ∗∗P < .01, ∗∗∗P < .001 significantly different from each condition of the MS-275 treatment (0, 0.5, 1, and 2 μM) in the absence of IL-6 or BMSC-CM; the Tukey-Kramer multiple comparison test.
PIM2 expression is regulated not only by the intrinsic axis, but also by extrinsic stimulations in MM cells. (A) MM.1S cells were treated with MS-275 at the indicated concentration in the presence of 10 ng/mL of IL-6 or patient-derived BMSC-CM (20% of total cell culture media) for 48 hours. The whole cell lysates extracted were subjected to immunoblotting with the indicated antibodies. (B-C) MM.1S cells were transduced with shLuc, shIRF4 (#1) (B), or shHDAC1 (#1) (C). Transduced cells were stimulated or cocultured with IL-6 (10 ng/mL) or BMSC-CM (20% of total cell culture media) for 48 hours, and whole cell lysates were then extracted. Lysates were subjected to immunoblotting using the indicated antibodies. β-Actin served as the loading control. Relative expression levels of each target, which are normalized to its loading control, are shown below for each immunoblotting image. (D) MM.1S cells were treated with MS-275 at the indicated concentration in the presence of IL-6 (10 ng/mL) or BMSC-CM (20% of total cell culture media) for 48 hours. Cell viability was assessed by the CCK-8 assay. Error bars show the SD of triplicates. ∗∗P < .01, ∗∗∗P < .001 significantly different from each condition of the MS-275 treatment (0, 0.5, 1, and 2 μM) in the absence of IL-6 or BMSC-CM; the Tukey-Kramer multiple comparison test.
The dual inhibition of PIM and class I HDACs exhibits significant anti-MM activity in vitro and in vivo
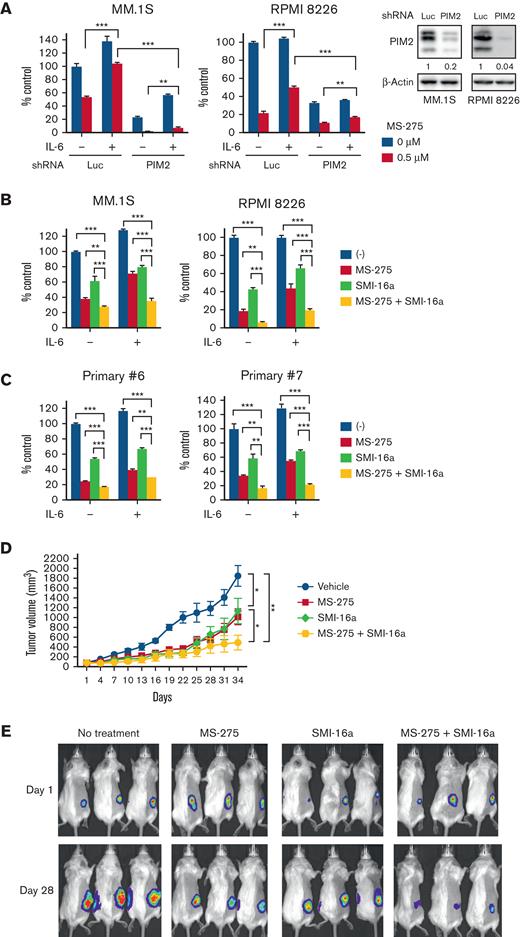
To overcome IL-6-mediated protective effects, we evaluated the biological impact of the downregulation of PIM2 in the presence of IL-6. The results obtained showed that the knockdown of PIM2 attenuated MS-275 resistance mediated by IL-6 in MM.1S and RPMI 8226 cells (Figure 6A).
