Key Points
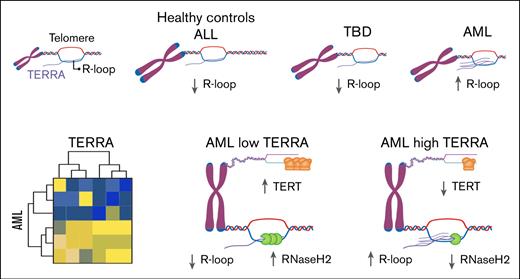
TERRA is repressed in primary human hematopoietic cells (in the bone marrow and peripheral blood), regardless of their telomere lengths.
TERRA is dysregulated in a subset of AML characterized by higher R-loop formation and low TERT and RNAseH2 expression.
Abstract
TERRA (telomeric repeat-containing RNA) is a class of long noncoding RNAs transcribed from subtelomeric and telomeric regions. TERRA binds to the subtelomeric and telomeric DNA–forming R-loops (DNA-RNA hybrids), which are involved in telomere maintenance and telomerase function, but the role of TERRA in human cells is not well characterized. Here, we comprehensively investigated for the first time TERRA expression in primary human hematopoietic cells from an exploratory cohort of patients with acute myeloid leukemia (AML), patients with acute lymphoblastic leukemia (ALL), patients with telomere biology disorder (TBD), and healthy subjects. TERRA expression was repressed in primary human hematopoietic cells, including healthy donors, patients with ALL, and patients with TBD, irrespective of their telomere length, except for AML. A second cohort comprising 88 patients with AML showed that TERRA was overexpressed in an AML subgroup also characterized by higher R-loop formation, low TERT and RNAseH2 expression, and a paucity of somatic splicing factor mutations. Telomere length did not correlate with TERRA expression levels. To assess the role of TERRA R-loops in AML, we induced R-loop depletion by increasing RNAseH1 expression in 2 AML cell lines. Decreased TERRA R-loops in AML cell lines resulted in increased chemosensitivity to cytarabine. Our findings indicate that TERRA is uniformly repressed in primary human hematopoietic cells but abnormally expressed in an AML subset with low telomerase.
Introduction
Telomeres are heterochromatic nucleoprotein complexes located at the ends of linear chromosomes.1 In mammals, they are composed of tandem repeats of 5′-TTAGGG-3′ capped with a 6-protein structure collectively called shelterin.2,3 Telomeres protect the chromosomes from erosion and prevent DNA recombination.4 When critically short, telomeres trigger cellular proliferative arrest and apoptosis signals.5 The inactivation of protective mechanisms, such as p53 and retinoblastoma tumor suppressor pathways, overrides short telomere signaling and allows continuous proliferation and, consequently, excessive telomere erosion. Extremely short telomeres become dysfunctional, causing a telomeric crisis characterized by genomic instability and rearrangements.6,7
Initially, telomeres were assumed to be silent genomic regions, but this premise has been challenged by the discovery of telomeric repeat-containing RNA (TERRA).8 TERRA is a long noncoding RNA transcribed from subtelomeric regions toward telomere ends, and thus, TERRA molecules consist of subtelomeric-derived sequences and UUAGGG repeats.8,9 TERRA binds to telomeric DNA, forming DNA-RNA hybrids called R-loops.10,11 TERRA and R-loops are involved in telomere protection and telomerase regulation.11-14
TERRA has different and divergent effects on telomerase activity, depending on the model used. In in vitro models, TERRA binds to the telomerase enzyme, acting as a competitive inhibitor of telomeric DNA, and functions in a noncompetitive fashion.9,15 In mouse embryonic stem cells (ESCs), TERRA knockdown increases telomerase activity but also causes telomeric DNA damage.16 In a study using telomerase-positive human cancer cell lines (HCCLs), recombinant TERRA overexpression was correlated with reduced telomerase activity.17 Conversely, others overexpressed TERRA in HCCLs by knocking out DNA methyltransferases and inducing telomeric transcription, which did not affect telomerase activity.18
TERRA also exerts different effects on telomere length using different approaches. In yeast, TERRA upregulation results in telomere shortening.19 Human patients with immunodeficiency with centromeric malies (ICF) syndrome type 1 carry biallelic germ line pathogenic variants in the DNMT3B gene.20 In ICF syndrome type 1, most distal subtelomeric regions are hypomethylated, resulting in increased TERRA transcription and R-loop formation.21,22 In ICF syndrome type 1 primary fibroblasts, increased TERRA expression is associated with DNA damage, telomere attrition, and premature senescence.21-23 In contrast, TERRA can promote telomere elongation by enabling telomerase activity or triggering alternative telomere lengthening (ALT). In yeast, telomere shortening promotes TERRA expression.12,24 TERRA accumulates at critically short telomeres, causing replication stress and DNA damage foci, triggering homologous direct repair, an ALT mechanism.11,25 In ALT HCCLs, TERRA transcription triggers replication stress and promotes telomere elongation through break-induced replication.25 This mechanism is regulated by the THO complex, which inhibits the formation of TERRA R-loops and represses chromatin sister-chromatid exchange in ALT HCCL cells.26 Additionally, TERRA interacts with telomerase, forming TERRA-telomerase clusters, which are recruited to short telomeres and stimulate telomerase-mediated elongation in cis.12,24,27
TERRA transcription is regulated by various pathways. DNMT3B methylates TERRA promoters during embryonic development, decreasing TERRA transcription.28,29 Additionally, both TRF2, a core shelterin component, and ATRX, a chromatin remodeler, inhibit TERRA transcription.16,30-32 TERRA levels are also controlled by RNaseH1 and RNaseH2, which degrade TERRA R-loops.
