Key Points
WGS reveals that precursors with low genomic complexity have a lower risk for progression to WM.
Molecular time analysis shows chr12 gain occurred early in development, whereas other gains occur later and may be associated with progression.
Abstract
The genomic landscape of Waldenström macroglobulinemia (WM) is characterized by somatic mutations in MYD88, present from the precursor stages. Using the comprehensive resolution of whole genome sequencing (WGS) in 14 CD19-selected primary WM samples; comparing clonal and subclonal mutations revealed that germinal center (GC) mutational signatures SBS9 (poly-eta) and SBS84 (AID) have sustained activity, suggesting that the interaction between WM and the GC continues over time. Expanding our cohort size with 33 targeted sequencing samples, we interrogated the WM copy number aberration (CNA) landscape and chronology. Of interest, CNA prevalence progressively increased in symptomatic WM and relapsed disease when compared with stable precursor stages, with stable precursors lacking genomic complexity. Two MYD88 wild-type WGS contained a clonal gain affecting chromosome 12, which is typically an early event in chronic lymphocytic leukemia. Molecular time analysis demonstrated that both chromosomal 12 gain events occurred early in cancer development whereas other CNA changes tend to occur later in the disease course and are often subclonal. In summary, WGS analysis in WM allows the demonstration of sustained GC activity over time and allows the reconstruction of the temporal evolution of specific genomic features. In addition, our data suggest that, although MYD88-mutations are central to WM clone establishment and can be observed in precursor disease, CNA may contribute to later phases, and may be used as a biomarker for progression risk from precursor conditions to symptomatic disease.
Introduction
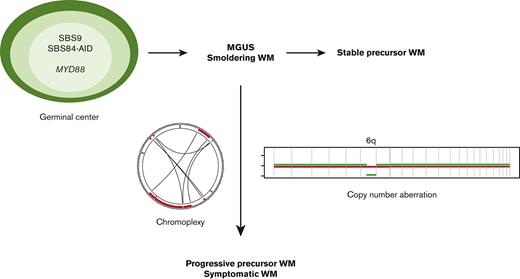
Waldenström macroglobulinemia (WM) is an immunoglobulin M (IgM)–producing indolent B-cell lymphoma, existing on a clinical spectrum from precursor states (IgM monoclonal gammopathy of uncertain significance [MGUS] and asymptomatic WM) to symptomatic disease.1 The genomic landscape of WM is characterized by recurrent somatic mutations in MYD88, producing increased signaling through Bruton’s tyrosine kinase (BTK) and nuclear factor kappa B activation, which promote WM growth and survival.1-3MYD88L265P mutations are not specific to WM, occurring in a majority of primary central nervous system and testicular lymphomas and a minority of marginal zone and nongerminal-center (GC) diffuse large B-cell lymphomas.4,5 Though highly prevalent in WM, MYD88L265P mutations are not sufficient for progression to symptomatic disease, occurring in IgM MGUS and both stable asymptomatic and symptomatic WM.4,6 This distribution suggests that MYD88 mutation is an early event in clonal development, potentially acquired before the first GC encounter,7 whereas other genomic events promote subsequent cancer progression.
WM wild-type (wt) for MYD88L265P has an estimated prevalence below 10%.8 The lack of MYD88L265P produces diagnostic uncertainty, and this entity is not well characterized genomically, with some tumor features overlapping with other low-grade lymphomas, including marginal zone lymphoma and chronic lymphocytic leukemia (CLL).