Class I HDAC and PIM inhibition cooperatively suppresses MM cell growth in vitro and in vivo. (A) MM.1S or RPMI 8226 cells were transduced with shLuc or shPIM2 (#1). Transduced cells were treated with or without MS-275 (0.5 μM) in the presence or absence of IL-6 (10 ng/mL) for 48 hours, and cell viability was then assessed by the CCK-8 assay. Lysates extracted from transduced cells after puromycin selection were subjected to immunoblotting using the indicated antibodies. β-Actin served as a loading control. Relative expression levels of each target, which are normalized to its loading control, are shown below for each immunoblotting image. Error bars show the SD of triplicates. ∗∗P < .01, ∗∗∗P < .001 significantly different from the shLuc or shPIM2 condition with the MS-275 treatment; the Tukey-Kramer multiple comparison test. (B-C) MM.1S, RPMI 8226 (B), and primary CD138-positive cells (C) were treated with or without MS-275 (0.5 μM), SMI-16a (50 μM), or their combination in the presence or absence of IL-6 (10 ng/mL) for 48 hours. Cell viability was assessed by the CCK-8 assay. Error bars show the SD of triplicates. ∗∗P < .01, ∗∗∗P < .001; the Tukey-Kramer multiple comparison test. (D) After the development of measurable tumors (>50 mm3), cohorts were treated for 3 weeks with the vehicle control (n = 8; blue line), 3.5 mg/kg MS-275 3 days a week (n = 8; red line), 20 mg/kg SMI-16a 5 days a week (n = 8; green line), or 3.5 mg/kg MS-275 3 days a week with 20 mg/kg SMI-16a 5 days a week (n = 9; yellow line). Tumor growth was monitored with caliper measurements every 3 days. Error bars show the SEM of tumor volumes in each group. ∗P < .05 (control vs MS-275, SMI-16a vs MS-275 plus SMI-16a), ∗∗P < .01 (control vs MS-275 plus SMI-16a) on day 34; the Tukey-Kramer multiple comparison test. (E) Images show representative in vivo images, ordered from left to right: vehicle control, MS-275, SMI-16a, and the combination group of MS-275 with SMI-16a at the time of treatment on days 1 and 28.
Class I HDAC and PIM inhibition cooperatively suppresses MM cell growth in vitro and in vivo. (A) MM.1S or RPMI 8226 cells were transduced with shLuc or shPIM2 (#1). Transduced cells were treated with or without MS-275 (0.5 μM) in the presence or absence of IL-6 (10 ng/mL) for 48 hours, and cell viability was then assessed by the CCK-8 assay. Lysates extracted from transduced cells after puromycin selection were subjected to immunoblotting using the indicated antibodies. β-Actin served as a loading control. Relative expression levels of each target, which are normalized to its loading control, are shown below for each immunoblotting image. Error bars show the SD of triplicates. ∗∗P < .01, ∗∗∗P < .001 significantly different from the shLuc or shPIM2 condition with the MS-275 treatment; the Tukey-Kramer multiple comparison test. (B-C) MM.1S, RPMI 8226 (B), and primary CD138-positive cells (C) were treated with or without MS-275 (0.5 μM), SMI-16a (50 μM), or their combination in the presence or absence of IL-6 (10 ng/mL) for 48 hours. Cell viability was assessed by the CCK-8 assay. Error bars show the SD of triplicates. ∗∗P < .01, ∗∗∗P < .001; the Tukey-Kramer multiple comparison test. (D) After the development of measurable tumors (>50 mm3), cohorts were treated for 3 weeks with the vehicle control (n = 8; blue line), 3.5 mg/kg MS-275 3 days a week (n = 8; red line), 20 mg/kg SMI-16a 5 days a week (n = 8; green line), or 3.5 mg/kg MS-275 3 days a week with 20 mg/kg SMI-16a 5 days a week (n = 9; yellow line). Tumor growth was monitored with caliper measurements every 3 days. Error bars show the SEM of tumor volumes in each group. ∗P < .05 (control vs MS-275, SMI-16a vs MS-275 plus SMI-16a), ∗∗P < .01 (control vs MS-275 plus SMI-16a) on day 34; the Tukey-Kramer multiple comparison test. (E) Images show representative in vivo images, ordered from left to right: vehicle control, MS-275, SMI-16a, and the combination group of MS-275 with SMI-16a at the time of treatment on days 1 and 28.
