Key Points
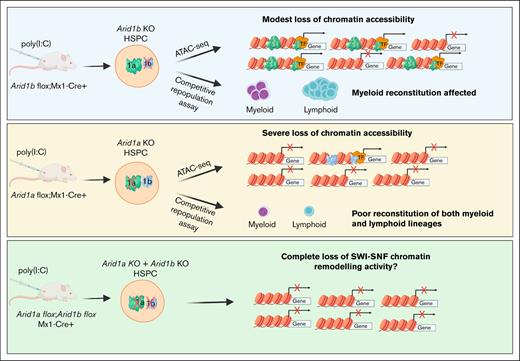
ARID1B regulates chromatin accessibility and gene expression in hematopoietic cells, and its loss affects myeloid reconstitution in mice.
Concurrent loss of ARID1A and ARID1B in the murine hematopoietic compartment leads to rapid mortality because of bone marrow failure.
Abstract
The adenosine triphosphate (ATP)–dependent chromatin remodeling complex, SWItch/Sucrose Non-Fermentable (SWI/SNF), has been implicated in normal hematopoiesis. The AT-rich interaction domain 1B (ARID1B) and its paralog, ARID1A, are mutually exclusive, DNA-interacting subunits of the BRG1/BRM-associated factor (BAF) subclass of SWI/SNF complex. Although the role of several SWI/SNF components in hematopoietic differentiation and stem cell maintenance has been reported, the function of ARID1B in hematopoietic development has not been defined. To this end, we generated a mouse model of Arid1b deficiency specifically in the hematopoietic compartment. Unlike the extensive phenotype observed in mice deficient in its paralog, ARID1A, Arid1b knockout (KO) mice exhibited a modest effect on steady-state hematopoiesis. Nonetheless, transplantation experiments showed that the reconstitution of myeloid cells in irradiated recipient mice was dependent on ARID1B. Furthermore, to assess the effect of the complete loss of ARID1 proteins in the BAF complex, we generated mice lacking both ARID1A and ARID1B in the hematopoietic compartment. The double-KO mice succumbed to acute bone marrow failure resulting from complete loss of BAF–mediated chromatin remodeling activity. Our Assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) analyses revealed that >80% of loci regulated by ARID1B were distinct from those regulated by ARID1A; and ARID1B controlled expression of genes crucial in myelopoiesis. Overall, loss of ARID1B affected chromatin dynamics in murine hematopoietic stem and progenitor cells, albeit to a lesser extent than cells lacking ARID1A.
Introduction
Adenosine triphosphate–dependent chromatin remodeling mediated by SWItch/Sucrose Non-Fermentable (SWI/SNF) complexes, involves regulation of gene transcription via mobilization of nucleosomes. The epigenetic regulation mediated by SWI/SNF complexes is critical for diverse cellular processes such as cell fate, differentiation, proliferation, and DNA repair.1,2
AT-rich interaction domain 1A (ARID1A; BAF250a) and its paralog, ARID1B (BAF250b), are mutually exclusive, ARID domain containing core subunits of the BRG1/BRM-associated factor (BAF) subclass of SWI/SNF chromatin remodeling complexes. Inactivating mutations in genes encoding for several SWI/SNF subunits, including the ARID proteins, have been identified at a high frequency in a variety of cancers.3-5 Although the role of ARID1A as a tumor suppressor is well defined, alterations of ARID1B are also frequently observed in a wide range of cancers, albeit at a lower frequency than ARID1A mutations. Somatic mutations, deletion, or loss of ARID1B expression are observed across several types of solid tumor.3-20 In addition, we have previously reported predominant loss-of-function mutations of ARID1B and ARID1A in newly diagnosed and relapsed acute promyelocytic leukemia, a distinct subtype of acute myeloid leukemia.21 Furthermore, in a murine MLL-AF9 model, deletion of ARID1B promoted leukemogenesis.22
Using a conditional knockout (KO) murine model, we have previously demonstrated that depletion of ARID1A resulted in global reduction in open chromatin regions, including decreased chromatin accessibility at promoter/enhancer regions of several key regulators of hematopoiesis. Loss of ARID1A in the hematopoietic compartment led to a multitude of defects that included elevated frequency of hematopoietic stem cells (HSCs), loss of their quiescence and reconstitution ability, as well as impaired differentiation of myeloid and lymphoid lineages.23 However, the role of its closely related homolog, ARID1B, in hematopoietic development has, to our knowledge, not been reported.
Here, to investigate the function of ARID1B in hematopoietic development, we generated mice with germ line and conditional deficiency of ARID1B. Although mice with germ line deletion of ARID1B are not viable, hematopoietic cell–specific deletion of ARID1B in adult mice does not overtly affect steady-state hematopoiesis. However, Arid1b KO HSCs exhibit impaired myeloid cell reconstitution in competitive transplantation experiments. Deficiency of ARID1B affected chromatin accessibility in hematopoietic stem and progenitor cells and led to downregulation of key hematopoiesis genes. We also demonstrate that dual loss of ARID1A and ARID1B leads to bone marrow (BM) failure, which causes rapid mortality in mice.
Materials and methods
Generation of Arid1b-deficient mice
C57BL/6N embryonic stem (ES) cells bearing the “KO-first” (KOF) allele of Arid1b (tm1a; EUCOMM), in which β-galactosidase and neomycin selection cassette are inserted in intron 4, and exon 5 is flanked by LoxP sites, were used to generate Arid1b-deficient mice. Targeted ES cells were microinjected into blastocysts and resulting chimera were mated with C57BL/6 mice. Black offspring were genotyped and mice with germ line transmission of targeted trap allele were used to establish a colony. Subsequently, mice with the targeted allele (tm1a) were crossed with the cytomegalovirus (CMV)-Cre strain to excise exon 5 along with the neomycin selection cassette to generate an Arid1b-null allele (tm1b). In addition, a conditional Arid1b allele (tm1c) was also generated by crossing mice bearing targeted allele (tm1a) with mice that express Flp recombinase (ACTB:FLPe), which resulted in the removal of β-galactosidase and neomycin selection cassette, leaving behind exon 5 flanked by LoxP sites. Hematopoietic cell–specific knock out of Arid1b was achieved using Mx1-Cre–mediated excision of exon 5 (tm1d). Deletion of exon 5 in Arid1bfl/fl;Mx1-Cre+ mice was induced by 5 doses of 300 μg of polyinosinic:polycytidylic acid (poly[I:C]; GE Healthcare) injected intraperitoneally, every alternate day.
To generate Arid1a/Arid1b double-KO mice, Arid1afl/fl;Mx1-Cre+ mice23 were first crossed with mice harboring conditional Arid1b allele (tm1c), and then intercrossed to generate homozygous conditional alleles for both Arid1a and Arid1b. poly(I:C) was injected thrice on alternate days in Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+ mice to induce Cre-recombinase–mediated deletion of both Arid1a and Arid1b.
