Key Points
Zbtb11 is crucial for HSC function in establishing and maintaining early murine hematopoiesis.
Absence of Zbtb11 in the hematopoietic compartment leads to dysregulation of niche and oxidative phosphorylation genes.
Abstract
Hematopoiesis produces diverse blood cell lineages to meet the basal needs and sudden demands of injury or infection. A rapid response to such challenges requires the expansion of specific lineages and a prompt return to balanced steady-state levels, necessitating tightly coordinated regulation. Previously we identified a requirement for the zinc finger and broad complex, tramtrak, bric-a-brac domain–containing 11 (ZBTB11) transcription factor in definitive hematopoiesis using a forward genetic screen for zebrafish myeloid mutants. To understand its relevance to mammalian systems, we extended these studies to mice. When Zbtb11 was deleted in the hematopoietic compartment, embryos died at embryonic day (E) 18.5 with hematopoietic failure. Zbtb11 hematopoietic knockout (Zbtb11hKO) hematopoietic stem cells (HSCs) were overabundantly specified from E14.5 to E17.5 compared with those in controls. Overspecification was accompanied by loss of stemness, inability to differentiate into committed progenitors and mature lineages in the fetal liver, failure to seed fetal bone marrow, and total hematopoietic failure. The Zbtb11hKO HSCs did not proliferate in vitro and were constrained in cell cycle progression, demonstrating the cell-intrinsic role of Zbtb11 in proliferation and cell cycle regulation in mammalian HSCs. Single-cell RNA sequencing analysis identified that Zbtb11-deficient HSCs were underrepresented in an erythroid-primed subpopulation and showed downregulation of oxidative phosphorylation pathways and dysregulation of genes associated with the hematopoietic niche. We identified a cell-intrinsic requirement for Zbtb11-mediated gene regulatory networks in sustaining a pool of maturation-capable HSCs and progenitor cells.
Introduction
Hematopoietic stem cells (HSCs) are characterized by the dual hallmarks of stemness: producing daughter HSCs (self-renewal) and/or daughter progenitor cells (differentiation).1-4 Progenitors proliferate and differentiate in response to specific signals that engender different blood lineages.2 Under steady-state hematopoiesis, adult HSCs can remain dormant for long periods before responding to the growth/differentiation stimuli. In contrast, developing HSCs emerge from the embryonic aorta, proliferate to seed hematopoietic niches, and generate a balanced hematopoietic output before exiting the cell cycle and awaiting a stimulus for re-entry.5 HSCs have limited capacity for continued cell cycling and eventually succumb to replicative stress, resulting in exhaustion.5 All of these processes are tightly regulated by the measured interplay between cytokines, growth factors, adhesion molecules, and transcription factors.6,7
ZBTB11 (zinc finger and broad complex, tramtrak, bric-a-brac [BTB] domain–containing protein 11), originally identified as a repressor of metallothionein 2A,8 is a member of the ZBTB superfamily of transcription factors. These proteins regulate crucial biological processes, including proliferation, morphogenesis, apoptosis, protein degradation,9,10 and hematopoiesis.11-17 We previously showed that ZBTB11 is required for cell cycle progression in zebrafish,18 suggesting its potential involvement in oncogenesis.19 ZBTB11 also plays an emerging role in response to viruses.20-22 Clinically, ZBTB11 mutations have recently been associated with intellectual disability,23-26 and interest in ZBTB11 function is burgeoning, with multifaceted roles emerging recently.27-29
A key role for Zbtb11 as a hematopoietic regulator was first identified in the zebrafish zbtb11-deficient mutant marsanne.18 In marsanne, all blood lineages, including HSCs, are initially formed, but after switching to HSC-dependent hematopoiesis, multilineage development fails, accompanied by the loss of HSCs.18 This suggests an essential role for Zbtb11 in HSC maintenance and continued HSC longevity; however, it is not known whether this function is conserved in mammals. To assess the role of Zbtb11 in mammalian hematopoiesis and HSC maintenance, we generated Zbtb11fl/fl mice crossed with vav-iCre30 to delete Zbtb11 from the hematopoietic compartment (Zbtb11hKO). In Zbtb11hKO mice, HSCs are specified in abundance yet do not generate adequate numbers of lineage-committed progenitors or mature differentiated blood cells, leading to hematopoietic failure and death by E18.5. Investigation of Zbtb11 target genes led us to identify novel Zbtb11-mediated gene regulatory axes that are pivotal for balancing hematopoietic output.