We then examined the combined treatment effects of MS-275 with the PIM inhibitor SMI-16a against MM cells. The combined treatment significantly induced apoptosis (supplemental Figure 6A) and cell growth inhibition in MM cell lines (Figure 6B) as well as primary MM cells (Figure 6C). We next examined whether the combined treatment induced a synergistic effect against MM cells using SynergyFinder 2.0. The combination induced the anti-MM cytotoxic effect, but the effect was additive in the absence of IL-6 (Synergy score: 3.63 in RPMI 8226 and −3.05 in MM.1S). Of note, the effect was synergistic in the presence of IL-6 (Synergy score: 10.59 in RPMI 8226 and 10.92 in MM.1S) (supplemental Figure 6B; supplemental Table 7), indicating induction of the synergistic anti-MM effect by targeting both class I HDACs and PIM2 in the presence of IL-6. We then performed in vivo experiments using a human myeloma cell xenograft murine model. Monotherapy with MS-275 or SMI-16a significantly inhibited tumor growth compared with the vehicle control. Importantly, anti-MM activity was higher in the combined treatment cohort than in each monotherapy group (Figure 6D-E), in addition to longer overall survival (supplemental Figure 7A) without significant body weight loss (supplemental Figure 7B). We also examined the efficacy of the combined treatment using the disease model in which tumor volumes were larger than 400 mm3 when treatment started. Although monotherapy with MS-275 or SMI-16a did not significantly induce growth inhibitory effects, their combination reduced the tumor growth rate and prolonged survival (supplemental Figure 7C-D). Therefore, the combined treatment of PIM with a class I HDAC inhibitor was more potent than monotherapy with a class I HDAC inhibitor in vitro and in vivo.
Discussion
HDACs regulate gene expression through the deacetylation of lysine at the histone tail,5,6 thereby mediating cellular homeostasis. However, the precise functions of HDACs in histone modifications in MM cells currently remain unclear, even though HDAC inhibitors have been shown to induce MM cell death associated with alterations in the expression of several genes.36-38 In this study, we demonstrated that HDAC1 negatively and positively regulated gene expression in MM cells. Specifically, the expression of the master TF IRF4 in MM cell survival was positively regulated by HDAC1 through the fine-tuning of histone acetylation levels (Figure 7). Previous reports showed that class I HDACs are essential isoforms for core regulatory transcription in cancer cells.35,39 Histone acetylation regulates the promotor-enhancer interaction and histone hyperacetylation by HDAC inhibitors rather impairs the proper promoter-enhancer interaction, which reduces RNA pol II binding and thereby transcription of target genes.35 By analogy, we assume that HDAC1 inhibition/knockdown induces histone hyperacetylation to reduce RNA pol II bindings in the promoter/enhancer regions of IRF4 gene, which mitigates IRF4 transcription in MM cells, although detailed studies are required to show alteration of the three-dimensional structures of the IRF4 region after HDAC inhibitor treatment.
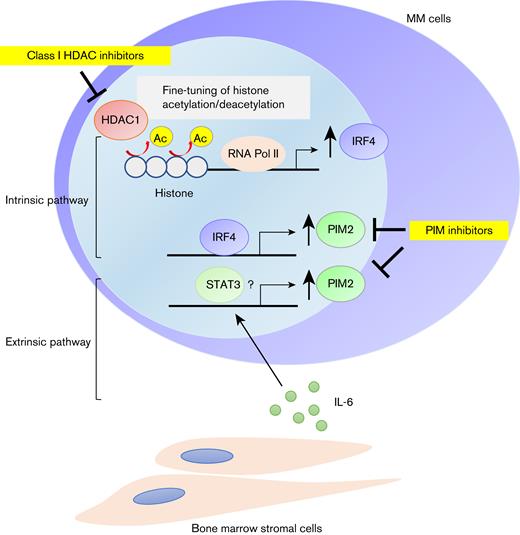
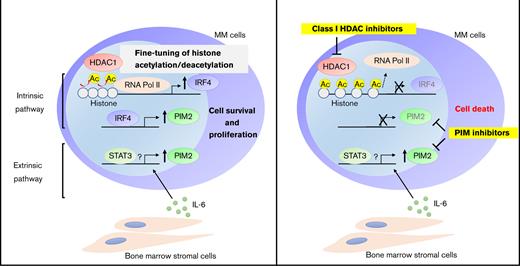
PIM2 is overexpressed in MM cells through the intrinsic HDAC1-IRF4 pathway and further enhanced by exogenous stimuli including IL-6. Schema of the regulatory mechanisms of PIM2 in MM cells.