All mice were maintained in the animal facility of the Comparative Medicine Centre, National University of Singapore (NUS). All mice experiments were performed per protocols approved by the NUS Institutional Animal Care and Use Committee.
Harvesting of cells for immunophenotypic analysis
For phenotypic analysis of Arid1bfl/fl;Mx1-Cre+ mice, poly(I:C) administration was initiated in 7- to 15-weeks-old male or female littermates. Mice were euthanized either 4 weeks or 16 to 20 weeks after the last poly(I:C) injection and the BM, spleen, and thymus were collected. BM cells were obtained by flushing femurs and tibias with phosphate-buffered saline (PBS) containing 5% fetal bovine serum (FBS). The spleen and thymus were homogenized between the frosted ends of the slides. All cell suspensions were passed through a cell strainer (30 μm) mounted on a 50-mL conical tube, and viability was determined using trypan blue exclusion. Cell suspensions were maintained on ice throughout different processing steps and staining for flow cytometric analysis.
For immunophenotyping of Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+ and control mice, poly(I:C) was injected every alternate day in 7- to 12-weeks-old male or female mice, which were euthanized 2 to 4 days after the third poly(I:C) injection. The BM, spleen, and thymus were processed as above.
Peripheral blood analysis
Blood was collected from the submandibular vein. Complete peripheral blood counts were analyzed using an Abbott Cell-Dyn 3700 hematology analyzer (Abbott Laboratories).
Flow cytometry and cell sorting
Cells were stained with antibodies diluted in PBS containing 2% FBS and incubated on ice protected from light. After washing, cells were resuspended in SYTOX Blue Dead Cell Stain (Thermo Scientific) and acquired on either LSR II flow cytometer (BD Biosciences) or FACSAria cell sorter (BD Biosciences). Data were analyzed using FACSDIVA software (BD Biosciences). A list of antibodies used for flow cytometry is included in supplemental Table 1.
Repopulation assays
For competitive reconstitution assays, BM cells from either Arid1bfl/fl;Mx1-Cre+ or Arid1bfl/fl;Mx1-Cre− mice (CD45.2+) were mixed in equal proportion with CD45.1-expressing B6.SJL (Ptprca Pepcb/BoyJ) competitor BM cells. Two million BM cells were injected into the tail vein of each lethally irradiated (10.5 Gy) B6.SJL recipient mouse. Donor engraftment was assessed in the peripheral blood 4 weeks after transplantation, after which deletion of Arid1b exon 5 was induced in recipient mice by 5 intraperitoneal doses of 300 μg poly(I:C), injected on alternate days. Reconstitution of different blood lineages was determined at 4-, 8-, 12- and 16-weeks after poly(I:C) injection in the peripheral blood of recipient mice. Blood leukocytes were stained with antibodies against CD3 (T cells), CD19 (B cells), CD11b, Gr1, F4/80 (granulocytes and monocytes), along with donor (CD45.2) and competitor (CD45.1) markers.
For noncompetitive repopulation assays, 2 × 106 BM cells from either Arid1bfl/fl;Mx1-Cre+ or Arid1bfl/fl;Mx1-Cre− mice (CD45.2+) were injected intravenously in lethally irradiated B6.SJL mice. Donor cell engraftment was verified 4 weeks after transplantation, after which poly(I:C) was administered to deplete Arid1b as described earlier. Peripheral blood counts were analyzed every 4 weeks after poly(I:C) injection.
RNA sequencing
RNA extracted from sorted Arid1b KO and wild-type (WT) granulocyte monocyte progenitors (GMPs) was used for library preparation using TruSeq RNA Sample Preparation kit (Illumina) per the manufacturer’s protocol.
Libraries were sequenced on a HiSeq 4000 and 100–base pair paired-end reads were aligned to the murine reference transcriptome (GRCm38/mm10) using Kallisto (version 0.46.0).24 Transcript level fragment counts were summarized to gene level using TxImport Bioconductor package, and differential analysis was performed using DESeq2 version 1.24.0.25,26 Gene expression was quantified in fragments per kilobase of transcript per million mapped reads units using DESeq2 “fpkm” command, and was used for all downstream analysis and plotting. All other test statistics and plotting were performed using R version 3.4.0. For gene set enrichment analysis, a GseaPreranked (version 4.3.2) analysis was done using genes ranked by their log2 fold changes.27 We used all “active transcripts” with a mean expression of 0.5 fragments per kilobase of transcript per million mapped reads in order to identify significantly enriched gene sets among MSigDB Hallmark and M2 mouse gene sets.
ATAC-sequencing
Assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) libraries were prepared from lineage marker–negative (Lin−)/c-kit–positive (Kit+) BM cells, as previously described.23,28 Transposed DNA libraries were sequenced on an HiSeq 4000. Quality of the paired-end 50–base pair reads was assessed using fastqc version 0.11.8, and reads were then aligned to the mm10 reference genome using Bowtie2 version 2.3.5 with the parameters --very-sensitive -X 2000 --no-mixed --no-discordant.29 Polymerase chain reaction duplicate reads were removed using Picard MarkDuplicates version 2.20.1 (http://broadinstitute.github.io/picard). The reads were filtered using SAMtools version 1.9 for a minimum mapping quality of 10 and only properly paired reads were retained.30 In addition, any reads aligning to the mitochondrial chromosome were removed. Peaks were called subsequently using MACS2 version 2.1.1.20160309 with the parameters --keep-dup all -f BAMPE -q 0.05, while simultaneously generating bedgraph files in reads-per-million scale.31 Peaks were filtered against the ENCODE blacklist regions (https://sites.google.com/site/anshulkundaje/projects/blacklists).32 The bedgraph files were converted to BigWig file format using the bedGraphToBigWig version 3774 utility.33 Differential binding analysis was performed using DESeq2 version 1.28.1 through the DiffBind version 2.16.2 bioconductor package with an false discovery rate cut-off of 0.01.25,34 Heat maps were generated using the deepTools version 3.5.1 package.35 Peaks were annotated using homer annotatePeaks.pl script.36 Genomic profile plots for ATAC-seq signals were generated using fluff version 3.0.2.37
Statistical analysis
Statistical analyses for all mice experiments were performed using GraphPad Prism 8 software.