The role of TERRA in telomere maintenance, telomerase activity, and heterochromatin formation is not entirely understood. Different cell lines, including yeast, mouse ESCs, and telomerase-negative and telomerase-positive HCCLs, have been used, which may explain the conflicting results. Moreover, studies using human primary cells are scarce.33
Here, we investigate for the first time TERRA expression in primary cells obtained from patients diagnosed with acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), and telomere biology disorder (TBD). We found that TERRA is not expressed in human leukocytes, but its expression is dysregulated in AML.
Methods
Patients
The study is composed of 2 patient cohorts. In the first cohort, we prospectively selected 17 patients with AML, 5 with ALL, and 9 with TBD seen at the Hematology Division, University Hospital, Ribeirão Preto Medical School, University of São Paulo. Eighteen healthy donors were studied as controls (14 donated peripheral blood [PB] and 4 donated bone marrow [BM]). In the second cohort, we retrospectively selected 98 consecutive patients with AML from the International Consortium on Acute Promyelocytic Leukemia (IC-APL) and the International Consortium on Acute Promyelocytic Leukemia (IC-AML) seen between December 2015 and January 2020. The study was approved by the local research ethics committees (Certificate of Presentation of Ethical Appreciation numbers 12386813.0.1001.5440; 05410818.5.0000.5440; 1557.0.004.000-05) and was conducted according to the Declaration of Helsinki. Informed consent for all patients was obtained before enrollment.
Eligibility criteria required a diagnosis of AML or ALL according to the World Health Organization criteria 2016.34 Eligibility criteria for TBD included short age-adjusted telomere length below the first percentile and/or the presence of germ line pathogenic variants in telomere biology genes associated with a clinical phenotype (BM failure, pulmonary fibrosis, or liver disease).
Cell lines and culture
HeLa (American Type Culture Collection), a telomerase-positive cell line, and VA-13 (American Type Culture Collection), an ALT cell line, were cultivated in Dulbecco modified Eagle medium (Gibco), supplemented with 10% fetal bovine serum. H1 (WiCell International Stem Cell Bank), a telomerase-positive cell line, was cultivated in ESC medium. NB4, an acute promyelocytic leukemia cell line, and THP1, an acute monocytic leukemia cell line, were cultured in RPMI 1640 medium (Gibco), supplemented with 10% and 20% fetal bovine serum, respectively. All cells were kindly donated by the Center for Cell-based Therapy and were maintained in a humidified incubator at 37°C with 5% CO2.
Telomere length measurement by qPCR
Telomere length by quantitative polymerase chain reaction (qPCR) was performed as previously described.35 Southern blot was performed to validate the qPCR telomere measurement. Primers used in this experiment are described in supplemental Table 1.
TERRA RT-qPCR
Real-time qPCR (RT-qPCR) for TERRA was performed as previously described.36 Briefly, total RNA was extracted from TRIzol LS (Ambion), treated with RNase-Free DNase Set (Qiagen), and purified with RNA Clean and Concentrator (Zymo Research) twice. Two micrograms of RNA were converted to complementary DNA (cDNA) using High-Capacity cDNA Reverse Transcription (Thermo Fisher), 10-μM TERRA oligonucleotide, and 1-μM β-actin. Quantitative PCR for TERRA 10q, 15q, 20q, XqYq, 17p, and XpYp was executed using Power SYBR Green PCR Master Mix (Applied Biosystems no. 4367659) in an Applied Biosystems 7500 Real-Time PCR System. Master mix and cDNA were applied to a 96-well reaction plate (Applied Biosystems MicroAmp Fast Optical 96-well reaction plate with barcode 0.1 mL). qPCR was conducted in duplicate as described by the manufacturer, and qPCR data were analyzed using the relative quantification method using β-actin as a housekeeping gene. The relative differences in TERRA expression between samples were calculated by applying the −2ΔΔCT method. Primers used in these experiments are described in supplemental Table 2.
TERT, TRF2, ATRX, RNaseH1, and RNaseH2 RT-qPCR
RT-qPCR for TERT, TRF2, ATRX, RNaseH1, and RNaseH2 was performed following the manufacturer’s protocol. Briefly, RT-PCR was performed with 1 μg of RNA using High-Capacity cDNA Reverse Transcription (Thermo Fisher). qPCR was conducted in duplicate as described by the manufacturer, and qPCR data were analyzed using the relative quantification method using glyceraldehyde-3-phosphate dehydrogenase as a housekeeping gene. TaqMan probes used in these experiments are described in supplemental Table 3.
