MDS/MPN with neutrophilia should be treated with alloHSCT whenever possible.
Cytogenetics and molecular mutations may help to diagnose MDS/MPN with neutrophilia but are not exclusive.
Visual Abstract
Myelodysplastic and myeloproliferative neoplasms (MDS/MPN) with neutrophilia, until recently called atypical chronic myeloid leukemia (aCML), being part of the MDS/MPN is a very rare disease with poor prognosis. Although emerging data reveal its cytogenetic and molecular profile, integrated survival and treatment data remain scarce. We analyzed a cohort of 347 adult patients diagnosed with MDS/MPN with neutrophilia, registered in the Netherlands Cancer Registry between 2001 and 2019. Our demographic baseline data align with other cohorts. We observed cytogenetic aberrations exclusively in patients aged >65 years, with trisomy 8 being the most common abnormality. We identified 16 distinct molecular mutations, with some patients (16/101) harboring up to 3 different mutations; ASXL1 being the most frequent one (22%). In a multivariable Cox regression analysis, only age, hemoglobin level and allogeneic hematopoietic stem cell transplant (alloHSCT) were associated with overall survival (aged >65 years; hazard ratio [HR] 1.85; P = .001 and alloHSCT HR, 0.51; P = .039). Because no other treatment modality seemed to affect survival and might cause toxicity, we propose that all patients eligible for alloHSCT should, whenever possible, receive an allogeneic transplant. It is imperative that we strive to improve outcomes for patients who are not eligible for alloHSCT. Tackling this challenge requires international collaborative efforts to conduct prospective intervention studies.
Introduction
Atypical chronic myeloid leukemia was introduced as a distinct disease entity in the 2001 World Health Organization (WHO) classification.1 This malignancy is classified within the myelodysplastic/myeloproliferative neoplasms (MDS/MPN) group, encompassing 4 entities.2 Dysplasia in the neutrophil lineage is most prominent among both myelodysplastic and myeloproliferative characteristics. Because of this feature, the fifth edition of the WHO classification of hematolymphoid tumors redefined it as “MDS/MPN with neutrophilia.”2 Besides hypercellularity with granulocytic predominance and granulocytic dysplasia, the disease is characterized by an elevated white blood cell count (≥13 × 109 cells per L) with immature myeloid cells comprising ≥10% of white blood cells, but low blasts (<20%) and monocytes (<10%). Finally, certain diagnostic criteria for other myeloid neoplasms may not be met (eg, MPNs [specifically, exclusion of a BCR::ABL1 fusion gene], myeloid neoplasms with eosinophilia and defining gene rearrangement, chronic myelomonocytic leukemia, or MDS/MPN with SF3B1 mutation and thrombocytosis). Unlike MPNs with driver mutations in JAK2, CALR, and MPL, MDS/MPN with neutrophilia frequently harbors alternative molecular mutations in the myeloid lineage,3-11 some of which are designed as desirable by the WHO (SETBP1 and ETNK1)2 and international consensus classifications (SETBP1 and ASXL1).12
MDS/MPN with neutrophilia is a very rare disease. It carries a poor median overall survival (OS) of 15 months (with a reported range of 12.4-37 months).4,10,11,13-18 Factors associated with worse survival include older age (>65 years), female sex, leukocytosis (>50 × 109 cells per L), anemia (≤10 g/dL), and circulating blasts.14 Without treatment, 30% to 40% of patients progress to acute myeloid leukemia.13,15-17,19,20 Reported treatments vary widely, resulting in diverse outcomes.13,18,20-22 Because of the low incidence, prospective (randomized) intervention studies are lacking, and treatment strategies are derived from other myeloid diseases.
Using a large cohort of patients with MDS/MPN with neutrophilia from the Netherlands Cancer Registry (NCR), our population-based study aims to validate known prognostic markers, discover novel ones, and provide evidence-based treatment recommendations.
Methods
The NCR
In the Netherlands, data from patients with newly diagnosed malignancies have been ascertained on a regional basis since the 1960s, achieving national coverage of at least 95% since 1989. Notifications of new malignancies are received from the Nationwide Network and Registry of Histopathology and Cytopathology and the National Registry of Hospital Discharges (ie, inpatient and outpatient discharges). Combining both sources is crucial, because pathologists in the Netherlands do not analyze bone marrow aspirations, which hematologists or clinical chemists perform. Upon notification of a case to the NCR, trained registrars of the NCR retrospectively collect a minimal set of data on patient and tumor characteristics and primary treatment from medical records. This information is collected within 9 to 12 months after diagnosis. More detailed information has been available for patients diagnosed from 2014 onwards, including baseline cytogenetic data (done by karyotyping), molecular analysis results at diagnosis (performed by diverse commercially available next-generation sequencing–based techniques analyzing at least ∼30 genes with a sensitivity of 1% to 5% depending on the year of testing), and the exact type of first-line treatment. The NCR uses the International Classification of Diseases for Oncology (ICD-O) to classify tumor topography and morphology. The patient’s vital status (ie, alive, deceased, or emigrated) is updated annually through linkage with the Nationwide Population Registries Network, which holds this information for all (legal) residents in the Netherlands.