Other recurrent somatic mutations described, predominantly detected by targeted sequencing, include CXCR4, ARID1A, CD79B, NOTCH2, MLL2, KMT2D, and TP53, many of which also occur in WM precursor states.2 Indeed, Paiva et al have demonstrated that there are almost no genes showing significantly different dysregulation when comparing IgM MGUS and asymptomatic WM to symptomatic WM.9 Clinically, although most mutations have an unclear impact on prognosis, once treatment is indicated, responses to BTK-signaling inhibitors are affected by MYD88 and CXCR4 mutational status.10,11
Specific copy number aberrations (CNA) are highly recurrent in WM, with del6q resulting in loss of genes that modulate BTK and NFκB signaling (IBTK, TNFAIP3, HIVEP3) as well as cell survival/apoptotic balance (BCLAF1, FOXO3).2,12 Del6q in WM precursors has been associated with a shorter time to requiring treatment and shorter progression-free survival once treatment is initiated.13
The increased resolution gained from WGS in comparison to targeted sequencing allows a comprehensive characterization of mutational processes (ie, mutational signatures) and structural variation (SV) contributing to cancer pathogenesis, in addition to detailing single base substitution (SBS) and CNA.14,15 In the context of multiple myeloma (MM), targeted and whole exome sequencing defined some genomic features predicting progression from precursor states to symptomatic MM, including KRAS/NRAS mutations, TP53 deficiency, and MYC aberrancy.16,17 WGS analysis has allowed us to extend these findings and demonstrate that precursor states that subsequently progress to MM already have MM-defining genomic events, including the complex SV chromothripsis, specific CNA, templated insertions, and APOBEC-mutational activity (SBS signatures SBS2/SBS13).18 WGS data, therefore, allow the distinction of 2 biologically and clinically distinct entities; either stable or progressive precursors.
WGS allows the differentiation between clonal and subclonal events, and in addition, in tumors with large chromosomal gains, can provide insight into the temporal acquisition of genomic clonal events. We have demonstrated in MM how utilizing the corrected ratio between clonal mutations being either duplicated (present on both alleles and therefore acquired before the gain) or nonduplicated (present on a single allele, acquired postgain) within large chromosomal gains provides a molecular time estimation.19-21 Using this approach, we’ve shown that hyperdiploid copy number profiles in MM may be acquired in multiple time windows.19,21
In this article, we aim to characterize the temporal relationship between SBS mutational signatures, CNA, and SV in WM, using the comprehensive resolution afforded by WGS. We examined WGS from 14 CD19-selected primary samples from patients with WM, together with 33 samples from WM having targeted sequencing available.22 Our data show that GC mutational signatures are present in both clonal and subclonal populations, demonstrating that GC activity is sustained over a period of time. Both SBS landscape and mutational drivers predict clinical stage poorly. In contrast, SV and CNA emerged as key drivers to later phases and disease progression. Finally, WGS from MYD88-wt cases suggests that B-cell post-GC precursor clones may develop early gains in chromosome 12, with a later trajectory leading to either CLL or MYD88-wt WM.
Methods
Patient samples
This study involved the use of human samples, which were collected after written informed consent was obtained. Institutional review boards at each site approved the study, with samples and data obtained and managed in accordance with the Declaration of Helsinki. Patient details are summarized in supplemental Table 1.
DNA was extracted from CD19+ cells purified from the bone marrows of 14 patients collected at the University of Athens, with matched normal samples collected from peripheral blood mononuclear cells. Samples were collected at different clinical time points: MGUS (n = 1), asymptomatic WM (n = 5), symptomatic (n = 7), and relapsed WM (n = 1), with sequencing depth detailed in supplemental Table 2.
Previously published targeted sequencing data were obtained from 33 patients with WM at Memorial Sloan Kettering Cancer Center,22,23 predominantly from bone marrow (n = 13) and lymph node biopsies (n = 10), with the remainder from extramedullary disease (n = 6), peripheral blood (n = 3), and cerebrospinal fluid (n = 1). Again, there was a range of clinical scenarios, comprising stable asymptomatic (n = 5), asymptomatic which progressed during follow-up (n = 2), symptomatic but untreated (n = 9), and treatment-exposed WM (n = 17) (supplemental Table 1).
Genomic sequencing
For each WGS sample, 500ng of genomic DNA was sheared and sequencing libraries were prepared using the KAPA Hyper Prep Kit (KAPA Biosystems). Samples were run on a NovaSeq 6000 in a 150bp/150bp paired-end runs, using the SBS v1 Kit and an S4 flow cell (Illumina). The average sequence coverage was 69 for tumor samples and 35 for normal samples (supplemental Table 2). All new sequencing data will be available on publication.