Vector cloning
The RNASEH1-green fluorescent protein (GFP) and GFP sequences (pEGFP-RNASEH1 no. 108699, pEGFP-N1-FLAG no. 60360, Addgene) were transferred to the lentiviral vector (pCDH-CMV-MCS-EF1α-Puro cDNA Dual Promoter Cloning and Expression Lentivector no. CD510B-1, System Biosciences). Both insert and vector were cut with NotI and NheI restriction enzymes (New England Biolabs). Following the manufacturer’s instructions, gel-purified fragments were ligated to the lentiviral backbone using T4 DNA ligase (New England Biolabs). One ShotTM Stbl3TM cells (Thermo Scientific) were transformed, and the resulting expression plasmid was purified by Qiagen Plasmid Maxi Kit (Qiagen). All sequences were confirmed by Sanger sequencing.
Viral particles production and transduction of cell lines
The viral particle process involved cotransfection of LentiX TM-293T cells with 3 DNA plasmids, including transfer plasmids (RNASEH1-GFP or GFP), psPAX2 (Addgene), and pMD2.G (Addgene) at a ratio of 3:2:1, respectively, using lipofectamine 3000 (Invitrogen), according to the manufacturer’s instructions. Supernatants containing lentiviral particles were collected 24 hours and 48 hours after transfection and filtered through a 0.45-μm filter membrane (Millipore). Then, the filtered supernatant was concentrated by high-speed centrifugation in OptimaTM XL-100 K ultracentrifuge (Beckman Coulter, rotor SW28) for 2 hours at 4°C with 25 000 rpm through a 20% sucrose cushion. The concentrated viruses were resuspended in precooled phosphate-buffered saline, aliquoted, and stored at −80 °C until use. The viral titer was determined by flow cytometry after transduction of the Jurkat cell line. The cell lines NB4 and THP1 were transduced with the viral particles (multiplicity of infection 5) in the presence of polybrene (Santa Cruz Biotechnology) at 8 mg/mL. The plate was then centrifuged for 90 minutes at 1000g, 32°C, and incubated. After 48 hours, cells were analyzed by flow cytometry to determine the frequency of GFP-positive cells.
Targeting NGS
Patients with AML from the IC-AML and IC-APL were screened for somatic variants in splicing factors, epigenetic modifiers, ASXL1, TP53, and RUNX1 by a customized targeting next-generation sequencing (NGS) panel (supplemental Table 4).
Somatic variants of enrolled patients were systematically analyzed and classified as pathogenic, likely pathogenic, variants of uncertain significance, likely benign, or benign according to the American College of Medical Genetics and Genomics criteria.37
Cell viability (MTT assay)
The MTT assay followed the protocol described by Mosmann (1983) with modifications and was conducted in triplicate.38 Cells were seeded in 96-well plates and incubated with different cytarabine concentrations (300, 600, 1200, 2400, 4800, 9600, and 19 200 nM) at 37°C for 48 hours. Subsequently, 10 μL of MTT solution (5 mg/mL) was added to each well and incubated at 37°C. After 4 hours, the formazan crystals were dissolved with 100 μL of acidified isopropanol, and the absorbance was measured at 570 nm using a microplate reader. The 50% inhibitory (or inhibition or infective) concentration values were calculated in GraphPad Prism v9 (GraphPad Software, Inc, La Jolla, CA).
Results
TERRA expression in human leukocytes and hematologic disorders
To explore TERRA expression in human primary hematopoietic cells, we analyzed samples from 17 patients with AML, 5 with ALL, 9 with TBD, and 18 healthy controls. Two cell lines, HeLa, a telomerase-dependent cell line, and VA-13, a telomerase-independent cell line, were used as controls. The patient’s clinical and molecular profiles are summarized in Table 1.