Results
Effects of germ line and conditional deletion of Arid1b in mice
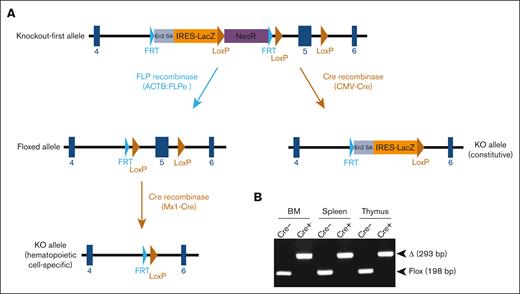
Mice with germ line transmission of the Arid1b KOF allele were obtained by microinjecting targeted ES cells into blastocysts. Heterozygous KOF mice were crossed with mice expressing Cre-recombinase to excise exon 5 of Arid1b gene, which results in an out-of-frame splicing between exons 4 and 6, thereby, leading to a constitutive KO allele (Figure 1A). In an intercross of heterozygous KO mice, the frequency of homozygous KO offspring was extremely low at weaning (1.1% instead of the expected 25.0%; 1/93 genotyped offspring) (Table 1), suggesting that constitutive depletion of Arid1b in mice was lethal, either during embryonic development or shortly postpartum. This necessitated generating a conditional KO allele of Arid1b, which was achieved by Flp recombinase–mediated excision of β-galactosidase and neomycin resistance genes in Arid1bKOF mice. This resulted in the excision of the gene-trap cassette flanked by FRT sites, leaving Arid1b exon 5 flanked by 2 LoxP sites (floxed allele; Figure 1A). Further crossing of mice carrying the floxed allele with mice expressing interferon-inducible Mx1-Cre transgene and induction of Cre-recombinase using poly(I:C) injection in Arid1bfl/fl;Mx1-Cre+ mice led to the excision of Arid1b exon 5 in a hematopoietic cell–specific manner (KO allele; Figure 1A-B).
Generation of mice deficient in ARID1B. (A) Mice harboring KOF Arid1b allele were initially produced and subsequently crossed with transgenic mice expressing either FLP recombinase or Cre-recombinase as depicted in the schematic. Mice with floxed Arid1b allele were injected with poly(I:C) to induce its deletion in a hematopoietic system–specific manner. (B) Polymerase chain reaction analysis to detect deletion of Arid1b alleles (exon 5) in BM leukocytes, splenocytes, and thymocytes of Arid1bf/f;Mx1-Cre+ and Arid1bf/f;Mx1-Cre− mice 16 weeks after poly(I:C) injection. Δ, deleted allele.
Generation of mice deficient in ARID1B. (A) Mice harboring KOF Arid1b allele were initially produced and subsequently crossed with transgenic mice expressing either FLP recombinase or Cre-recombinase as depicted in the schematic. Mice with floxed Arid1b allele were injected with poly(I:C) to induce its deletion in a hematopoietic system–specific manner. (B) Polymerase chain reaction analysis to detect deletion of Arid1b alleles (exon 5) in BM leukocytes, splenocytes, and thymocytes of Arid1bf/f;Mx1-Cre+ and Arid1bf/f;Mx1-Cre− mice 16 weeks after poly(I:C) injection. Δ, deleted allele.
Number and frequency of pups of different genotypes obtained in intercrosses of heterozygous Arid1b-KO mice
| . | Expected frequency, % . | Number of offspring obtained . | Frequency of offspring obtained, % . |
|---|---|---|---|
| WT | 25.0 | 37 | 39.8 |
| Heterozygous KO | 50.0 | 55 | 59.1 |
| Homozygous KO | 25.0 | 1 | 1.1 |
| . | Expected frequency, % . | Number of offspring obtained . | Frequency of offspring obtained, % . |
|---|---|---|---|
| WT | 25.0 | 37 | 39.8 |
| Heterozygous KO | 50.0 | 55 | 59.1 |
| Homozygous KO | 25.0 | 1 | 1.1 |
Hematopoietic cell–specific deletion of Arid1b in adult mice marginally disrupts steady-state hematopoiesis
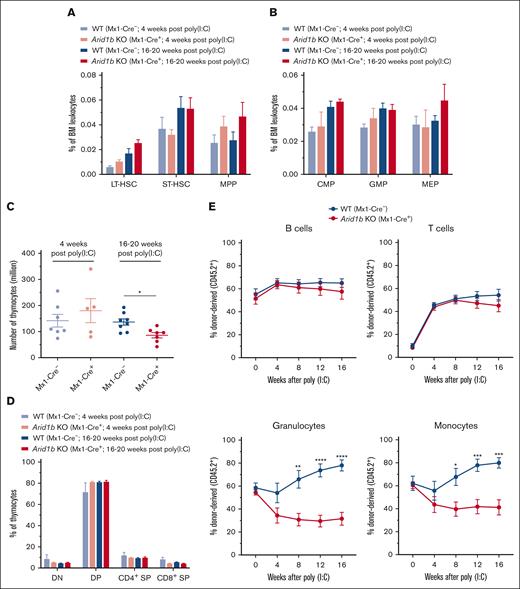
We initially assessed the frequency of HSCs in Arid1bfl/fl;Mx1-Cre+ and Arid1bfl/fl;Mx1-Cre− mice 4 weeks after administration of poly(I:C). This time point was chosen because we had previously identified extensive hematopoietic defects in conditional Arid1a-deficient (Arid1afl/fl;Mx1-Cre+) mice after 4 weeks of poly(I:C) injection.23 At 4 weeks after poly(I:C) injection, depletion of Arid1b did not significantly affect either total BM cellularity or the frequencies of Lin−Sca1+Kit+ (LSK) cells as well as HSC subsets contained within the LSK population, although a trend toward increased long-term (LT)-HSCs (CD34−Flt3− LSK) and multipotent progenitors (MPPs) (CD34+Flt3+ LSK) populations was observed in ARID1B-deficient mice (Figure 2A; supplemental Figures 1A-B and 2A). The frequencies of major myeloid precursors, common myeloid progenitors (CMPs; Lin−Kit+Sca1−CD34+FcγRII/IIIlo), GMPs (Lin−Kit+Sca1−CD34+FcγRII/IIIhi), and megakaryocyte erythrocyte progenitors (MEPs; Lin−Kit+Sca1−CD34−FcγRII/III−) were also largely unchanged in Arid1b KO mice (Figure 2B; supplemental Figure 2B). To preclude that 4-week duration was inadequate to manifest the effect of loss of ARID1B, we assessed hematopoietic development at 16 to 20 weeks after administration of poly(I:C). However, no significant difference was evident in BM cellularity and frequencies of HSC subsets and myeloid precursor in Arid1b KO mice even after this prolonged duration, albeit a trend toward increased frequency of LT-HSCs and MPPs was evident in Arid1bfl/fl;Mx1-Cre+ mice at 16 to 20 weeks after administration of poly(I:C) (Figure 2A-B; supplemental Figures 1A-B and 2A-B). In addition, terminal granulopoiesis in the BM, defined by surface expression of CD11b and Gr1, was unaltered in the absence of ARID1B at both time points (supplemental Figure 1C). Furthermore, our immunophenotypic analyses showed that the development of both erythroid and B-cell lineages was unaffected by loss of ARID1B at both 4 weeks and 16 to 20 weeks after poly(I:C) injection (supplemental Figure 1D-E). The consequences of loss of ARID1B on T-cell development were also assessed. Although thymus cellularity was not altered at 4 weeks after poly(I:C) injection, a significant reduction in the number of thymocytes was found at 16 weeks after Arid1b deletion (P < .05; Figure 2C). Despite the reduced thymus size, the frequencies of major thymocyte subsets, CD4−CD8− double-negative, CD4+CD8+ double-positive, and CD4+ and CD8+ single-positive cells, were still maintained (Figure 2D; supplemental Figure 2C). Given the largely normal hematopoietic development observed in Arid1bfl/fl;Mx1-Cre+ mice even 16 to 20 weeks after inducing recombination of the Arid1b allele, total splenocyte counts and proportion of major blood lineages in the spleen were also unaltered (supplemental Figure 1F-G). This apparent lack of a major hematopoietic phenotype was not a result of an incomplete deletion of Arid1b exon 5, because efficient Mx1-Cre–mediated recombination was confirmed in the BM, spleen, and thymus of Cre+ mice (Figure 1B).