Reviewing process
In the Netherlands, clinical hematologists and pathologist have well-organized consultation structures. All new patients are supposed to be discussed within a multidisciplinary consultation team (MDCT) with at least 2 hematologists (minimally 1 working in an academic center), a clinical chemist, and a pathologist to pinpoint the diagnosis and discuss the best available therapy for each patient. Centers are asked to report the percentage of patients discussed in these MDCT before the annual quality rankings are published. On top of the MDCT, Dutch pathologists have their own consultation structure, in which merely all rare diseases are discussed within regional panels. Because MDS/MPN with neutrophilia is extremely rare and difficult to differentiate from MDS, MPN, and other overlapping syndromes, these cases are discussed within these panels, or patients are referred to academic centers were a complete revision of the histopathology and laboratory data takes place.
Study population
Our study cohort includes all patients diagnosed with MDS/MPN with neutrophilia in the Netherlands between 1 January 2001 and 31 December 2019, identified from the NCR using ICD-O morphology code 9876 (notably, chronic neutrophilic leukemia (CNL) has a separate code and was not part of this study). This ICD-O code was introduced in 2001 when MDS/MPN with neutrophilia (initially termed atypical chronic myeloid leukemia) was formally recognized as a specific entity in the WHO 2001 classification. Therefore, data before 2001 could not be used. Possible cases were not traceable, most probably classified as CML (ICD-O morphology code 9863). Data for 2020 and 2021 were excluded due to the short follow-up period.
According to the Central Committee on Research involving Human Subjects, this type of observational, noninterventional study does not require approval from an ethics committee in the Netherlands. The Privacy Review Board of the NCR approved using anonymous data for this study. The study was conducted in accordance with the Declaration of Helsinki.
Primary treatment
In the overall series, primary treatment was categorized as (1) best supportive care (BSC) only, (2) antineoplastic therapy without an allogeneic hematopoietic stem cell transplantation (alloHSCT), and (3) alloHSCT with or without preceding induction therapy. This information was related to 2 age groups at diagnosis (≤65 and >65 years) and stratified by calendar period of diagnosis (2001-2013 and 2014-2019). The latter calendar period was chosen due to the availability of more detailed data from 2014 onwards. The categories for primary treatment for patients diagnosed from 2014 onwards were defined as (1) BSC only, (2) hydroxyurea, (3) hypomethylating agents, (4) tyrosine kinase inhibitors, (5) intensive induction chemotherapy, with or without an alloHSCT, and (6) upfront alloHSCT. This information was presented according to the 2 age groups described above.
Statistical analysis
Descriptive statistics were used to present patient and treatment characteristics. The Pearson χ2 test and the Kruskal-Wallis test were used to compare categorical covariates and nonnormal distributed continuous covariates, respectively.
OS was the study end point, defined as the time from diagnosis until all-cause death or last follow-up data (ie, 1 February 2022). The methodology as per Kaplan-Meier was applied to estimate OS. The log-rank test was used to evaluate whether the OS distribution differed across the following covariates for the overall cohort: sex, calendar period of diagnosis, age at diagnosis, and primary therapy in the 3 broad groupings. Furthermore, the log-rank test was used to assess the prognostic implication of the presence of (i) cytogenetic abnormalities and (ii) mutational profiles, subdivided into (a) epigenetic (ASXL1, TET2, EZH2, and DNMT3A); (b) signaling (JAK2, CALR, KIT, IDH2, and KRAS/NRAS); and (c) splicing (SRSF2) mutations, as well as the number of molecular mutations.
Descriptive statistics for OS, with associated 95% confidence intervals (CIs), were presented as median OS and the projected 1-, 5-, and 10-year OS. These statistics were presented for the overall cohort, stratified by age at diagnosis, sex, and calendar period of diagnosis. Moreover, these statistics were presented for patients diagnosed during years 2014 to 2019 according to the presence of cytogenetic abnormalities and mutational aberrations, as described above. Notably, the projected 10-year OS for this cohort could not be computed because the follow-up period was not long enough.