The targeted sequencing data were produced using the MSK-IMPACT Heme gene panel,22 with all data previously published23,24 and accessible in the cBioportal for Cancer Genomics (http://cbioportal.org/msk-impact).25 Copy number aberration had been previously defined by Fraction and Allelic Copy number Estimation from Tumor/normal Sequencing.26
Bioinformatic analysis
All WGS bioinformatics analyses were performed using our in-house pipeline Isabl.27 Short-insert paired-end reads were aligned to the reference human genome (GRCh37) using Burrows-Wheeler Aligner (v0.7.8). Somatic mutations were identified by CaVEman.28 To detect genes and hotspots under positive selection in our cohort we applied the dndscv method. This algorithm was designed to detect genes under positive selection, even in small datasets (https://github.com/im3sanger/dndscv).29 The background mutation rate across each gene is estimated from synonymous mutations within the gene using the variation of mutation rates across genes. The detection of positive selection is further enhanced by controlling the input in restricted hypothesis testing to a set of known relevant oncogenes rather than all available genes (ie, the MSK-IMPACT Heme gene panel22).
Copy number analysis and tumor purity (ie, cancer cell fraction) were evaluated using Battenberg (https://github.com/Wedge-Oxford/battenberg), with significantly aberrant regions identified using GISTIC2.0 (v2.0.23, https://www.genepattern.org). SVs were defined by merging calls from SvABA,30 BRASS (https://github.com/cancerit/BRASS), and GRIDSS analyses (https://github.com/PapenfussLab/gridss). Complex events were defined and annotated as previously described.21 The phylogenetic tree of each case was reconstructed using the Dirichlet process (https://github.com/Wedge-Oxford/dpclust).
To estimate the activity of mutational signatures, we followed our recently published workflow based on 3 steps: de novo extraction, assignment, and fitting.31,32 For the first step, we ran SigProfiler.31 Then all extracted signatures were compared with the latest COSMIC reference (https://cancer.sanger.ac.uk/cosmic/signatures/SBS/) in order to define which known mutational processes were active in our cohort. Finally, we applied mmsig (https://github.com/UM-Myeloma-Genomics/mmsig), a fitting algorithm designed for MM, to confirm the presence and estimate the contribution of each mutational signature in each sample.33 Confidence intervals were generated by drawing 1000 mutational profiles from the multinomial distribution, each time repeating the signature fitting procedure, and finally taking the 2.5th and 97.5th percentile for each signature. To explore the contribution of each mutational signature over time, we explore all Dirichlet process clusters with more than 30 mutations using mmsig as described above.
The relative timing of each multichromosomal gain event was estimated using the R package mol_time (https://github.com/UM-Myeloma-Genomics/mol_time).19,21 Correcting the ratio between duplicated mutations (variant allele frequency around 66%, acquired before the chromosomal duplication) and nonduplicated mutations (variant allele frequency around 33%, acquired on either the nonduplicated allele or on one of the 2 duplicated ones). This approach allows for estimation of the relative molecular time of acquisition of all large (>1 Mb) chromosomal gains with adequate clonal mutations as estimated by the Dirichlet process.
Data analysis and statistics
Analysis was carried out in R version 3.6.1. Key software tools noted throughout the workflow (including SigProfiler, mmsig, dndscv and mol_time) are publicly available.
Results
Experimental data and design
We performed WGS on 14 primary bone marrow samples from patients with WM at various clinical stages, including IgM monoclonal gammopathy (MGUS, n = 1), asymptomatic (n = 5), symptomatic (n = 7), and relapsed WM (n = 1). These data were compared with targeted sequencing on 33 patients with WM, of whom 5 were stable precursors, using the MSK-IMPACT Heme gene panel (400-570 genes depending on assay version).22 Patient characteristics are shown in supplemental Table 1, with progression-free survival of asymptomatic patients detailed in supplemental Figure 1 (median, 2.2 years; interquartile range [IQR], 2.1-2.3 years).
Mutational landscape in WM reveals sustained GC interaction
WGS from 14 patients with WM identified a median of 2806 clonal SBS per sample (IQR, 1870-3079). 12 out of 14 (85%) samples harbored MYD88 mutations.
To identify recurrent nonsynonymous mutations occurring at a higher than expected rate by chance alone, we ran dndscv,29 performing analysis against both an unrestricted background and a restricted hypothesis for 570 known oncogenic drivers, selected because of their inclusion in the MSK-IMPACT Heme panel. Twenty mutations in 5 driver genes were extracted, which in addition to MYD88 included CXCR4 (3/14), CD79B (2/14), HIST1H1D (2/14) and HIST1H1D (2/14) (supplemental Table 3). Examination of known oncogenic hotspots using dndscv revealed 4 significant hotspots in the 14 WGS; MYD88 (L265P), TP53 (W91∗), PIM1 (K24N), and EZH2 (Y646F).