Loss of ARID1B affects reconstitution of myeloid cells in competitive repopulation assays. (A) Frequencies of LT-HSC (CD34−Flt3− LSK), short-term (ST)-HSC (CD34+Flt3− LSK), and MPP (CD34+Flt3+ LSK) populations observed in the BM of Arid1bfl/fl;Mx1-Cre+ and Arid1bfl/fl;Mx1-Cre− mice, either 4 or 16 to 20 weeks after poly(I:C) injection; 4 weeks: n = 4 for control, and n = 3 for KO mice; 16 to 20 weeks: n = 5 for control, and n = 4 for KO mice. (B) Percentages of myeloid precursors, common myeloid progenitors (CMPs), granulocyte monocyte progenitors (GMPs), and megakaryocyte erythrocyte progenitors (MEPs), in the BM of Arid1bfl/fl;Mx1-Cre+ and Arid1bfl/fl;Mx1-Cre− mice after poly(I:C) injection; 4 weeks: n = 4 for control, and n = 2 for KO mice; 16 to 20 weeks: n = 6 for control, and n = 5 for KO mice. (C) Total thymocyte count in ARID1B-deficent or control mice at either 4 or 16 to 20 weeks after poly(I:C)-mediated deletion of Arid1b. (D) Proportion of double-positive (DP), double-negative (DN), CD4+ single-positive (SP), and CD8+ SP populations in Arid1bfl/fl;Mx1-Cre+ and Arid1bfl/fl;Mx1-Cre− mice, either 4 or 16 to 20 weeks after poly(I:C) injection; 4 weeks: n = 7 for control, and n = 5 for KO mice; 16 to 20 weeks: n = 8 for control, and n = 7 for KO mice. (E) Donor chimerism (CD45.2+) in the peripheral blood of recipient mice in competitive repopulation assays. Lethally irradiated recipients received transplantation with either Arid1bfl/fl;Mx1-Cre+ or Arid1bfl/fl;Mx1-Cre− BM mixed in equal proportion with competitor BM (CD45.1+) and deletion of Arid1b was induced 4 weeks after transplantation using poly(I:C). Proportion of donor-derived B cells, T cells, granulocytes, and monocytes were determined every 4 weeks in the peripheral blood of recipient mice (n = 7 for Arid1bfl/fl;Mx1-Cre− and n = 8 for Arid1bfl/fl;Mx1-Cre+). Data are represented as mean ± standard error of the mean (SEM). Statistical difference between WT and KO mice was calculated using unpaired t test and is denoted as ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
Loss of ARID1B affects reconstitution of myeloid cells in competitive repopulation assays. (A) Frequencies of LT-HSC (CD34−Flt3− LSK), short-term (ST)-HSC (CD34+Flt3− LSK), and MPP (CD34+Flt3+ LSK) populations observed in the BM of Arid1bfl/fl;Mx1-Cre+ and Arid1bfl/fl;Mx1-Cre− mice, either 4 or 16 to 20 weeks after poly(I:C) injection; 4 weeks: n = 4 for control, and n = 3 for KO mice; 16 to 20 weeks: n = 5 for control, and n = 4 for KO mice. (B) Percentages of myeloid precursors, common myeloid progenitors (CMPs), granulocyte monocyte progenitors (GMPs), and megakaryocyte erythrocyte progenitors (MEPs), in the BM of Arid1bfl/fl;Mx1-Cre+ and Arid1bfl/fl;Mx1-Cre− mice after poly(I:C) injection; 4 weeks: n = 4 for control, and n = 2 for KO mice; 16 to 20 weeks: n = 6 for control, and n = 5 for KO mice. (C) Total thymocyte count in ARID1B-deficent or control mice at either 4 or 16 to 20 weeks after poly(I:C)-mediated deletion of Arid1b. (D) Proportion of double-positive (DP), double-negative (DN), CD4+ single-positive (SP), and CD8+ SP populations in Arid1bfl/fl;Mx1-Cre+ and Arid1bfl/fl;Mx1-Cre− mice, either 4 or 16 to 20 weeks after poly(I:C) injection; 4 weeks: n = 7 for control, and n = 5 for KO mice; 16 to 20 weeks: n = 8 for control, and n = 7 for KO mice. (E) Donor chimerism (CD45.2+) in the peripheral blood of recipient mice in competitive repopulation assays. Lethally irradiated recipients received transplantation with either Arid1bfl/fl;Mx1-Cre+ or Arid1bfl/fl;Mx1-Cre− BM mixed in equal proportion with competitor BM (CD45.1+) and deletion of Arid1b was induced 4 weeks after transplantation using poly(I:C). Proportion of donor-derived B cells, T cells, granulocytes, and monocytes were determined every 4 weeks in the peripheral blood of recipient mice (n = 7 for Arid1bfl/fl;Mx1-Cre− and n = 8 for Arid1bfl/fl;Mx1-Cre+). Data are represented as mean ± standard error of the mean (SEM). Statistical difference between WT and KO mice was calculated using unpaired t test and is denoted as ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
ARID1B deficiency impairs repopulation of myeloid cells
Although our comprehensive phenotypic analyses demonstrated that depletion of ARID1B in hematopoietic cells of adult mice did not notably affect steady-state hematopoiesis, we went ahead and evaluated the ability of ARID1B-deficient HSCs to reestablish multilineage hematopoietic cell reconstitution in lethally irradiated mice, either in a competitive or noncompetitive setting. ARID1B deletion was induced after the establishment of BM chimeras from either Arid1bfl/fl;Mx1-Cre+ or Arid1bfl/fl;Mx1-Cre− mice. We detected a similar engraftment ability of BM cells from both genotypes 4 weeks after transplantation, after which poly(I:C) was administered to recipient mice to induce deletion of Arid1b. In competitive repopulation assays, Arid1b KO HSCs reconstituted lymphoid lineages (B and T cells) in recipient mice as efficiently as WT cells at all the time points tested; however, the repopulation of myeloid lineages (both granulocytes and monocytes) was significantly impaired compared with the WT HSCs; granulocytes: P < .01 at 8 weeks and P < .0001 at both 12 and 16 weeks after poly(I:C); monocytes: P < .05 at 8 weeks and P < .001 at both 12 and 16 weeks after poly(I:C) (Figure 2E; supplemental Figure 3).