Baseline characteristics of the initial cohort
| Group . | . | . | . | . | . |
|---|---|---|---|---|---|
| Health controls | ID | Sex | Age | ||
| PB | PB1 | M | 28 | ||
| PB2 | F | 25 | |||
| PB3 | F | 41 | |||
| PB4 | F | 71 | |||
| PB5 | F | 62 | |||
| PB6 | M | 33 | |||
| PB7 | F | 26 | |||
| PB8 | F | 27 | |||
| PB9 | M | 26 | |||
| PB10 | F | 58 | |||
| PB11 | F | 56 | |||
| PB12 | F | 28 | |||
| PB13 | F | 31 | |||
| PB14 | F | 26 | |||
| BM | BM1 | F | 75 | ||
| BM2 | F | 28 | |||
| BM3 | M | 38 | |||
| BM4 | M | 54 | |||
| Acute leukemias | ID | Sex | Age | Blasts (BM) | |
| ALL | ALL1 | M | 20 | 96 | |
| ALL2 | M | 12 | 46 | ||
| ALL3 | M | 1 | 93 | ||
| ALL4 | F | 3 | 92 | ||
| ALL5 | F | 19 | 99 | ||
| AML | AML1 | M | 26 | 90 | |
| AML2 | F | 90 | 90 | ||
| AML3 | F | 64 | 80 | ||
| AML4 | F | 84 | 31 | ||
| AML5 | F | 56 | 90 | ||
| AML6 | M | 23 | 30 | ||
| AML7 | F | 58 | 80 | ||
| AML8 | F | 1 | 72 | ||
| AML9 | M | 74 | 87 | ||
| AML10 | F | 72 | 90 | ||
| AML11 | F | 55 | 95 | ||
| AML12 | F | 36 | 93 | ||
| AML13 | M | 75 | 33 | ||
| AML14 | F | 51 | 75 | ||
| AML15 | F | 45 | 77 | ||
| AML16 | M | 65 | 46 | ||
| AML17 | M | 52 | 90 | ||
| TBD | ID | Sex | Age | Telomere length (percentile) | Germ line variant |
| TBD | TBD1 | F | 20 | <1 | TINF2/p.R282C |
| TBD2 | M | 10 | <10 | DKC1/p.A353V | |
| TBD3 | F | 16 | <10 | TERT/p.R865H | |
| TBD4 | M | 40 | <1 | Not identified | |
| TBD5 | F | 35 | <1 | Not identified | |
| TBD6 | F | 64 | <1 | Not identified | |
| TBD7 | M | 57 | <10 | TERC/r.C94T | |
| TBD8 | M | 59 | 30 | TERT/p.R865H | |
| TBD9 | M | 27 | <1 | TERT/p.R865H | |
| Group . | . | . | . | . | . |
|---|---|---|---|---|---|
| Health controls | ID | Sex | Age | ||
| PB | PB1 | M | 28 | ||
| PB2 | F | 25 | |||
| PB3 | F | 41 | |||
| PB4 | F | 71 | |||
| PB5 | F | 62 | |||
| PB6 | M | 33 | |||
| PB7 | F | 26 | |||
| PB8 | F | 27 | |||
| PB9 | M | 26 | |||
| PB10 | F | 58 | |||
| PB11 | F | 56 | |||
| PB12 | F | 28 | |||
| PB13 | F | 31 | |||
| PB14 | F | 26 | |||
| BM | BM1 | F | 75 | ||
| BM2 | F | 28 | |||
| BM3 | M | 38 | |||
| BM4 | M | 54 | |||
| Acute leukemias | ID | Sex | Age | Blasts (BM) | |
| ALL | ALL1 | M | 20 | 96 | |
| ALL2 | M | 12 | 46 | ||
| ALL3 | M | 1 | 93 | ||
| ALL4 | F | 3 | 92 | ||
| ALL5 | F | 19 | 99 | ||
| AML | AML1 | M | 26 | 90 | |
| AML2 | F | 90 | 90 | ||
| AML3 | F | 64 | 80 | ||
| AML4 | F | 84 | 31 | ||
| AML5 | F | 56 | 90 | ||
| AML6 | M | 23 | 30 | ||
| AML7 | F | 58 | 80 | ||
| AML8 | F | 1 | 72 | ||
| AML9 | M | 74 | 87 | ||
| AML10 | F | 72 | 90 | ||
| AML11 | F | 55 | 95 | ||
| AML12 | F | 36 | 93 | ||
| AML13 | M | 75 | 33 | ||
| AML14 | F | 51 | 75 | ||
| AML15 | F | 45 | 77 | ||
| AML16 | M | 65 | 46 | ||
| AML17 | M | 52 | 90 | ||
| TBD | ID | Sex | Age | Telomere length (percentile) | Germ line variant |
| TBD | TBD1 | F | 20 | <1 | TINF2/p.R282C |
| TBD2 | M | 10 | <10 | DKC1/p.A353V | |
| TBD3 | F | 16 | <10 | TERT/p.R865H | |
| TBD4 | M | 40 | <1 | Not identified | |
| TBD5 | F | 35 | <1 | Not identified | |
| TBD6 | F | 64 | <1 | Not identified | |
| TBD7 | M | 57 | <10 | TERC/r.C94T | |
| TBD8 | M | 59 | 30 | TERT/p.R865H | |
| TBD9 | M | 27 | <1 | TERT/p.R865H | |
F, female; M, male.