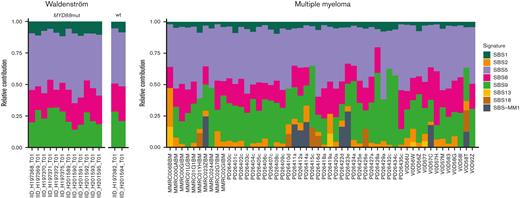
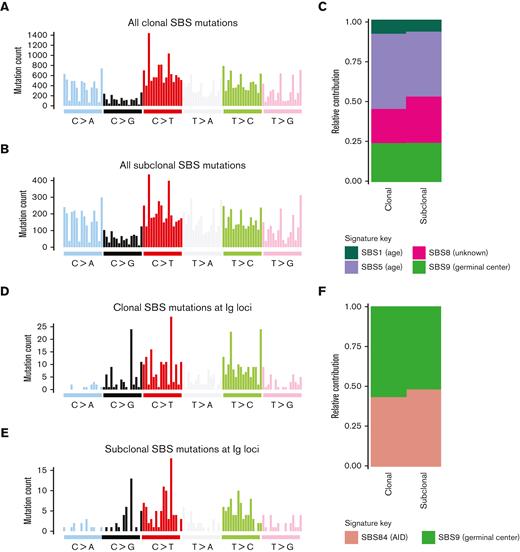
Using the ability of WGS to incorporate noncoding DNA and to investigate which mutational processes are involved in shaping the genomic landscape of WM we performed a mutational signature analysis.31-33 Four previously reported SBS signatures were detected (Figure 1): SBS1 and SBS5 (associated with aging), SBS9 (reflecting poly-eta activity in the GC), and SBS8 (not yet associated with a known biological process). In line with previous evidence in other malignancies,21 the contribution of age-related signatures SBS1/SBS5 was directly correlated with the age at presentation (R2 = 0.44, P = .014). When comparing SBS signatures in WM with those observed in MM (Figure 1,19,32,34), we note a striking absence of APOBEC-mutational activity (SBS2/SBS13) in all samples.35 The SBS signature contribution was similar when examining total clonal and subclonal mutations (Figure 2A-C). The signature SBS9 demonstrated sustained GC activity, as evidenced by the same proportion of mutations attributable to SBS9 after collapsing all mutations at each of the clonal and subclonal level (24%, Figure 2C). Examining the 96-class SBS profile and signatures extracted from SBS specific to the immunoglobulin loci, we observed evidence of clustered SBS84 (a signature associated with activation-induced cytidine deaminase activity). This reflects somatic hypermutation within the GC with SBS84 accounting for 48% of signature contribution from subclonal mutations within immunoglobulin genes (Figure 2D-F). Overall, these data suggest that the interaction between WM and the GC is sustained over time, similar to the early phase of MM19 and other hematologic malignancies.36
Clonal and subclonal contribution of SBS signatures in WM. 96-class profile of genome-wide SBS mutations at the (A) clonal and (B) subclonal level. (C) SBS mutational signatures obtained after collapsing all genome-wide mutations at the clonal and subclonal level. (D-E) 96-class profile of SBS mutations specific to the immunoglobulin loci at the (D) clonal and (E) subclonal level. (F) SBS mutational signatures obtained after collapsing all mutations within immunoglobulin loci at the clonal and subclonal level.
Clonal and subclonal contribution of SBS signatures in WM. 96-class profile of genome-wide SBS mutations at the (A) clonal and (B) subclonal level. (C) SBS mutational signatures obtained after collapsing all genome-wide mutations at the clonal and subclonal level. (D-E) 96-class profile of SBS mutations specific to the immunoglobulin loci at the (D) clonal and (E) subclonal level. (F) SBS mutational signatures obtained after collapsing all mutations within immunoglobulin loci at the clonal and subclonal level.