Furthermore, in noncompetitive repopulation assays, Arid1b deletion was induced 4 weeks after transplantation of the BM. Loss of ARID1B did not affect the peripheral blood cell counts in recipient mice up to 16 weeks after poly(I:C) injection (supplemental Figure 4). Collectively, these data indicate that ARID1B contributes to myeloid development in vivo, an effect manifested only under competitive repopulation stress in lethally irradiated mice.
ARID1B regulates expression and chromatin accessibility of genes critical for hematopoiesis
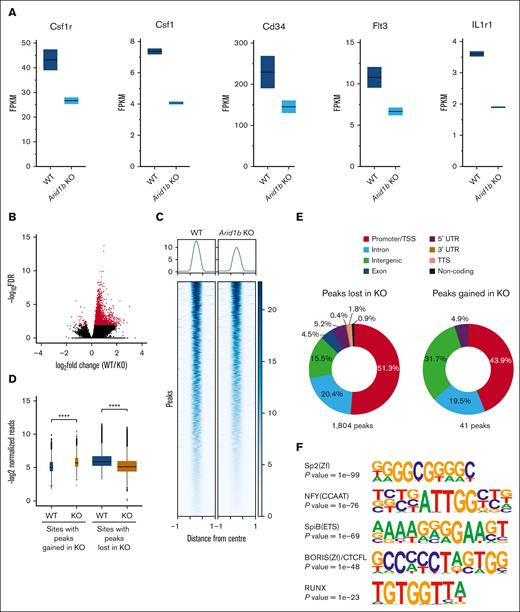
To gain molecular insights into the function of ARID1B in the myeloid compartment, we did RNA sequencing on sorted GMPs from mice administered with poly(I:C) for 16 weeks. In total, 85 genes were differentially expressed between WT and Arid1b KO GMP cells, with 67 genes downregulated and 18 genes upregulated in cells lacking ARID1B (supplemental Figure 5A; supplemental Table 2). Notably, the consequences of ARID1B loss on gene expression were less evident compared with differentially expressed genes observed upon deficiency of ARID1A, as we have previously reported.23 Nonetheless, the expression of key genes essential in hematopoietic development, Csf1r, Csf1, Cd34, Flt3, and Il1r1, were suppressed upon loss of ARID1B (Figure 3A). Interestingly, binding of BRG1, a catalytic component of the SWI/SNF complex, was evident at all these gene loci in chromatin immunoprecipitation sequencing (ChIP-seq) of both RN2 cells and murine macrophages38-40 (supplemental Figure 5B), suggesting their direct regulation by SWI/SNF complex. Interestingly, binding of ARID1B was also observed at 4 of these 5 genes in human (K562 ChIP-seq) cells32,38,39 (supplemental Figure 5C). Furthermore, using Enrichr,41-43 we noticed binding of well-known myeloid transcription factors at genomic loci of the downregulated genes (supplemental Figure 5D), suggesting their coregulation by myeloid transcription factors and ARID1B-containing SWI/SNF complex. Moreover, gene set enrichment analysis (GSEA) revealed that gene sets associated with hematopoiesis, myeloid development, cell cycle, as well as inflammatory response were enriched while comparing Arid1b KO GMPs with the WT GMPs (supplemental Figure 6; supplemental Table 3).
Loss of ARID1B perturbs expression and chromatin accessibility in murine hematopoietic stem and progenitor cells. (A) Expression levels of key genes involved in hematopoiesis (Csf1r, Csf1, Cd34, Flt3, and Il1r1) in WT and Arid1b KO GMPs. (B) ATAC-seq peaks identified in WT and Arid1b−/− Lin−Kit+ BM cells. Peaks that are enriched significantly (false discovery rate of <0.01) in either genotype are depicted in red. (C) Heat map shows ATAC-seq peaks (±1 kilo base) identified using DiffBind as significantly closed in ARID1B-deficient cells. (D) Box plots show normalized read counts in the open chromatin regions enriched in either WT or KO cells. The width of the box plots is an indicator of the number of enriched sites for each group. Statistical significance was calculated using Wilcoxon rank-sum test. (E) Distribution of ATAC-seq peaks either lost or gained in Arid1b KO Lin−Kit+ BM cells. (F) Top 5 Homer de novo motifs enriched in genomic loci with significantly reduced ATAC-seq signal in Arid1b KO cells. A hypergeometric test was used to calculate the statistical significance of the enrichment of the identified motif in the input sequences compared with a background model. TTS; transcriptional termination site; UTR, untranslated region.
Loss of ARID1B perturbs expression and chromatin accessibility in murine hematopoietic stem and progenitor cells. (A) Expression levels of key genes involved in hematopoiesis (Csf1r, Csf1, Cd34, Flt3, and Il1r1) in WT and Arid1b KO GMPs. (B) ATAC-seq peaks identified in WT and Arid1b−/− Lin−Kit+ BM cells. Peaks that are enriched significantly (false discovery rate of <0.01) in either genotype are depicted in red. (C) Heat map shows ATAC-seq peaks (±1 kilo base) identified using DiffBind as significantly closed in ARID1B-deficient cells. (D) Box plots show normalized read counts in the open chromatin regions enriched in either WT or KO cells. The width of the box plots is an indicator of the number of enriched sites for each group. Statistical significance was calculated using Wilcoxon rank-sum test. (E) Distribution of ATAC-seq peaks either lost or gained in Arid1b KO Lin−Kit+ BM cells. (F) Top 5 Homer de novo motifs enriched in genomic loci with significantly reduced ATAC-seq signal in Arid1b KO cells. A hypergeometric test was used to calculate the statistical significance of the enrichment of the identified motif in the input sequences compared with a background model. TTS; transcriptional termination site; UTR, untranslated region.
Given that ARID1B is a core component of the chromatin remodeling BAF complex, we performed ATAC-seq on sorted Lin−Kit+ myeloid BM precursors from poly(I:C)-treated Arid1blfl/fl;Mx1-Cre+ and Arid1bfl/fl;Mx1-Cre− mice. We observed high concordance among the ATAC-seq peaks obtained in the 2 replicates of each genotype (supplemental Figure 7A). As reported previously,23 the majority of ATAC-seq peaks were localized at intergenic and intronic regions with ∼15% of peaks present around transcriptional start sites (TSS)/promoter regions in both WT and KO cells (supplemental Figure 7B).
Loss of ARID1B caused an overall decrease in open chromatin with 1804 ATAC-seq peaks lost in the KO cells whereas only 41 peaks were gained compared with the WT cells (Figure 3B-D; supplemental Table 4). Approximately half of the regions with reduced chromatin accessibility corresponded with TSS/promoter sites, with intronic and intergenic regions also affected in cells deficient in ARID1B (Figure 3E). This indicates targeted loss of promoter accessibility in cells lacking ARID1B. Some examples of loss of open chromatin in Arid1b KO cells at TSS/promoters and other genomic locations are shown in supplemental Figure 8. Gene Ontology analysis of genes with lost ATAC-seq peaks in Arid1b KO cells revealed their involvement in a wide variety of biological processes (supplemental Figure 9). Importantly, motif search of ATAC-seq peaks lost in Arid1b KO cells identified enrichment of binding sites for RUNX, ETS family, and other transcription factors crucial in hematopoiesis (Figure 3F).