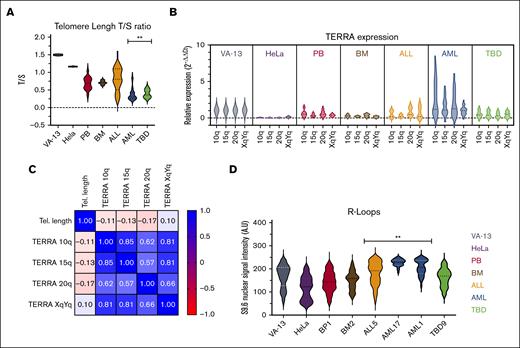
The telomere length was determined by qPCR, and the telomere repeat copy number/single gene copy number relation was calculated. Southern blot analyses validated the qPCR telomere length results. Southern blot was performed in 17 samples with short and long telomeres in different diagnoses (AML, ALL, TBD, and healthy controls). Bland-Altman analysis showed good correspondence between both methods without bias. Additionally, there was a good correlation between qPCR and Southern blot (r2 = 0.7) (supplemental Figure 1). Although the ALL samples showed telomere lengths comparable to those of healthy controls, the TBD and AML samples had very short telomeres (Figure 1A). We then measured the expression of TERRA in 10q, 15q, 20q, and XqYq. VA-13 exhibited the highest TERRA levels,39,40 whereas TERRA expression was very low in HeLa cells, healthy donors (BM and PB samples), patients with ALL, and those with TBD in comparison to VA-13 (P < .05). AML was the only group with high TERRA expression compared with healthy donors (P < .01) (Figure 1B). The AML group could be further divided into 2 subsets based on TERRA expression.
TERRA is constitutively expressed in low levels in patients with TBD, patients with ALL, and healthy donors. Patients with AML present a distinct pattern. (A) Violin plots of telomere length in the initial cohort. The patients with AML and TBD demonstrated the shortest telomere repeat copy number/single gene copy number (T/S) ratios compared with the BM and PB controls. (B) Violin plot of TERRA expression in the initial cohort. Patients with ALL and TBD demonstrate a similar level of TERRA expression compared with the BM and PB controls. (C) Spearman correlation matrix of TERRA 10q, 15q, 20q, XqYq, and telomere length among all cases included. TERRA expression is not upregulated by telomere shortening (P > .05). (D) Violin plot of R-loops in 8 samples. AML and ALL demonstrated high R-loops compared with the BM and PB controls.
TERRA is constitutively expressed in low levels in patients with TBD, patients with ALL, and healthy donors. Patients with AML present a distinct pattern. (A) Violin plots of telomere length in the initial cohort. The patients with AML and TBD demonstrated the shortest telomere repeat copy number/single gene copy number (T/S) ratios compared with the BM and PB controls. (B) Violin plot of TERRA expression in the initial cohort. Patients with ALL and TBD demonstrate a similar level of TERRA expression compared with the BM and PB controls. (C) Spearman correlation matrix of TERRA 10q, 15q, 20q, XqYq, and telomere length among all cases included. TERRA expression is not upregulated by telomere shortening (P > .05). (D) Violin plot of R-loops in 8 samples. AML and ALL demonstrated high R-loops compared with the BM and PB controls.
No correlation was observed between telomere length and TERRA expression in human primary leukocytes (Figure 1C). When the groups were compared, patients with TBD and patients with AML had the shortest telomeres, but only a subset of those with AML upregulated TERRA.
Because TERRA binds to DNA, forming R-loops, we selected 8 samples from different groups to quantify the R-loops using the S9.6 antibody. Both AML and ALL samples showed R-loop accumulation compared with healthy donors (P < .01) (Figure 1D; supplemental Figure 2).
These findings demonstrate that TERRA is not expressed in human primary hematopoietic cells regardless of their telomere length, but in neoplastic conditions, a subset of AML leukemic cells abnormally upregulate TERRA and form DNA-RNA hybrids.
TERRA expression pattern in AML
In the initial AML cohort, 2 subgroups were identified based on TERRA expression. To further understand the role of TERRA in AML, we selected 98 AML retrospective samples from the IC-APL and IC-AML clinical studies diagnosed between December 2015 and January 2020. Ten patients were excluded because of sample quality or lack of clinical information (Figure 2). The characteristics of the 88 patients studied are summarized in Table 2.
In this study, 98 patients were screened, but 10 were initially excluded. The patients were classified into 2 groups based on TERRA expression. Two patients from the high TERRA AMLs and 7 from the low TERRA AMLs were excluded from the NGS panel. AML cohort diagram.
In this study, 98 patients were screened, but 10 were initially excluded. The patients were classified into 2 groups based on TERRA expression. Two patients from the high TERRA AMLs and 7 from the low TERRA AMLs were excluded from the NGS panel. AML cohort diagram.