SV and CNA in WM predict progression to symptomatic disease
WGS allows for an accurate definition of SV and complex events, which we have previously demonstrated to be critical for the pathogenesis and clinical outcomes of MM and aggressive lymphomas.37-39 Using IgCaller40 to reconstruct the VDJ and class-switch recombination of each patient immunoglobulin gene, we confirmed that all WGS samples had a productive immunoglobulin H rearrangement and either a productive immunoglobulin K or immunoglobulin L rearrangement consistent with known WM biology41 (supplemental Table 4). Although, as expected, we documented an absence of oncogenic translocations involving BCL2, CCND1 or MYC, 2 potentially oncogenic translocations were included in the IgCaller output, (t[2;22], IGL - no partner gene identified, and t[4;22], LYAR-IGL), neither of which have clear biological or prognostic impact in WM. These 2 translocations both occurred in MYD88-mut cases with symptomatic WM.
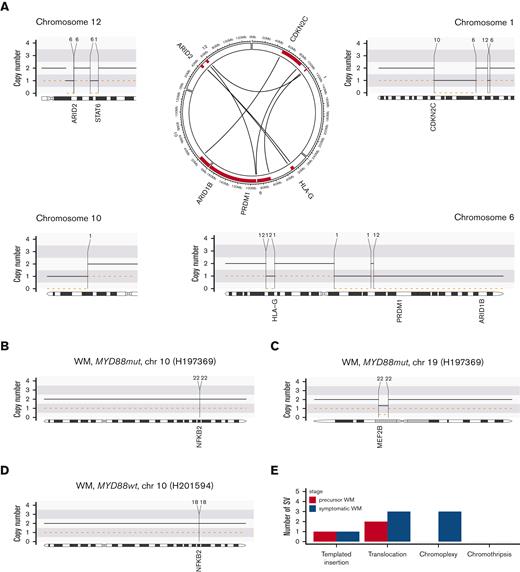
In contrast to MM,37 in this WM WGS cohort we found a low prevalence of complex SV with no chromothripsis detected and a single chromoplexy event found in 3 patients (21%, Figure 3A and supplemental Figure 2), all of whom had symptomatic WM. Chromoplexy is characterized by a concatenation of multiple SV on different chromosomes leading to multiple deletions.14,15 These kinds of events are particularly relevant because they allow the tumor cell to acquire multiple genomic drivers at the same time.21 Specifically, in sample H201589 (Figure 3A), copy number loss related to a chromoplexy event affected genes including CDKN2C, PRDM1, HLA-G, ARID2, and ARID1B. In sample H197369 (Figure 3B-C, supplemental Figure 2) chromoplexy affected driver genes NFKB2 and MEF2B, whereas in sample H201593, chromoplexy is linked to del6q. We also note a reciprocal translocation affecting NFKB2 in a further patient (H201594, Figure 3D). Overall, there was a low incidence of any SV and fewer SV in asymptomatic precursor samples compared with those having symptomatic disease (Figure 3E).
Structural variants in WM. (A) Example of a chromoplexy event between chromosomes 1, 6, 10, and 12. (B-D) Examples of structural variants affecting driver genes: translocations affecting NFKB2 (B/C) and MEF2B (D). (E) Total number of structural variants by disease stage.
Structural variants in WM. (A) Example of a chromoplexy event between chromosomes 1, 6, 10, and 12. (B-D) Examples of structural variants affecting driver genes: translocations affecting NFKB2 (B/C) and MEF2B (D). (E) Total number of structural variants by disease stage.