Dual loss of ARID1A and ARID1B in hematopoietic compartment is lethal in mice
We have previously demonstrated that ARID1A deficiency results in multiple aberrations in steady-state hematopoiesis and that multilineage reconstitution ability of Arid1a KO HSCs is severely impaired.23 This study shows that its close homolog, ARID1B, instead, plays a limited role in hematopoiesis, and that Arid1b KO HSCs have defective reconstitution of myeloid cells. Given that ARID1A and ARID1B are mutually exclusive subunits of the BAF complex, the presence of at least 1 of the homologs is understood to be essential for its chromatin remodeling activity.
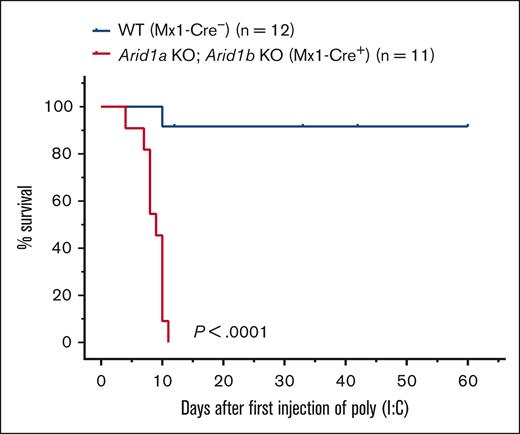
To understand the consequences of complete loss of BAF complex–mediated chromatin remodeling activity in hematopoietic cells, we generated mice deficient in both ARID proteins. Mice harboring a floxed allele of Arid1a and Arid1b along with an Mx1-Cre transgene were generated, and complete deletion of ARID1A and ARID1B was achieved using either 3 or 5 injections of poly(I:C) in Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+ mice whereas Arid1afl/fl;Arid1bfl/fl;Mx1-Cre− mice were used as controls. All 11 Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+ mice died within 11 days of first injection of poly(I:C), whereas only 1 of 12 control mice died after administration of poly(I:C) (P < .0001; Figure 4). This demonstrated that complete loss of BAF complex–mediated chromatin remodeling activity in hematopoietic cells cannot be tolerated in mice.
Acute deletion of both ARID1A and ARID1B in hematopoietic compartment in adult mice leads to rapid mortality. Kaplan-Meier curves show survival of double-KO (Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+) and control (Arid1afl/fl;Arid1bfl/fl;Mx1-Cre−) mice. Mice were administered poly(I:C) to induce recombination of Arid1a and Arid1b alleles in Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+ mice. Study Day 0 is the day of administration of first dose of poly(I:C). P < .0001 calculated using log-rank (Mantel-Cox) test.
Acute deletion of both ARID1A and ARID1B in hematopoietic compartment in adult mice leads to rapid mortality. Kaplan-Meier curves show survival of double-KO (Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+) and control (Arid1afl/fl;Arid1bfl/fl;Mx1-Cre−) mice. Mice were administered poly(I:C) to induce recombination of Arid1a and Arid1b alleles in Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+ mice. Study Day 0 is the day of administration of first dose of poly(I:C). P < .0001 calculated using log-rank (Mantel-Cox) test.
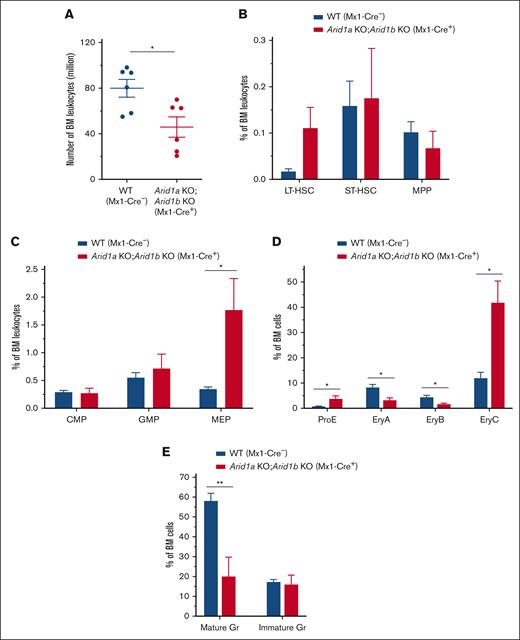
To understand how hematopoiesis was affected after acute loss of both ARID1A and ARID1B in adult mice, we assessed the BM 2 to 4 days after third injection of poly(I:C) in Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+ mice compared with that in Arid1a+/fl;Arid1bfl/fl;Mx1-Cre− or Arid1afl/fl;Arid1bfl/fl;Mx1-Cre− mice used as controls. Concurrent depletion of both ARID1A and ARID1B caused a rapid loss of BM cellularity (P < .05) with a trend toward increased frequency and numbers of LT-HSCs, whereas the number of MPPs was significantly reduced in the double-KO mice (P < .01; Figure 5A-B; supplemental Figures 10A and 11A). In addition, a significant increase in the frequency and number of the -MEP population (P < .05) and a decrease in number of CMPs (P < .05) was observed in the BM of Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+ mice (Figure 5C; supplemental Figures 10B and 11B). Arid1a and Arid1b double-deficient mice were also characterized by impaired erythroid and myeloid differentiation (Figure 5D-E; supplemental Figures 10C-D and 11C-D). This rapid hematopoietic degeneration likely leads to mortality, suggesting that dual loss of ARID1A and ARID1B cannot be tolerated in mice. Although loss of ARID1A alone greatly impairs hematopoietic differentiation and stem cell function,23 presence of ARID1B can sustain the vital chromatin remodeling activity; however, further depletion of ARID1B is likely detrimental to BAF–mediated chromatin remodeling.
Dual loss of ARID1A and ARID1B in hematopoietic compartment results in swift failure of hematopoietic development in mice. (A) BM cellularity in ARID1A and ARID1B double-deficient mice enumerated 2 to 4 days after third injection of poly(I:C). Cell counts represent number of leukocytes obtained from 2 femurs and 2 tibias for each mouse. (B) Percentages of HSC subsets, LT-HSCs, ST-HSCs, and MPP cells, in the BM of Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+ and control mice analyzed at 2 to 4 days after third injection of poly(I:C); n = 5 for control, and n = 5 for double-KO mice. (C-D) Frequencies of myeloid (C) and erythroid precursors (D) in flow cytometric evaluation of the BM of ARID1A and ARID1B double-deficient and WT mice assessed at 2 to 4 days after third injection of poly(I:C); proE, CD71+TER119lo; EryA, CD71+TER119+FSChi; EryB, CD71+TER119+FSClo; and EryC, CD71−TER119+FSClo; n = 5 for control, and n = 5 for double-KO mice. (E) Percentages of mature (CD11b+Gr1hi) and immature (CD11b+Gr1med) granulocytes in the BM of WT and Arid1b KO mice; n = 5 for control, and n = 5 for double-KO mice. Control animals in these experiments comprised of either Arid1a+/fl;Arid1bfl/fl;Mx1-Cre− or Arid1afl/fl;Arid1bfl/fl;Mx1-Cre− mice. Data are represented as mean ± SEM. Statistical difference between WT and KO mice was calculated using unpaired t test and is denoted as ∗P < .05.