For patients diagnosed during years 2001 to 2019, multivariable Cox regression was performed to assess the relationship between a set of predictor variables (ie, age at diagnosis, sex, and calendar period of diagnosis) and the time to an event of interest (ie, all-cause death). The model estimates the hazard ratio, with associated 95% CIs, for each level of the categorical variable, which is a measure of how much the risk of the event-of-interest (ie, all-cause death) changes for a specific level of the predictor variable, compared with the reference level, while adjusting for the other predictor variables in the model. This model was established to assess which levels of the predictor variables are associated with a higher or lower risk of death, and how these variables influence the outcome over time. Next, to assess how primary therapy influences the relationship among the other 3 variables described above treatment, we added primary therapy into the model described above. The proportional hazard assumption was tested based on Schoenfeld residuals.
For patients diagnosed during years 2014 to 2019, for whom detailed information was available in the NCR on blood and bone marrow features and cytogenetic and molecular analysis, we performed univariable and multivariable Cox regression. These analyses were performed to assess, in an exploratory fashion, how demographic characteristics (ie, age at diagnosis and sex), blood and bone marrow features, and cytogenetic and molecular alterations influence OS.
As for the multivariable analysis, we started with a reduced model, in which variables were entered with a forward selection method, after adjusting for the weight of the variables already selected according to their significance level. The reduced model was accomplished when the P value for entering an additional variable was <.10. In the full model, all the above-mentioned variables were simultaneously adjusted. The likelihood ratio test compared the reduced model’s fit to the full model. When the difference between the 2 models is statistically significant (ie, a P value of <.05), the full model fits the data significantly better than the reduced model.
A P value of <.05 indicated statistical significance. Stata Statistical Software, Release 17.0 (StataCorp, College Station, TX) was used for the analysis.
Results
Demographic data
Between 2001 and 2019, 347 patients were diagnosed and registered with MDS/MPN with neutrophilia in the NCR. Basic patient characteristics, according to the calendar period of diagnosis (2001-2013 vs 2014-2019), are presented in supplemental Table 1. Overall, 65% of patients were male, and 71% were aged >65 years. Ages at diagnosis ranged from 22 to 95 years, and the median age at diagnosis was 72 years (interquartile range [IQR] of age, 64-95 years). The sex and age distribution were similar across both calendar periods (P > .05 for both comparisons). Five patients were known to have a prior hematological malignancy. No one was found to have a prior MPN. Two patients were diagnosed with MDS. The remaining 3 patients had lymphoma.
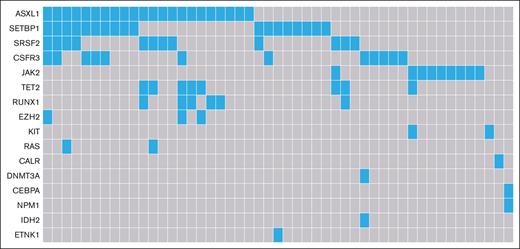
Table 1 displays detailed baseline characteristics for 110 patients diagnosed during years 2014 to 2019. Cytogenetic testing was performed in 89% of patients, with only 15% having cytogenetic abnormalities (including 1 patient with -Y). Only patients aged >65 years had cytogenetics abnormalities, of which trisomy 8 was the most common aberration (6/15 patients). For those with a cytogenetic abnormality, the median age at diagnosis was 73 years, with an IQR of 69 to 76 years. Molecular analysis was available for 92% of patients (Table 1). Overall, 16 different mutations were found in 49 patients, with up to ≥3 mutations in the same patient (Table 1; Figure 1) and the most common mutations being ASXL1 and SETBP1. Ten patients (20%) harbored the combination of an ASXL1 and a SETBP1 mutation. Moreover, mutations in CSF3R were frequently observed in our cohort (24% [12/49 patients]). SRSF2 is the only mutated gene involved in the splicing machinery (in 24% of the patients), of which 75% harbor additional mutations.