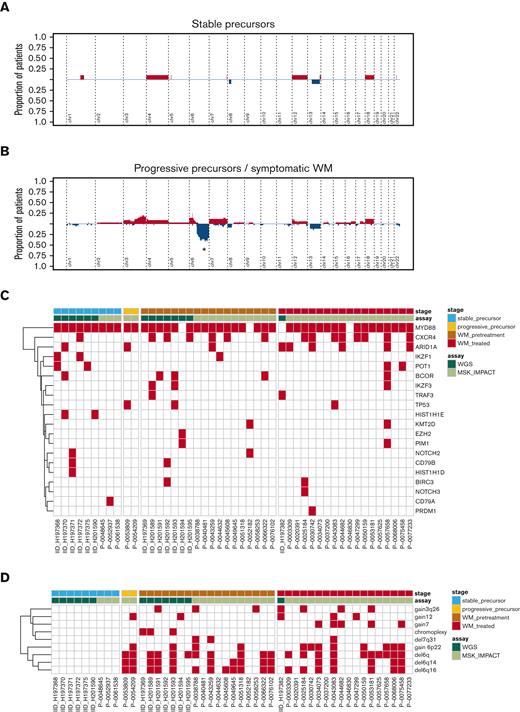
Targeted sequencing is not able to accurately define SV but allows for the definition of CNA. To explore WM CNA features in a larger cohort, we examined the WGS data together with 33 patients with WM in whom targeted sequencing was available (MSK-IMPACT Heme gene panel [400-570 genes depending on assay version],22 data via cBioportal,25 clinical information in supplemental Table 1). Running GISTIC2.0 we identified focal genomic regions recurrently involved by gains (n = 2) and deletions (n = 3, supplemental Table 5). CNA analysis demonstrated that some samples harbored typical CNA features such as del6q, whereas other tumors had minimal CNA. Cumulative CN profiles comparing samples from stable WM precursors to those with symptomatic or relapsed disease demonstrated significantly increased CN with disease progression, with 6q being significant from GISTIC2.0 analysis, being only detected in symptomatic and relapsed WM and absent among stable WM precursors (Figure 4A-B).13
Copy number profiles from Waldenström's macroglobulinemia. Cumulative copy number profiles across samples with (A) IgM MGUS or symptomatic precursor WM, and (B) symptomatic and treatment-exposed Waldenström's macroglobulinemia. Samples from patients with asymptomatic disease that later progressed were considered together with samples from symptomatic patients. In (A/B), red, gain; blue, deletion. ∗ = peak is significantly different between group according to GISTIC analysis. (C) Heatmap of mutations according to clinical stage. (D) Heatmap of copy number and structural variants according to clinical stage.
Copy number profiles from Waldenström's macroglobulinemia. Cumulative copy number profiles across samples with (A) IgM MGUS or symptomatic precursor WM, and (B) symptomatic and treatment-exposed Waldenström's macroglobulinemia. Samples from patients with asymptomatic disease that later progressed were considered together with samples from symptomatic patients. In (A/B), red, gain; blue, deletion. ∗ = peak is significantly different between group according to GISTIC analysis. (C) Heatmap of mutations according to clinical stage. (D) Heatmap of copy number and structural variants according to clinical stage.
We examined the SBS landscape of known oncogenic drivers, drivers extracted in this WGS cohort by dndscv, and mutations previously described in WM literature, and found that they poorly predict clinical stage (Figure 4C). Investigating the genomic landscape of the 2 most recurrent genomic drivers after MYD88 mutation (ie, CXCR4 mutation and del6q), no significant differences were observed after correcting for the clinical stage (supplemental Figure 3).
In contrast to the mutational spectra, the combination of CNA with SV provided clear clustering by clinical stage (Figure 4D), with patients having precursor disease that subsequently progressed to symptomatic WM having CNA similar to those having already developed symptomatic disease. These data suggest that, although MYD88-mutations are central to WM clone establishment and can be observed in stable precursor disease, CNA and SV may contribute to later phases and disease progression, with certain CNA at the asymptomatic phase predicting subsequent progression.13
Molecular time in WM
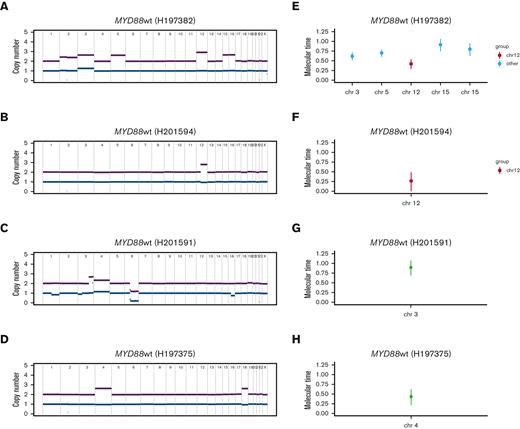
The proportion of WM wt for MYD88 has been estimated to be less than 10%.8,12 2 out of 14 of the WGS and 2 out of 33 of the targeted sequencing samples were MYD88-wt (8.5% in total), collected from patients producing large amounts of IgM paraprotein (each >2800 mg/dL) and having other mutations previously described in WM, including ARID1A, PIM1, TRAF3, NOTCH2, and KMT2D (supplemental Table 1). We hypothesized that the resolution of WGS may define additional genomic features relevant to disease pathogenesis, in particular for patients with MYD88-wt. Of interest, the 2 MYD88-wt WGS samples both contained a clonal gain affecting chromosome 12 (Figure 5A-B), which is typically an early event in lymphoproliferative disorders such as CLL.20 Other clonal gains affected chromosomes 3, 5, 15, and 16 in the sample of a patient with MYD88-wt, with a gain of 3 q and gains of chromosomes 4 and 18 in 2 MYD88-mutant samples (Figure 5A-D). To estimate the chronological order of these large chromosomal duplications, we ran a molecular time analysis, which uses the corrected ratio between duplicated and nonduplicated clonal mutations within large chromosomal gains in WGS data.19 This demonstrated that these 2 chromosomal gain events affecting chromosome 12 occurred early in cancer development (molecular time <0.5, Figure 5E-F), similar to the timing of chromosome 12 gains observed in the PCAWG CLL data set20 (median molecular time 0.36, IQR 0.17-0.44, supplemental Figure 4). Multiple other CNA changes occurred later in the disease course (molecular time >0.5) and tended to be subclonal (Figure 5E-H). We observed that the GC center signature SBS9 is present both in early and late molecular time windows, and in both pregain and postgain populations, suggesting that these large chromosomal gains were acquired within the GC.