Baseline characteristics of the AML cohort
| Variable . | Number . |
|---|---|
| Patients included | 88 |
| Age, mean (range), y | 43.6 (18-74) |
| Sex (%) | |
| Male | 50 |
| Female | 50 |
| AML type (%) | |
| Acute promyelocytic leukemia | 27 |
| AML with mutated NPM1 | 15 |
| AML with RUNX1-RUNXT1 | 9 |
| AML with CBFB-MYH11 | 5 |
| AML with biallelic mutation of CEBPA | 2 |
| Other AML with recurrent genetic abnormalities | 2 |
| AML not otherwise specified | 40 |
| Non-APL AML risk stratification (ELN 2017, %) | |
| Favorable | 34 |
| Intermediate | 36 |
| Adverse | 30 |
| APL risk stratification (GIMEMA/PETHEMA 2000, %) | |
| Favorable | 4 |
| Intermediate | 15 |
| Adverse | 81 |
| Variable . | Number . |
|---|---|
| Patients included | 88 |
| Age, mean (range), y | 43.6 (18-74) |
| Sex (%) | |
| Male | 50 |
| Female | 50 |
| AML type (%) | |
| Acute promyelocytic leukemia | 27 |
| AML with mutated NPM1 | 15 |
| AML with RUNX1-RUNXT1 | 9 |
| AML with CBFB-MYH11 | 5 |
| AML with biallelic mutation of CEBPA | 2 |
| Other AML with recurrent genetic abnormalities | 2 |
| AML not otherwise specified | 40 |
| Non-APL AML risk stratification (ELN 2017, %) | |
| Favorable | 34 |
| Intermediate | 36 |
| Adverse | 30 |
| APL risk stratification (GIMEMA/PETHEMA 2000, %) | |
| Favorable | 4 |
| Intermediate | 15 |
| Adverse | 81 |
ELN, European LeukemiaNet; GIMEMA, Gruppo Italiano Malattie EMatologiche dell’Adulto; PETHEMA, Programa de Estudio y Tratamiento de las Hemopatías Malignas.
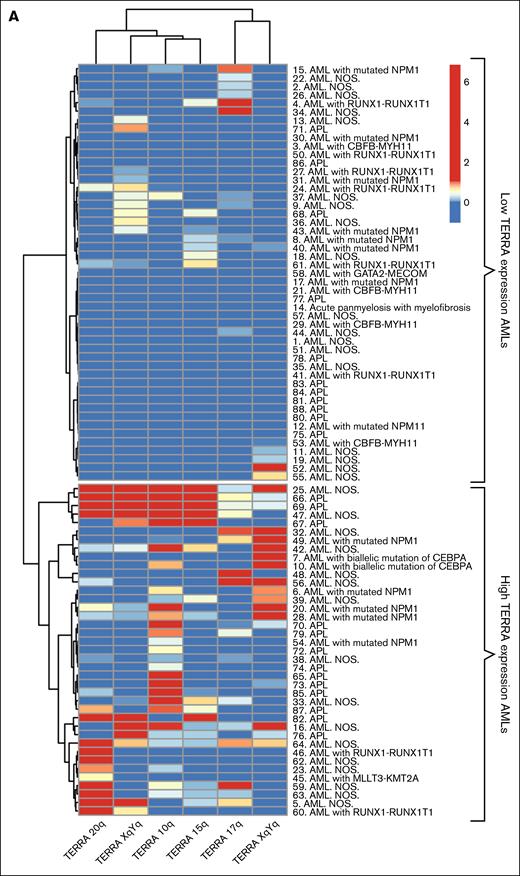
We first investigated whether the 2 AML subgroups based on TERRA expression could be confirmed in the second cohort. We applied an unsupervised hierarchical cluster algorithm based on the expression of 6 different TERRAs to the AML cohort.41 The hierarchical cluster algorithm confirmed the occurrence of 2 clusters, one with high TERRA expression and the other with low TERRA expression (Figure 3A).
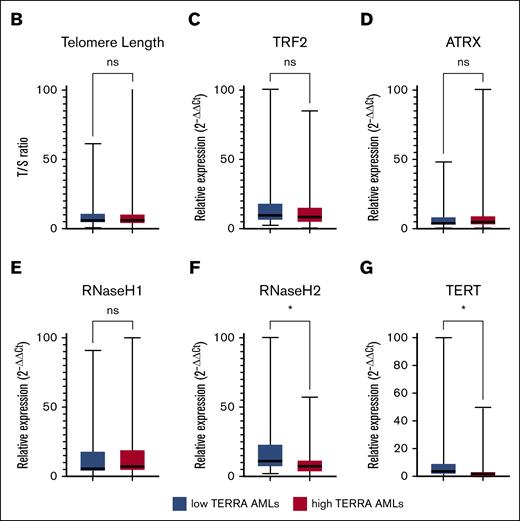
Patients with AML can be categorized in 2 groups based on TERRA expression. (A) Unsupervised hierarchical clusterization of AML samples. Both rows and columns are clustered using Euclidean distance. Two groups were formed, high-TERRA expression AMLs and low-TERRA expression AMLs. (B) Box plot of telomere length T/S ratio. (C) Box plot of TRF2 expression. (D) Box plot of ATRX expression. (E) Box plot of RNaseH1. (F) Box plot of RNaseH2. (G) Box plot of TERT expression. ∗P < .05. ns, nonsignificant.
Patients with AML can be categorized in 2 groups based on TERRA expression. (A) Unsupervised hierarchical clusterization of AML samples. Both rows and columns are clustered using Euclidean distance. Two groups were formed, high-TERRA expression AMLs and low-TERRA expression AMLs. (B) Box plot of telomere length T/S ratio. (C) Box plot of TRF2 expression. (D) Box plot of ATRX expression. (E) Box plot of RNaseH1. (F) Box plot of RNaseH2. (G) Box plot of TERT expression. ∗P < .05. ns, nonsignificant.