Dual loss of ARID1A and ARID1B in hematopoietic compartment results in swift failure of hematopoietic development in mice. (A) BM cellularity in ARID1A and ARID1B double-deficient mice enumerated 2 to 4 days after third injection of poly(I:C). Cell counts represent number of leukocytes obtained from 2 femurs and 2 tibias for each mouse. (B) Percentages of HSC subsets, LT-HSCs, ST-HSCs, and MPP cells, in the BM of Arid1afl/fl;Arid1bfl/fl;Mx1-Cre+ and control mice analyzed at 2 to 4 days after third injection of poly(I:C); n = 5 for control, and n = 5 for double-KO mice. (C-D) Frequencies of myeloid (C) and erythroid precursors (D) in flow cytometric evaluation of the BM of ARID1A and ARID1B double-deficient and WT mice assessed at 2 to 4 days after third injection of poly(I:C); proE, CD71+TER119lo; EryA, CD71+TER119+FSChi; EryB, CD71+TER119+FSClo; and EryC, CD71−TER119+FSClo; n = 5 for control, and n = 5 for double-KO mice. (E) Percentages of mature (CD11b+Gr1hi) and immature (CD11b+Gr1med) granulocytes in the BM of WT and Arid1b KO mice; n = 5 for control, and n = 5 for double-KO mice. Control animals in these experiments comprised of either Arid1a+/fl;Arid1bfl/fl;Mx1-Cre− or Arid1afl/fl;Arid1bfl/fl;Mx1-Cre− mice. Data are represented as mean ± SEM. Statistical difference between WT and KO mice was calculated using unpaired t test and is denoted as ∗P < .05.
ARID1A and ARID1B regulate chromatin accessibility at largely distinct loci
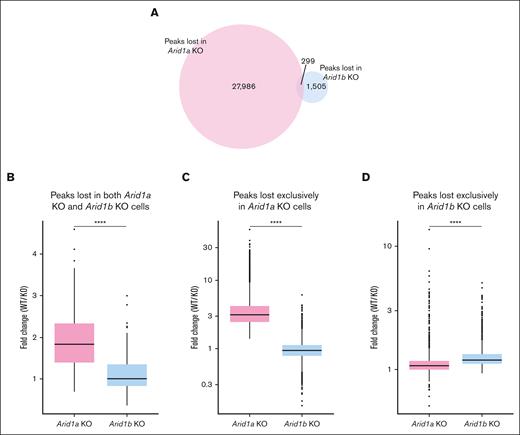
Next, we assessed how the 2 paralogs, ARID1A and ARID1B, differ in maintaining the chromatin structure in murine hematopoietic stem and progenitor cells by comparing the ATAC-seq peaks lost in Arid1b-KO cells (this study) vs those lost in Arid1a-KO cells.23 We noted that the number of peaks lost in Arid1a-deficient cells (28 285) was notably higher compared with those lost in cells lacking Arid1b (1804) (Figure 6A). This was not caused by any technical or procedural differences because the total number of peaks called in both data sets were similar (76 147 and 68 379 in Arid1a KO + WT data set and Arid1b KO + WT data set, respectively). Moreover, a high proportion of ATAC-seq peaks (62 204) overlapped between the 2 data sets (supplemental Figure 12A). Among those loci with significantly closed chromatin states compared with WT cells, only 299 ATAC-seq peaks were common between cells lacking ARID1A and those lacking ARID1B (Figure 6A). These findings demonstrate that ARID1A and ARID1B regulate chromatin architecture at largely nonoverlapping loci. Notably, the decrease in peak intensity was more pronounced in Arid1a-KO cells compared with Arid1b-KO cells (Figure 6B-D; supplemental Figures 12B and 13). Interestingly, despite low overlap of lost ATAC-seq peaks between Arid1a-KO and Arid1b-KO cells, >50% of genes assigned to lost peaks in Arid1b-KO cells were also identified among lost peaks in Arid1a-KO cells (supplemental Figure 12C). Overall, the ATAC-seq data revealed that, among the 2 paralogs, ARID1A plays a more dominant role in maintaining chromatin dynamics in murine hematopoietic cells.
ARID1B deficiency affects chromatin structure less notably compared with loss of ARID1A. (A) Venn diagram shows number of distinct and overlapping ATAC-seq peaks lost in Arid1a-KO and Arid1b-KO cells. (B) Fold change (WT vs KO) in normalized read counts for ATAC-seq peaks significantly lost in both Arid1a-KO and Arid1b-KO cells. (C-D) Box plots depict fold change (WT vs KO) in normalized signal values for ATAC-seq peaks significantly lost exclusively in either (C) Arid1a-KO cells or (D) Arid1b-KO cells. Statistical significance was calculated using Wilcoxon rank-sum test.
ARID1B deficiency affects chromatin structure less notably compared with loss of ARID1A. (A) Venn diagram shows number of distinct and overlapping ATAC-seq peaks lost in Arid1a-KO and Arid1b-KO cells. (B) Fold change (WT vs KO) in normalized read counts for ATAC-seq peaks significantly lost in both Arid1a-KO and Arid1b-KO cells. (C-D) Box plots depict fold change (WT vs KO) in normalized signal values for ATAC-seq peaks significantly lost exclusively in either (C) Arid1a-KO cells or (D) Arid1b-KO cells. Statistical significance was calculated using Wilcoxon rank-sum test.
Discussion
Chromatin remodeling mediated by the SWI/SNF complex is essential for hematopoietic development. Previous studies using genetically engineered mice have elucidated the indispensable role played by several components of the SWI/SNF complex.44-53 We, and others, have demonstrated that ARID1A is crucial for both embryonic and adult hematopoiesis.23,54 In this study, using a conditional KO allele of Arid1b, we show that the loss of ARID1B in the hematopoietic compartment is well tolerated compared with loss of its paralog, ARID1A. The only discernible effect observed upon deletion of Arid1b in adult mice, with established hematopoiesis, was significantly reduced thymus cellularity 16 to 20 weeks after poly(I:C) administration. Despite overall lower thymocyte counts, the frequencies of populations that define T-cell maturation stages were maintained in the absence of ARID1B. This indicates potential paucity in seeding of the thymus with BM HSC–derived progenitors in mice lacking ARID1B, and further studies are needed to discern this hypothesis. One limitation of our study is that the deletion of the Arid1b allele is dependent on the interferon-inducible Mx1-Cre deleter strain, which necessitates poly(I:C) injections. Although this inducible system is widely used in experimental hematology, leakiness of Cre-recombinase activity and poly(I:C)-induced transient changes in cellular phenotype have been reported.55 The experimental time points (≥4 weeks) and appropriate controls (simultaneous poly(I:C) injection in Mx1-Cre− mice) ensured that we avoided these potential pitfalls of the Mx1-Cre recombination system.