Molecular timing analysis. Copy number profiles from; (A) MYD88-wt sample, demonstrating gains in chromosomes 3, 5, 12, 15, 16, (B) MYD88-wt sample with gain in chromosome 12, (C) MYD88-mutant sample with gain of 3q and (D) MYD88-mutant sample with gains of chromosomes 4 and 18. (E-H) Molecular timing analysis for the same 4 patient samples, using the ratio of duplicated:nonduplicated mutations within clonal chromosomal gains. Red, molecular time in chromosome 12 gains; aqua, gains in other chromosomes in patients harboring a chromosome 12 gain; black, other gains.
Molecular timing analysis. Copy number profiles from; (A) MYD88-wt sample, demonstrating gains in chromosomes 3, 5, 12, 15, 16, (B) MYD88-wt sample with gain in chromosome 12, (C) MYD88-mutant sample with gain of 3q and (D) MYD88-mutant sample with gains of chromosomes 4 and 18. (E-H) Molecular timing analysis for the same 4 patient samples, using the ratio of duplicated:nonduplicated mutations within clonal chromosomal gains. Red, molecular time in chromosome 12 gains; aqua, gains in other chromosomes in patients harboring a chromosome 12 gain; black, other gains.
Considering the targeted sequencing data, chromosome 12 gains were observed in 3 MYD88-mt samples; 2 from patients with symptomatic WM and 1 in a patient who subsequently progressed to symptomatic disease. Targeted sequencing does not allow for molecular time analysis, but these data support the hypothesis that CNA predicts later progression. Despite small numbers, taken altogether, our data suggest that precursor B-cell clones may develop gains in chromosome 12 early in cancer development within the GC, with a later trajectory leading to either CLL, MYD88-mt WM or MYD88-wt disease. Furthermore, they suggest that both deletions and large chromosomal gains tend to play a late driver role and might be used as biomarkers to predict for clinical progression in WM precursor conditions.
Discussion
The discovery of MYD88 L265P mutations, highly prevalent throughout the disease spectrum of IgM monoclonal gammopathies, revealed this mutation to be an early event in clonal development.2 Here, we have used the comprehensive resolution of WGS to examine for mutational signatures, CNA and structural variants which may contribute to disease development and progression to symptomatic WM.
Traditionally, WM was regarded as a post-GC malignancy, having undergone somatic hypermutation but not class-switch recombination. Recently, Dhodapkar et al published that the establishment of the malignant clone in WM is preceded by an expansion of nonmalignant extrafollicular B cells, with aberrant oncogenic signaling involving MYD88 in pre–B progenitor cells,7 in line with previous early evidence by Paiva et al.9 Here, examining the 96-class SBS profile and mutational signatures obtained from clonal and subclonal SBS mutations, we demonstrate that the malignant WM clone continues to interact with the GC over time, with the same proportion of mutations attributable to SBS9 being observed in both the clonal and subclonal fractions.