Baseline patient characteristics, from 2014 to 2019
| Characteristics . | 2014-2019 cohort . | |
|---|---|---|
| N . | (%) . | |
| Total number of patients | 110 | (100) |
| Demographics | ||
| Sex | ||
| Male | 78 | (71) |
| Female | 32 | (29) |
| Age, y | ||
| Median (IQR) | 73 (67-78) | |
| ≤65 | 22 | (20) |
| >65 | 88 | (80) |
| Blood counts, median (IQR) | ||
| Hb, g/dL∗ | 11.0 (9.0-12.4) | |
| Platelets, ×109/L∗ | 150 (95-273) | |
| Neutrophils, ×109/L† | 31.8 (18.6-60) | |
| Monocytosis (>1 × 109/L)∗ | 59 | (54) |
| Blasts, %‡ | 1 (1-4) | |
| Bone marrow features | ||
| Blast data available, n/N (%) | 93/110 (85%) | |
| Median (IQR), % | 2 (1-4) | |
| <5% | 72 | (77) |
| ≥5% | 21 | (23) |
| Fibrosis data available, n/N (%) | 87/110 (79%) | |
| Fibrosis present§ | 66 | (76) |
| Cytogenetics | ||
| Available | 98 | (89) |
| Normal karyotype | 83 | (85) |
| Abnormal karyotype‖ | 15 | (15) |
| Trisomy‖ | 10 | (10) |
| Molecular genetics | ||
| Available | 101 | (92) |
| ASXL1 mutation | 22 | (22) |
| SETBP1 mutation | 18 | (18) |
| SRSF2 mutation | 12 | (12) |
| CSFR3 mutation | 12 | (12) |
| JAK2 mutation | 9 | (9) |
| TET2 mutation | 8 | (8) |
| RUNX1 mutation | 6 | (6) |
| EZH2 mutation | 3 | (3) |
| Other mutations¶ | 8 | (8) |
| Number of mutations | ||
| No mutations | 52 | (51) |
| One mutation | 22 | (22) |
| Two mutations | 11 | (11) |
| Three or more mutations | 16 | (16) |
| First-line treatment | ||
| BSC only | 33 | (30) |
| Antineoplastic therapy without alloSCT | 62 | (56) |
| Antineoplastic therapy with alloSCT | 15 | (14) |
| Death during follow-up | 92 | (84) |
| Median follow-up, mo (IQR) | 16.8 (9.4-33.1) | |
| Characteristics . | 2014-2019 cohort . | |
|---|---|---|
| N . | (%) . | |
| Total number of patients | 110 | (100) |
| Demographics | ||
| Sex | ||
| Male | 78 | (71) |
| Female | 32 | (29) |
| Age, y | ||
| Median (IQR) | 73 (67-78) | |
| ≤65 | 22 | (20) |
| >65 | 88 | (80) |
| Blood counts, median (IQR) | ||
| Hb, g/dL∗ | 11.0 (9.0-12.4) | |
| Platelets, ×109/L∗ | 150 (95-273) | |
| Neutrophils, ×109/L† | 31.8 (18.6-60) | |
| Monocytosis (>1 × 109/L)∗ | 59 | (54) |
| Blasts, %‡ | 1 (1-4) | |
| Bone marrow features | ||
| Blast data available, n/N (%) | 93/110 (85%) | |
| Median (IQR), % | 2 (1-4) | |
| <5% | 72 | (77) |
| ≥5% | 21 | (23) |
| Fibrosis data available, n/N (%) | 87/110 (79%) | |
| Fibrosis present§ | 66 | (76) |
| Cytogenetics | ||
| Available | 98 | (89) |
| Normal karyotype | 83 | (85) |
| Abnormal karyotype‖ | 15 | (15) |
| Trisomy‖ | 10 | (10) |
| Molecular genetics | ||
| Available | 101 | (92) |
| ASXL1 mutation | 22 | (22) |
| SETBP1 mutation | 18 | (18) |
| SRSF2 mutation | 12 | (12) |
| CSFR3 mutation | 12 | (12) |
| JAK2 mutation | 9 | (9) |
| TET2 mutation | 8 | (8) |
| RUNX1 mutation | 6 | (6) |
| EZH2 mutation | 3 | (3) |
| Other mutations¶ | 8 | (8) |
| Number of mutations | ||
| No mutations | 52 | (51) |
| One mutation | 22 | (22) |
| Two mutations | 11 | (11) |
| Three or more mutations | 16 | (16) |
| First-line treatment | ||
| BSC only | 33 | (30) |
| Antineoplastic therapy without alloSCT | 62 | (56) |
| Antineoplastic therapy with alloSCT | 15 | (14) |
| Death during follow-up | 92 | (84) |
| Median follow-up, mo (IQR) | 16.8 (9.4-33.1) | |
alloSCT, allogeneic stem cell transplantation.
Missing in 1 patient.
Missing in 13 patients.
Missing in 18 patients.
Fibrosis data: 21 (19%) patients had no bone marrow fibrosis. 47 had grade 1 (42%), 18 had grade 2 (16%), 1 had grade 3 (1%), and 23 had an unknown grade (21%).
Abnormal karyotype includes 10 trisomies (ie, +8 [n = 6], +13 [n = 2], +21 [n = 1], and add(10p) [n = 1]). There remaining 5 patients had either a −20, -Y, del(13p), der(5p), or t(16;17). One patient had 2 abnormalities: add (10p) and i(17q). Notably, none of the patients had a complex karyotype.