PIM2 is overexpressed in MM cells through the intrinsic HDAC1-IRF4 pathway and further enhanced by exogenous stimuli including IL-6. Schema of the regulatory mechanisms of PIM2 in MM cells.
Although the nonselective HDAC inhibitor panobinostat has already been approved in clinical practice by the FDA, its adverse events such as general fatigue and muscle weakness have been issued. Based on such a background, class-selective HDAC inhibitors such as romidepsin and tucidinostat, which mainly inhibit class I HDACs, have been clinically developed in the treatment of peripheral T-cell lymphoma.40,41 This study reveals that class I HDACs, especially HDAC1, regulates the intrinsic axis of IRF4-PIM2 through fine-tuning histone acetylation in MM cells, suggesting the usefulness of class I-selective HDAC inhibitors in MM. We further show that BM microenvironment factor IL-6 up-regulates PIM2 expression, thereby mitigating the HDAC inhibitor-induced cytotoxicity. In contrast, PIM inhibitors overcome this, providing the rationale for targeting both the intrinsic and extrinsic regulatory mechanisms of PIM2 in MM cells (Figure 7). The sustained expression of PIM2 in the BM microenvironment may explain the limited efficacy of monotherapy with HDAC inhibitors for MM.42-44 HDAC inhibitors in combination with PIM inhibitors may augment the effect of HDAC inhibitors in clinical practice.
Because PIM2 plays important roles in drug resistance in MM cells and bone metabolism in ambient cells in BM, we adopted the PIM inhibitor SMI-16a in combination with the class I HDAC inhibitor MS-275 and observed significant growth inhibitory effects, even in the presence of IL-6. Several PIM inhibitors are currently available. The safety and efficacy of monotherapy with the ATP-competitive pan-PIM kinase inhibitor PIM447 has been demonstrated in patients with MM in a phase I clinical trial.45-47 Further studies are warranted on protective effects in bone metabolism in MM using the strategy of the combined inhibition of PIM and class I HDACs.
In summary, this study demonstrated a role for HDAC1 in the regulation of the transcriptionally activated master TF IRF4 in MM cells. Moreover, the downregulation of IRF4 by the inhibition of HDAC1 reduced the expression of PIM2 in MM cells, suggesting the existence of the HDAC1-IRF4-PIM2 intrinsic axis in MM cells. The extrinsic axis of the IL-6-mediated upregulation of PIM2 is a pivotal MM-protective signaling pathway against HDAC1 inhibition-induced MM cell death. Therefore, the dual inhibition of class I HDACs and PIM kinases represents a novel rationale for combined treatment in the context of the BM microenvironment that will improve the outcomes of patients with MM.
Acknowledgments
The authors thank the Center for Cancer Computational Biology, Dana-Farber Cancer Institute for assistance with the RNA-seq analysis.
This work was supported in part by JSPS KAKENHI grant numbers JP18K16118 (T.H.), JP17KK0169 (J.T.), JP17H05104 and JP19K22719 (M.H.), JP18K08329 (M.A.), the program of the Joint Usage/Research Center for Developmental Medicine, Inter-University Research Network for Trans-Omics Medicine, Institute of Molecular Embryology and Genetics, Kumamoto University (H.O), and the Research Clusters program of Tokushima University (1803003, M.A.). T.H. was also supported by the Kanae Foundation for the Promotion of Medical Science (4615062), the Japanese Society of Hematology Research Grants (17344, 18262, and 19203), and a Multiple Myeloma Research Grant from Myeloma Patients and Families, Japan. The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Authorship
Contribution: T.H. and H.O. designed the study; T.H., H.O., and A.O. performed experiments and analyzed data; H.O. generated data from ChIP-seq; M.N. and S.S. synthesized SMI-16a; T.H., R.S., M.O., K.S., T.M., M.T., S.F., S.N., H.M., K.K., and S.O. provided clinical samples; H.O., J.T., M.H., T.H., and M.A. supervised study design and experiments; T.H. wrote the manuscript; and T.H., H.O., T.H., and M.A. reviewed and edited the manuscript.