An overall maintenance of steady-state hematopoiesis in mice lacking ARID1B suggests that ARID1A can potentially compensate for ARID1B loss during hematopoietic development. However, in BM repopulation assays, ARID1B acts in a cell-autonomous manner and its deficiency leads to impaired reconstitution of myeloid cells whereas lymphoid lineage was not affected. This suggests that ARID1B has nonoverlapping, albeit limited, roles in hematopoietic differentiation compared with ARID1A. Similarly, hematopoietic cell–specific depletion of the Polybromo component, Arid2, did not affect steady-state hematopoiesis in mice but reduced lymphoid cell reconstitution in BM transplantation assays.44 Despite its limited function in murine hematopoiesis, ARID1B is essential to maintain homeostasis because Mx1-Cre–mediated depletion of both ARID1A and ARID1B but not ARID1A alone leads to rapid mortality in mice.23
A more pronounced role of ARID1A in hematopoiesis is supported by its prominent role in maintaining chromatin architecture compared with ARID1B. Our ATAC-seq data revealed that loss of ARID1A extensively impacts chromatin accessibility whereas ARID1B deficiency has a modest effect on chromatin structure in Lin−Kit+ BM cells. ATAC-seq experiments performed on WT and Arid1b-KO Lin−Kit+ BM cells facilitated direct comparison of chromatin organization with ARID1A-deficient Lin−Kit+ BM cells.23 However, because the RNA-seq was performed on GMP cells (a subset of Lin−Kit+ BM cells), a caveat of our study is that the gene expression changes caused by ARID1B deficiency cannot be precisely correlated with altered chromatin state specifically in the GMP cells.
Wang et al have shown using ChIP-seq that ARID1B occupies only a subset of ARID1A binding sites in a murine hepatocyte cell line, and its loss affected expression of fewer genes compared with loss of ARID1A in murine liver, which explained a more prominent role of ARID1A in transcriptional control.56 Similarly, ATAC-seq of colorectal cells revealed a dominant role of ARID1A compared with ARID1B in maintaining chromatin accessibility at enhancers.57 The phenotype observed in the hematopoietic compartment in our studies with Arid1a- and Arid1b-KO mice is suggestive of a similar predominance of ARID1A over ARID1B in regulating chromatin architecture in hematopoietic cells. The disparity between the function of ARID1A and ARID1B in hematopoietic development may also be explained by generally higher expression levels of ARID1A in the hematopoietic progenitor and mature cell types (supplemental Figure 14).
In the absence of ARID1A, ARID1B is likely essential for either the formation or stabilization of an intact BAF complex. Indeed, ARID1B was shown to be essential for survival of ARID1A-mutant cancer cell lines, and this paralog dependency suggests that ARID1B is a synthetic lethal target in ARID1A-mutant cancers.58,59
The differing phenotypes arising from the loss of either ARID1A or ARID1B can be attributed to the functionally distinct complexes that these 2 proteins form. Also, differing kinetics of expression of ARID1A and ARID1B in the cell cycle has been reported.60 Furthermore, a study showed that ARID1A-containing but not ARID1B-containing SWI/SNF complexes are crucial for normal cell cycle arrest in differentiating cells exiting the cell cycle.61 The study also showed that although loss of ARID1A in a preosteoblast cell line led to failure to repress E2F target genes, depletion of ARID1B did not affect E2F-responsive genes during osteoblast differentiation.61 A more significant role of ARID1B in a different tissue context can be envisioned. For instance, ARID1B is key in human brain development and function, and its haploinsufficiency causes autism spectrum disorder and intellectual disability including Coffin-Siris syndrome.62-67Arid1b heterozygous KO mice display abnormal cognitive and social behaviors. Mice lacking one Arid1b allele exhibit impaired spatial learning, recognition memory, and reference memory, thus mirroring behavioral characteristics of intellectual disability and autism.68-71
Although the loss of one paralog would favor the formation of BAF complexes containing the other ARID1 subunit, dual loss of both ARID1A and ARID1B is expected to extensively disrupt the chromatin remodeling function of BAF complexes. This is evident from our murine model in which acute depletion of both Arid1a and Arid1b in hematopoietic cells resulted in immediate failure of hematopoietic development, which likely led to lethality in mice. A previous study suggested that loss of both ARID1 paralogs can also potentially perturb the polybromo BAF complex, which is another class of SWI/SNF complexes, possibly because of the redistribution of residual BAF complexes.56 Concurrent inactivation of ARID1A and ARID1B is also associated with dedifferentiated endometrial carcinomas and their combined deletion in mice results in aggressive carcinogenesis.56,72
Taken together, this study uncovers how ARID1B deficiency alone or concurrent loss of both ARID1A and ARID1B affects hematopoietic development and chromatin dynamics in mice.
Acknowledgments
The authors thank the staff of Comparative Medicine, National University of Singapore for their support in mice maintenance and experiments. The authors also acknowledge expert help and support from the fluorescence-activated cell sorting (FACS) facility at Cancer Science Institute of Singapore (CSI), Singapore. The authors thank the Melamed Family and Reuben Yeroushalmi for their generous support.
This work was funded by the Leukemia and Lymphoma Society, the Singapore Ministry of Health’s National Medical Research Council (NMRC) under its Singapore Translational Research (STaR) Investigator Award to H.P.K. (NMRC/STaR/0021/2014), the NMRC Centre grant awarded to the National University Cancer Institute, Singapore (NMRC/CG/012/2013), National Research Foundation, Singapore, and National Medical Research Council, Singapore under its NMRC Centre Grant Programme (NMRC/CG/CG21Apr1005) and the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiatives.
Authorship
Contribution: V.M. conceived the study, designed and performed research, analyzed data, and wrote the manuscript; P.S. designed and performed research, analyzed data, and wrote the manuscript; P.D. performed bioinformatics and statistical analyses and wrote the manuscript; T.W.W., L.H., and Z.C. performed research, analyzed data, and performed genotyping; H.B.M.N. maintained mouse colonies and performed genotyping; S.J., Y.S., and M.Z.H. performed blastocyst injections to generate chimeras from targeted ES cells; H.P.K. conceived and supervised the study, interpreted the data, and wrote the manuscript; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pavithra Shyamsunder, Programme in Cancer and Stem Cell Biology, Duke-NUS Medical School, 8 College Rd, Singapore 169857; e-mail: pavithra.shyamsunder@duke-nus.edu.sg.
References
Author notes
∗V.M. and P.S. contributed equally to this study.
Data are available on request from the corresponding author, Pavithra Shyamsunder (pavithra.shyamsunder@duke-nus.edu.sg).
The full-text version of this article contains a data supplement.