Includes patients with the following mutations: CALR (n = 1), DNMT3A (n = 1), KIT (n = 2), CEPBA (n = 1), NPM1 (n = 1), IDH2 (n = 1), ETNK1 (n = 1), and KRAS/NRAS mutation (n = 2). Notably, a patient can harbor several mutations.
Mutational landscape of patients with MDS/MPN with neutrophilia in the Netherlands, from 2014 to 2019. Of the 101 patients with data available of their molecular profile, 49 patients had aberrations (22 with 1 single mutation, 11 with 2 mutations and 16 with ≥3 mutations).
Mutational landscape of patients with MDS/MPN with neutrophilia in the Netherlands, from 2014 to 2019. Of the 101 patients with data available of their molecular profile, 49 patients had aberrations (22 with 1 single mutation, 11 with 2 mutations and 16 with ≥3 mutations).
Treatment
Figure 2A presents data on primary therapy for the entire cohort, according to the calendar period of diagnosis and age at diagnosis. The only noteworthy and significant trend objectified was the increased use of alloHSCT in patients aged ≤65 years, from 16% (12/77) to 50% (11/22) between years from 2001 to 2013 and from 2014 to 2019 (P = .003). The proportion of patients not receiving antineoplastic therapy during years 2001 to 2013 was 17% (13/77) in the younger age group and 33% (53/160) for patients aged >65 years. This proportion remained unchanged over time. Generally, the use of antineoplastic therapy in patients aged >65 years did not increase significantly over time. However, the application of alloHSCT gradually emerged in this age group diagnosed during years 2014 to 2019 (4/88 = 5%) because it was not applied during the period 2001 to 2013.
Primary treatment of patients with MDS/MPN with neutrophilia in the Netherlands, from 2001 to 2019. (A) The results of primary therapy in broad categories are shown according to age at diagnosis and calendar period of diagnosis for patients diagnosed during the calendar period 2001 to 2019 (N = 347). The absolute number of patients within a specific calendar period and age group is shown above the bar. (B) The specific type of primary therapy according to age at diagnosis are shown for patients diagnosed during the calendar period 2014 to 2019 (N = 110). The absolute number of patients within a specific age group is shown above the graph bar. For both panels, the absolute number of patients with a specific treatment group is shown in the bar when space allows for readability. Of the 10 patients who received intensive chemotherapy, 8 (80%) were consolidated with an alloHSCT (7 in patients aged ≤65 years and 1 in a patient aged >65 years). One patient received an alloHSCT after treatment with imatinib. Lastly, 6 patients received an upfront alloHSCT (4 in-patients aged ≤65 years and 2 in-patients aged >65 years). Four patients, all aged >65, received treatment with a hypomethylating agent, of whom 3 received azacitidine and 1 received decitabine. Nine patients received treatment with a targeted agent, of whom 4 received dasatinib (1 in a patient aged ≤65 years and 3 in patients aged >65 years), 3 received imatinib (all in those aged >65 years), and 2 received ruxolitinib (1 in each age group). HMA, hypomethylating agent; HU, hydroxyurea; TKI, tyrosine kinase inhibitor.
Primary treatment of patients with MDS/MPN with neutrophilia in the Netherlands, from 2001 to 2019. (A) The results of primary therapy in broad categories are shown according to age at diagnosis and calendar period of diagnosis for patients diagnosed during the calendar period 2001 to 2019 (N = 347). The absolute number of patients within a specific calendar period and age group is shown above the bar. (B) The specific type of primary therapy according to age at diagnosis are shown for patients diagnosed during the calendar period 2014 to 2019 (N = 110). The absolute number of patients within a specific age group is shown above the graph bar. For both panels, the absolute number of patients with a specific treatment group is shown in the bar when space allows for readability. Of the 10 patients who received intensive chemotherapy, 8 (80%) were consolidated with an alloHSCT (7 in patients aged ≤65 years and 1 in a patient aged >65 years). One patient received an alloHSCT after treatment with imatinib. Lastly, 6 patients received an upfront alloHSCT (4 in-patients aged ≤65 years and 2 in-patients aged >65 years). Four patients, all aged >65, received treatment with a hypomethylating agent, of whom 3 received azacitidine and 1 received decitabine. Nine patients received treatment with a targeted agent, of whom 4 received dasatinib (1 in a patient aged ≤65 years and 3 in patients aged >65 years), 3 received imatinib (all in those aged >65 years), and 2 received ruxolitinib (1 in each age group). HMA, hypomethylating agent; HU, hydroxyurea; TKI, tyrosine kinase inhibitor.