Conflict-of-interest disclosure: M.A. received research funding from Chugai Pharmaceutical, Sanofi K.K., Pfizer Seiyaku K.K., Kyowa Hakko Kirin, MSD K.K., GSK, Nippon Shinyaku, Astellas Pharma, Takeda Pharmaceutical, Teijin Pharma, and Ono Pharmaceutical; and honoraria from Janssen. The remaining authors declare no competing financial interests.
Correspondence: Takeshi Harada, Department of Hematology, Endocrinology, and Metabolism, Tokushima University Graduate School of Biomedical Sciences, 3-18-15 Kuramoto-cho, Tokushima 770-8503, Japan; e-mail: takeshi_harada@tokushima-u.ac.jp; and Hiroto Ohguchi, Division of Disease Epigenetics, Institute of Resource Development and Analysis, Kumamoto University, 2-2-1 Honjo, Chuo-ku, Kumamoto 860-0811, Japan; e-mail: ohguchi@kumamoto-u.ac.jp.
References
Author notes
RNA-seq raw data are deposited in the Gene Express Omnibus (GEO) database under the accession code GSE193298.
For data sharing, contact the corresponding author, Takeshi Harada (takeshi_harada@tokushima-u.ac.jp).
The full-text version of this article contains a data supplement.

![HDAC1 epigenetically regulates IRF4 expression by fine-tuning histone acetylation in MM cells. (A) HDAC1 enrichment around H3K27Ac sites (left) and RNA Pol II binding sites (right) for all genes was analyzed using publicly available ChIP-seq data (GSM2302869 [HDAC1], GSM894083 [H3K27Ac], and GSM1070127 [RNA Pol II]). Heatmaps of HDAC1 levels at H3K27Ac sites or RNA Pol II-binding sites in MM.1S cells are shown (bottom). Each row indicated ±5 kb centered on the H3K27Ac or RNA Pol II sites. The mean signal in the same intervals is plotted (top). (B) RNA-seq data of HDAC1-knockdown RPMI 8226 cells (supplemental Table 1) allocated HDAC1-related (violet dots) and -nonrelated (gray dots) genes based on ChIP-seq data (GSM2302869). Genes shown as a volcano plot selected based on fold changes (x-axis) with adjusted P (y-axis). (C) The distribution of HDAC1, H3K27Ac, and RNA Pol II binding at the IRF4 locus in MM.1S cells was analyzed (GSM2302869 [HDAC1], GSM894083 [H3K27Ac], and GSM1070127 [RNA Pol II]). The x-axis shows the genomic position. (D-E) MM.1S and RPMI 8226 cells were treated with 1 μM of MS-275 for 24 hours and were then subjected to ChIP-Q-PCR for (D) H3K27Ac levels around the IRF4 gene or (E) RNA Pol II binding around the TSS of the IRF4 gene. Results were normalized to control immunoglobulin G (IgG) in each gene position. Error bars show the SD of triplicates. ∗∗P < .01, ∗∗∗P < .001 significantly different from the condition without MS-275 at each gene position; the Student t test. Ns, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/6/10.1182_bloodadvances.2022007155/4/m_blooda_adv-2022-007155-gr3.jpeg?Expires=1769193238&Signature=JFPgu28IogCdEXE2b3Hgc3sqYPzMm22uYK1SnYGKXnq7yV17q57-eYYoqhhS9VCFpc8eY6jJFuDitHsaisE3tpwUMWV9bZKpeNlkyyYwuo80dpqPlIGr8xAq1~MqGDpI27VC6~OWdE60de0YlJoU8pq-YQ6G5FqTgYegV9x9JmaQhkjtJJlq-tS7qE3R5xoz4S9Sz0YiW-tsOXSalLwstnMmkW6M3Ly7m2mERiiCYW6gUK~9e54uPNORfk5LORp0bHm9zEmzzq4WaOrNTOa4nCvklyYvHBdhCwdoapucdQ51pZLdxhRlogfEvtUfjBqadPeTI13ylEqTlSTkXkic2Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)