To understand what characteristics were associated with each subgroup, we investigated whether telomere length and TERRA regulators were correlated. Telomere length was uniformly short in AML samples and did not correlate with TERRA expression, as suggested in the initial cohort (Figure 3B). The expression of both TRF2 and ATRX were similar in the 2 AML subsets (Figure 3C-D). We next assessed the expression of RnaseH1 and RnaseH2, which are associated with R-loop removal. Similar RnaseH1 expression was observed in the 2 groups (Figure 3E), but RnaseH2 expression was reduced in the high-TERRA AMLs (P < .05; Figure 3F). Additionally, TERT transcription was significantly lower in the high-TERRA expression subgroup than in the low-TERRA expression samples, suggesting low telomerase activity in this group (P < .05; Figure 3G).
We designed an NGS panel to investigate the genomic signature associated with TERRA upregulation. Seventy-nine patients were sequenced (Figure 2), and we found that the splicing factor somatic mutations were underrepresented in the high-TERRA AMLs (P < .05; Table 3). No correlation was observed with AML subtype, AML risk stratification, progression-free survival, complete response rates, or overall survival (data not shown).
Somatic mutations in the AML cohort
| . | High-TERRA AML, n (%) . | Low-TERRA AML, n (%) . | Fisher test (P value) . |
|---|---|---|---|
| Patients sequenced | 37 | 42 | — |
| Splicing factors | 3 (8) | 12 (28) | P < .05 |
| Epigenetic modifiers | 13 (35) | 12 (28) | n.s. |
| RUNX1 | 1 (3) | 6 (14) | n.s. |
| ASXL1 | 0 (−) | 2 (5) | n.s. |
| TP53 | 0 (−) | 3 (7) | n.s. |
| . | High-TERRA AML, n (%) . | Low-TERRA AML, n (%) . | Fisher test (P value) . |
|---|---|---|---|
| Patients sequenced | 37 | 42 | — |
| Splicing factors | 3 (8) | 12 (28) | P < .05 |
| Epigenetic modifiers | 13 (35) | 12 (28) | n.s. |
| RUNX1 | 1 (3) | 6 (14) | n.s. |
| ASXL1 | 0 (−) | 2 (5) | n.s. |
| TP53 | 0 (−) | 3 (7) | n.s. |
n.s., nonsignificant. Boldface indicates not all patients included in the initial cohort were sequenced.
In aggregate, these results showed that AML cases can be categorized into 2 subgroups according to TERRA expression. The high-TERRA AML cases are characterized by low TERT and RNAseH2 expression levels and a low number of somatic mutations in splicing factors.
Additionally, we investigated whether TERRA R-loops may serve as a protective function in AML. RNaseH1 is an endonuclease that degrades TERRA R-loops. NB4 and THP1 cell lines were infected with RNaseH1-GFP–containing lentivirus or an empty-GFP vector. The 2 transduced RNaseH1 cell lines demonstrated high RNaseH1 expression, a decreased number of R-loops, and increased chemosensitivity to cytarabine (supplemental Figures 3 and 4).
Discussion
In this study, we show for the first time the TERRA physiology in primary human hematopoietic cells. We found that TERRA is not expressed in human leukocytes irrespective of their telomere length, indicating that telomere length does not regulate TERRA expression in human primary cells. We also found that TERRA is uniquely dysregulated in AML leukemic cells with low telomerase and RNaseH2 expression and forming TERRA R-loops. In AML cases with high TERRA levels, somatic splicing factor mutations were infrequent compared with those with low TERRA.
TERRA exerts key cellular functions, including telomerase activity modulation, telomere length regulation, chromatin structure organization, and gene expression control.42-44 Previous studies on TERRA expression in human cells were performed using cancer cell lines.20-23,25 Here, we assessed TERRA expression in primary human hematopoietic cells (malignant and nonmalignant) and found that TERRA is uniformly repressed in normal primary human hematopoietic cells, including mature cells in PB and more immature cells in the BM. These findings contrast with other models, which showed that telomere shortening triggers TERRA expression12,24; this was not observed in primary hematopoietic cells. In addition, TERRA expression did not seem to be triggered by telomere shortening in AML cells either because the AML group had short telomeres uniformly, but only a subset expressed TERRA. Patients with TBD also had short telomeres but no TERRA upregulation. The upregulation of TERRA was not a common event in neoplastic transformation, as patients with ALL and some of the patients with AML had low TERRA levels.
TERRA overexpression was specific to a subgroup of AML cells characterized by low TERT and RNaseH2 expression, suggesting that TERRA may serve to maintain telomere length in cells with low telomerase. The mechanism of TERRA overexpression, however, remains unclear. ICF type 1 is caused by loss-of-function germ line variants in the DNMT3B gene, resulting in hypomethylated subtelomeric regions and high TERRA levels.20 In AML samples, TERRAs transcribed from subtelomeric regions with or without CpG islands were upregulated, suggesting that epigenetic TERRA regulation is not likely responsible for TERRA expression in AML. Furthermore, TERRA expression did not correlate with somatic mutations in epigenetic regulator genes.