Figure 2B shows detailed data on primary therapy for 110 patients diagnosed during years from 2014 to 2019. Patients aged ≤65 years (n = 22) most commonly received intensive chemotherapy (8/22; 36%), followed by hydroxyurea, upfront alloHSCT, BSC, and kinase inhibitors. As for patients aged >65 years (n = 88), hydroxyurea (50%) was the most common primary therapy, followed by BSC and kinase inhibitors; only a few patients received hypomethylating agents, intensive chemotherapy, or upfront alloHSCT.
Detailed baseline information about the 15 patients treated with an alloHSCT between 2014 and 2019 are given in the supplemental information (supplemental Table 5). The median age was 58 years, with an IQR of 54 to 66 years. The minimum and maximum age were 47 and 70 years, respectively. In this cohort, 6 of 15 patients received an alloHSCT upfront, whereas the remaining 9 received an alloHSCT after intensive induction chemotherapy (n = 8) or after treatment with a tyrosine kinase inhibitor (n = 1). Of these 15 patients, 7 received a reduced intensity conditioning before their alloHSCT; for the remaining 8 patients, the conditioning type was unknown (or not retrievable).
Survival
At a median follow-up of 15.8 months (IQR, 5.3-33.9), 86% of patients diagnosed during years between 2001 and 2019 died. The median OS for the overall cohort was 15.8 months (95% CI, 13.8-17.2; Figure 3A), with no significant difference in OS across the sexes (P = .385; Figure 3B) and calendar periods (P = .923; Figure 3C). However, age (P < .001; Figure 3D) and primary therapy (P < .001; Figure 3E) had a prognostic effect on OS. The OS (regardless of 1, 5, or 10 years) in patients aged ≤65 years was higher than in their older counterparts (supplemental Table 2). Patients who received an alloHSCT had the highest OS, with a 2-year OS of 59% (95% CI, 39-75). Multivariable analysis confirmed the above-mentioned findings (supplemental Table 3). The 5- and 10-year OS after an alloHSCT was exactly the same as was the 2-year OS of 59% (95% CI, 39-75). Notably, although hampered by low numbers, there was no OS difference between the group of patients with an upfront alloHSCT and the ones who received induction treatment before alloHSCT (P for log-rank test = .688; supplemental Figure 1).
OS of patients with MDS/MPN with neutrophilia in the Netherlands, from 2001 to 2019. The panels show the OS of patients with MDS/MPN with neutrophilia diagnosed during 2001 to 2019 (A) overall and according to (B) sex, (C) calendar period of diagnosis, (D) age at diagnosis, and (E) primary therapy in broad categories. The median OS and the 1-, 5-, and 10-year OS according to the characteristics shown in panels A-E are presented in supplemental Table 2. alloSCT, allogeneic stem cell transplantation; Tx, therapy; w/o, without.
OS of patients with MDS/MPN with neutrophilia in the Netherlands, from 2001 to 2019. The panels show the OS of patients with MDS/MPN with neutrophilia diagnosed during 2001 to 2019 (A) overall and according to (B) sex, (C) calendar period of diagnosis, (D) age at diagnosis, and (E) primary therapy in broad categories. The median OS and the 1-, 5-, and 10-year OS according to the characteristics shown in panels A-E are presented in supplemental Table 2. alloSCT, allogeneic stem cell transplantation; Tx, therapy; w/o, without.
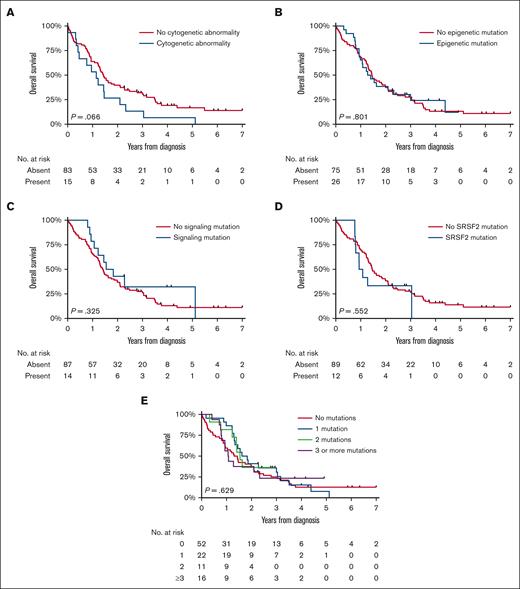
An exploratory survival analysis was conducted in 110 patients diagnosed during years between 2014 and 2019, with detailed data on prognostic factors available (Table 1). The median follow-up of this cohort was 16.8 months (IQR, 9.4-33.1 months), during which 92 (85%) patients died. OS distributions based on cytogenetic abnormalities and the type and number of molecular aberrations appeared similar (Figure 4), a finding corroborated in multivariable analysis for patients with available molecular and cytogenetic data (n = 90; supplemental Table 4). It is important to note that 1 patient with an unknown hemoglobin (Hb) level was excluded from this analysis, leaving 89 patients for the Cox regression analysis. This examination further established age as a significant prognostic factor. In addition, patients with Hb levels below 10 g/dL demonstrated significantly poorer outcomes in the multivariable Cox regression analysis, emphasizing the importance of this parameter in predicting survival outcomes. A separate exploratory analysis encompassing 10 patients with JAK2 (n = 9) and CALR (n = 1) mutations vs 91 without these driver mutations, showed no significant survival difference (P for log-rank = .164). Furthermore, stratifications by age and restriction to alloHSCT recipients also indicated no prognostic divergence.