In order to explore the biological relevance of CNA, we demonstrate the increasing prevalence of CNA in symptomatic WM and relapsed disease when compared with stable precursor stages. Of note, biopsies collected in asymptomatic patients who later progressed to symptomatic disease contained CNA similar to biopsies collected in symptomatic disease. In addition, complex SV was only observed in patients with symptomatic disease. Although some genomic features may be selected for under therapeutic pressure, CNA and SV clustered untreated symptomatic disease together with posttreatment biopsies and distinct from stable precursor states. Similarly to MM,18,19 these data suggest that WM precursors demonstrating low genomic complexity, lacking CNA and complex SV, are biologically lower risk for progression to symptomatic disease. This pathophysiology is best appreciated with the increased resolution of WGS data.
Samples from 2 MYD88-wt WGS contained a clonal gain affecting chromosome 12, which is typically an early event in CLL, as demonstrated in the PCAWG WGS data set.20 Molecular time analysis demonstrated that these 2 chromosomal gain events occurred early in cancer development whereas other CNA changes occurred later in the disease course. Though observed in only 2 samples, these data demonstrate the additional power of WGS in elucidating biology and suggest that precursor clones may develop gains in chromosome 12 early in cancer development, with multiple potential trajectories, not restricted to CLL.
Considering potential limitations of this study, the relationship between disease history and the timing of sample collection is difficult to evaluate. It is possible that biopsies from stable WM precursors were collected earlier in the disease life history than the progressive ones. Were this to be the case, it would mean that those clones haven’t yet acquired large aneuploidies and SV required for tumor progression, and our observations remain valid. We have also included relapsed WM in our study, and as noted above, whereas certain genomic features may be selected as a consequence of exposure to anticancer therapy, CNA and SV clustered symptomatic disease distinct from stable precursor states before the effect of treatment becomes relevant. Although the limited sample size doesn't allow for definitive conclusions, the data presented and exploratory analysis provide both additional support to previous reports1,2,8,13,16 and proof-of-principle that the use of WGS and specific computational methods allow for a comprehensive characterization of the WM genomic landscape.
In conclusion, we have 3 main findings from applying advanced analytical techniques to WGS data in WM. First, WGS analysis allows the demonstration of sustained GC activity in WM over time, supporting an ongoing interaction of the GC with the WM clone. Second, whereas MYD88-mutations are central to WM clone establishment and precursor clones may develop gains in chromosome 12 early in cancer development within the GC, additional late CNA may contribute to symptomatic disease progression. Finally, despite the limited number of available samples, our data suggest that in WM, similarly to MM,18 leveraging the comprehensive genomic characterization provided by WGS, it is possible to differentiate stable WM precursors from those destined to progress over time. Following validation of these findings in a larger cohort, future prognostication in WM precursor disease may be improved by the integration of CN and SV data.
Acknowledgments
This work was supported by the MSK Steven Greenberg Lymphoma Research Awards, Sylvester Comprehensive Cancer Center NCI Core Grant (P30 CA 240139), and the Memorial Sloan Kettering Cancer Center NCI Core Grant (P30 CA 008748). F.M. and K.M. are supported by the American Society of Hematology. F.M. and O.L. are supported by the Riney Family Foundation.
Authorship
Contribution: F.M. and T.D. designed and supervised the study, collected and analyzed the data, and wrote the paper; O.L. and L.P. designed and supervised the study, collected the data, and wrote the paper; K.M and T.B. collected and analyzed the data and wrote the paper; E.K. collected the data and wrote the paper; S.L., C.F., K.A., A.D., A.L., and S.U., collected the data; and B.Z., V.Y., A.D., and E.P. analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Francesco Maura, Myeloma Division, Sylvester Comprehensive Cancer Center, University of Miami, Miami, FL 33136; e-mail: fxm557@med.miami.edu; and Meletios-Athanasios Dimopoulos, National and Kapodistrian University of Athens, Athens, Greece; e-mail: mdimop@med.uoa.gr.
References
Author notes
WGS data will be made available upon publication, please contact: Kylee H. Maclachlan (maclachk@mskcc.org). Targeted sequencing data are available via the cBioportal for Cancer Genomics (http://cbioportal.org/msk-impact).
Presented in abstract form at the 63rd annual meeting of the American Society of Hematology, Atlanta, GA, 11-14 December 2021.
The full-text version of this article contains a data supplement.
K.H.M. and T.B. contributed equally to this work.
C.O.L., M.L.P., F.M., and M.A.D. contributed equally to this work.