In myelodysplastic syndrome, high levels of R-loops correlate with splicing factor mutations.45 The authors suggest that increased R-loops result from faulty transcription owing to splicing factor mutations in myelodysplastic syndrome. In contrast, in our cohort, somatic variants in splicing factors were uncommon in high-TERRA AML samples. Our study involved primary AML only, and 27% of cases were APL. The proportion of de novo AML with the diagnosis of APL in our cohort is higher than in non–Latin American studies, but similar to studies performed in Latin America.46,47 The high proportion of AML with monocytic differentiation is unusual but characteristic of the IC-AML study.48 The difference in epidemiology may explain, at least in part, the difference in the genetic signature related to R-loop formation.
AML and ALL exhibited increased R-loops levels, but TERRA was overexpressed only in a subset of AML. In these AML cases, TERRA may serve to maintain telomere length by enabling ALT or helping telomerase activity in short telomeres.11,12,24,25,27 To test the importance of TERRA R-loops in AML, we induced RNaseH1 expression in 2 AML cell lines to deplete R-loops. The RNaseH1 expression was associated with reduced R-loops and increased chemotoxicity, further suggesting that TERRA may act to maintain chromatin stability and telomere integrity. In AML, high TERRA levels are correlated with an increased number of R-loops, which can be used as a therapeutical target. In myeloid neoplasms, poly(ADP-ribose) polymerase inhibitors preferentially kill cells with R-loop accumulation.49 Thus, our findings suggest that poly(ADP-ribose) polymerase inhibitors may be tested in AML cases with high TERRA expression.
The monitoring of measurable residual disease (MRD) in AML is challenging. Flow cytometry is the usual method for detecting MRD, but it has limitations.50,51 qPCR-based molecular MRD assessment is recommended for a subset of AML cases, but specific markers for most AML subtypes are not identified.51 Circulating RNA may be used as an MRD marker in AML.52 Given that TERRA is not expressed by normal hematopoietic stem cells, including mature blood cells and immature marrow cells, we speculate that TERRA may serve as a specific marker for MRD in cases in which TERRA expression is high at diagnosis. The detection of TERRA during follow-up in this scenario may indicate the presence of abnormal cells. Thus, TERRA may be further evaluated in future studies as a target for MRD in patients with AML.
Our study has limitations. First, we could not identify the mechanism of TERRA overexpression in AML. Second, the number of patients included is relatively limited, which prevented us from studying TERRA in specific subtypes of AML, such as core binding factor leukemias, KMT2A-rearranged leukemias, and secondary leukemias. However, we identified characteristics associated with high TERRA expression in AML. Third, since the commencement of this study, new TERRA regulators have been described, such as THOC and RTEL, but these were not assessed in this study.26 Finally, we could not distinguish whether the depleted R-loops in the transduced cell lines were mostly TERRA R-loops. However, RNAseH1 has been successfully used before to deplete TERRA.53,54
The role of TERRA in human cells remains unknown. TERRA is not expressed in hematopoietic cells (either in the BM or PB). However, it is activated in AML cells. TERRA seems to be an atavistic mechanism that may be activated in very restricted circumstances, including cancer, and is involved in telomere length maintenance and telomerase regulation.
In summary, our study shows that TERRA expression is uniformly repressed in human primary leukocytes and is not affected by telomere length. However, TERRA is overexpressed in a subset of patients with AML with low TERT and RNAseH2 expression. Based on our results, we propose that TERRA depletion and R-loops should be investigated as targets for therapy in AML with high TERRA.
Acknowledgments
This study was supported by São Paulo Research Foundation (FAPESP) grants 13/08135-2 (E.M.R., F.T., and R.T.C.) and 17/09428-4 (F.S.D.).
Authorship
Contribution: L.F.B.C., L.C.Z., and R.T.C. made substantial contributions to the conception and design of the work, literature search, figures, data interpretation, and writing; L.F.B.C., L.C.Z., F.S.D., V.S.d.C., B.A.S., A.L.P., D.F., L.E.B.d.S., B.S.T., M.I.A., F.T., L.L.d.F.P., and E.M.R. contributed to patient care and acquisition, analysis, and interpretation of data, or performed the experiments; L.F.B.C., F.S.D., and R.T.C. drafted the work or revised it critically for important intellectual content; and L.F.B.C. and R.T.C. contributed to the final approval of the version to be published.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rodrigo T. Calado, Department of Medical Imaging, Hematology, and Oncology, Ribeirão Preto Medical School, University of São Paulo, Av. Bandeirantes, 3900, Sala 743, 7º andar, HCRP, Ribeirão Preto, SP 14048-900, Brazil; e-mail: rtcalado@usp.br.
References
Author notes
∗L.F.B.C. and L.C.Z. contributed equally to the study.
All data are available on request from the corresponding author, Rodrigo T. Calado (rtcalado@usp.br).
The full-text version of this article contains a data supplement.