OS of patients with MDS/MPN with neutrophilia in the Netherlands, from 2014 to 2019. The panels show the OS of patients with MDS/MPN with neutrophilia diagnosed during 2014 to 2019 according to the presence of (A) cytogenetic abnormalities, (B) epigenetic mutations, (C) signaling mutations, and (D) SRSF2 mutations and (E) the number of molecular mutations. The median OS and the 1-, 5-, and 10-year OS according to the characteristics shown in panels A-E are presented in supplemental Table 2.
OS of patients with MDS/MPN with neutrophilia in the Netherlands, from 2014 to 2019. The panels show the OS of patients with MDS/MPN with neutrophilia diagnosed during 2014 to 2019 according to the presence of (A) cytogenetic abnormalities, (B) epigenetic mutations, (C) signaling mutations, and (D) SRSF2 mutations and (E) the number of molecular mutations. The median OS and the 1-, 5-, and 10-year OS according to the characteristics shown in panels A-E are presented in supplemental Table 2.
Discussion
In this nationwide, population-based study, we present one of the largest historical cohorts of patients diagnosed with MDS/MPN with neutrophilia, including one-third (110 patients) of patient cohort suitable for in-depth analysis. Baseline characteristics, such as blood counts and bone marrow features, were in line with previously published cohorts,3,10,11,14,16,18,19 verifying representativeness and generalizability of our cohort.
For a population-based data set, the availability of cytogenetic data is quite satisfying. A cytogenetic analysis was performed in the large majority (89%), including 80% of older individuals and 30% who did not receive antineoplastic treatments. The alterations found were consistent with published data. Interestingly, in our cohort, these cytogenetic alterations were less frequently present (14% compared with 33%-42% in other cohorts).3,5,21 To figure our underlying reasons, we compared our demographic and laboratory data where possible with cohorts in the literature. Cohorts seem to be comparable without any striking discrepancies. We could not find confounding factors suggesting selection bias. Interestingly, cytogenetic alterations exclusively emerged in patients aged >65 years, reflecting higher genomic instability at older age.
Molecular analysis could be performed in 92% of patients diagnosed between 2014 and 2019. Detected mutations were typical for myeloid neoplasms, with ASXL1 (45%) and SETBP1 (37%) being the most prominent. Furthermore, molecular (co)mutations were identified in SRSF2, CSF3R, TET2, RUNX1, EZH2 RAS, and ETNK1 genes, aligning with current literature.5,10,18,21 The most common comutation, found in 20% of patients, was ASXL1 and SETBP1. Palomo et al, who described mutational landscapes for various adult MDS/MPN, reported the combination of ASXL1 and SETBP1 mutations as typical for MDS/MPN with neutrophilia.5 Our cohort’s diagnostic accuracy was supported by the presence of only 2 patients (out of 101) with features more suggestive of chronic myelomonocytic leukemia, according to Palomo’s data set. The accurate reader of our and merely all previously published data5,10,11,18,21 may recognize that a few cases with mutations in JAK2, CALR, and MPL are part of these MDS/MPN with neutrophilia cohorts. Mutations in these genes are uncommon in MDS/MPN with neutrophilia1,2,12 and should prompt morphologic examination to rule out alternative diagnoses. The same is to be said about CSF3R mutations and MDS/MPN-RS-T with SR3B1 mutations.1,2 In contrast, mutations such as SETBP1, ASXL1,12 and ETNK12 are desirable. The question arises of how to analyze data in the scope of these mutations whether desirable or not and the discrepancies between WHO and International Consensus Classification. Should one leave them out or include them, knowing that other mandatory parameters are in line with the definition criteria and that expert panels decided that no other diagnosis fitted better? Moreover, several patients with the comutation ASXL1 and SETBP1 considered to be typical for MDS/MPN with neutrophilia5 harbored CSF3R as well. The question is, which mutation should prevail in a developing field where more and more is learned about clonal evolution.
Interestingly, no survival differences were observed among calendar periods. However, Hb level (<10 g/dL), age, and primary therapy significantly affected survival. As expected, younger patients (≤65 years) had significantly better outcomes than their older counterparts (hazard ratio, 2.55; P < .007). After adjusting for primary therapy, age remained a prognostic factor, suggesting that treatment allocation differences did not entirely explain age-based OS disparities. The only baseline difference among age groups was cytogenetic aberrations, which exclusively occurred in older patients. This finding suggests that genomic instability may contribute to survival differences. However, survival analysis concerning cytogenetics and molecular mutations (clustered by their function and number) did not reveal significant differences, likely because of small numbers. However, this analysis hinted in favor of the absence of chromosomal aberrations and molecular mutations, which requires validation in forthcoming studies.
Recently, molecular profiles and outcomes of MDS/MPN with neutrophilia and CNL have been compared and discussed.10,11 However, we cannot contribute those to this discussion because we did not analyze CNL data.
Currently, no standard therapy exists for MDS/MPN with neutrophilia. The disease’s rarity makes conducting any (randomized) studies challenging. Nevertheless, different treatment strategies were applied and reported, eloquently summarized by Crisa et al21 and Gotlib.22 Our unique, population-based data set enables the comparison of different strategies for the entire cohort of 347 patients. BSC, antineoplastic therapy without alloHSCT, and alloHSCT (with or without induction) were compared. Survival was not affected by antineoplastic therapy without an alloHSCT or BSC, whereas alloHSCT yielded a significant survival benefit. Although results favoring alloHSCT are highly significant, this finding should be interpreted cautiously owing to the likely selection of fit patients eligible for an alloHSCT and lead-time bias (ie, patients living long enough to receive an alloHSCT). Nevertheless, our cohort encompasses patients up to the age of 70 years and the plateau in OS after 2 years after diagnosis in patients who received allograft suggests potential long-lasting remissions or cure. Apparently, somehow, cells from patients with MDS/MPN with neutrophilia are immunogenic, allowing immunological control over the disease. Lastly, our results show better OS than that of the most extensive alloHSCT study in MDS/MPN with neutrophilia.20 More specifically, we reached better OS estimates, namely a median OS that was not reached, with a plateau at 2 years and a projected 2-year OS of 59%. However, a direct comparison is difficult because of differing age limits and limited detailed data in our study.
Our study possesses several notable strengths, such as using a nationwide, population-based cohort, which effectively minimizes the selection and referral biases that frequently affect more traditional multicenter studies in MDS/MPN with neutrophilia. In addition, we have extensive data available for individual patients, bolstering the validity of our findings. Although we are candid about the study’s limitations, they should not detract from its relevance. These limitations include the absence of treatment data beyond 1 year after diagnosis and the discussion about mutational patterns. In addition, the transformation rates to acute myeloid leukemia were not consistently available throughout the registry.
In conclusion, our nationwide, population-based study presents a very large and most comprehensive data set of patients with MDS/MPN with neutrophilia, including diagnostics, cytogenetics, molecular, treatment, and survival data. We confirmed previous findings and provided a comprehensive picture of population outcomes. Lastly, our results indicate that, to date, only alloHSCT significantly improves OS. It might be interesting to use the power of global collaboration in combining various registries worldwide into a unified, large-scale data set, enabling more meaningful population-based studies in MDS/MPN with neutrophilia and ultimately enhancing patient care.
Acknowledgments
The authors thank Priscilla Kamphuis for designing Figure 1 depicting the landscape of molecular mutations. The authors thank the registration clerks of the Netherlands Cancer Registry (NCR) for their dedicated data collection. Established in 1989, the nationwide population-based NCR is maintained and hosted by the Netherlands Comprehensive Cancer Organization.
Authorship
Contribution: S.K.K. designed the study, analyzed the data, and wrote the manuscript; G.A.H. helped design the study and reviewed the manuscript; O.V. collected data and reviewed the manuscript; H.C.K-N. helped analyze the data and wrote the manuscript; and A.G.D. helped design the study, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Saskia K. Klein, Department of Hematology, University Medical Center Groningen, University of Groningen, PO Box 30.001, 9700 RB Groningen, The Netherlands; email: s.k.klein@umcg.nl.
References
Author notes
Additional data may be found in the supplemental section, and original data may be available upon reasonable request from the corresponding author, Saskia K. Klein (s.k.klein@umcg.nl).
The full-text version of this article contains a data supplement.