Materials and methods
Animals
Vav-iCre (B6.Cg-Tg[Vav1-iCre]A2Kio/J) mice (The Jackson Laboratory, Bar Harbor, ME)31 were crossed with blue fluorescent protein (BFP) mice, and both sexes were bred at the Monash Animal Research Platform (Monash University, Clayton, VIC, Australia) using standard husbandry practices. Enhanced flippase (FLPe)-deleter mice were purchased from and crossed at the Monash Animal Research Platform. Experiments were performed according to the protocols approved by the relevant animal ethics and institute biosafety committees.
Zbtb11hKO mouse
Zbtb11hKO mice were generated in-house at the Gene Recombineering and Monash Gene Technology Facility. Targeting constructs were generated (supplemental Figure 1A), and 2 verified targeted clones were used to generate germ line-transmitting knockout lines by pronuclear injection into C57BL/6J blastocysts, according to standard protocols.32 Before analysis, the neomycin selection cassette was removed by crossing with a ubiquitous FLPe-deleter mouse line and mice with an excised neocassette were crossed as vav-iCre × BFP. Because no phenotypic difference was observed between heterozygotes and wild-type (WT), these were included together in the WT/control groups.
Flow cytometry
Timed matings and isolation of the fetal liver (FL), bone marrow (BM), spleen, and long bones have been described previously.33 FL or BM cells were immunolabeled with a lineage cocktail (anti-B220, anti-CD3, and anti-Gr1), anti–Sca-1, anti–c-Kit, anti-CD48, anti-CD150, anti-CD127, anti–Fms-like receptor tyrosine kinase 3 (anti-Flt3), anti-CD34, and anti-CD16/32 antibodies. Cells were analyzed as hematopoietic stem and progenitor cells (Lin−Sca-1+c-Kit+ [LSK]), HSC (Lin−Sca-1+c-Kit+CD150+CD48−), myeloerythroid progenitors (Lin−CD127−Sca-1−c-Kit+CD34−CD16/32−), granulocyte–macrophage progenitors (Lin−CD127−Sca-1−c-Kit+CD34+CD16/32+), common myeloid progenitors (CMPs; Lin−CD127−Sca-1−cKit+CD34+CD16/32−), common lymphoid progenitors (Lin−CD127+Sca-1+c-Kit+Flt3+), lymphoid-primed multipotent progenitors (LSKCD34+Flt3+), differentiated myeloid cells (Gr1/Mac-1+), and differentiated lymphoid cells (B220+ and CD3+). Flow cytometry was performed using an LSRII flow cytometer (BD Biosciences) as previously described.34 To assess the cell cycle, 1 to 2 ×106 FL cells were immunolabeled with an anti-Gr1 lineage cocktail and anti–Sca-1/-c-Kit, antibodies, washed, and then stained with anti-Ki67-BV786 (BD Biosciences) and Hoechst 33342 (20 μg/mL) using Fixation/Permeabilization Kit (Thermo Fisher Scientific) according to manufacturer’s instructions.
Transplantation homing assay
Sorted E16.5 FL HSC were labeled with the fluorescent tracking dye carboxy fluorescein diacetate succinimidyl ester (5(6)-carboxyfluorescein diacetate N-succinimidyl ester when incorporated; Molecular Probes, Eugene OR) for Zbtb11hKO and seminaphtorhodafluor-1-acetoxymethylester (Invitrogen) for controls to use as donor cells, as previously described.34 Briefly, a total of 4000 to 8000 donor cells were transplanted into each nonirradiated WT neonate recipient via intraperitoneal injection with an additional nonablated 200 000 whole-BM carrier cells. Transplanted cell numbers were standardized as follows: twice as many control vs Zbtb11hKO HSCs were injected to account for the doubling of FL HSC in Zbtb11hKO mice compared with that in controls. Homing efficiency of transplanted donor HSCs was assessed after 15.5 hours.34 Independent experiments were performed using cells from pooled Zbtb11hKO pups each time.
BM sections
E17.5 femurs were dissected,33 embedded in Tissue-Tek Optimal Cutting Temperature compound (ProSciTech), and sectioned on a cryostat. Sections were fixed and stained with hematoxylin and eosin, and slides were scanned using an Olympus dotSlide microscope to reconstruct a complete image.
Single-cell RNA sequencing
A total of 186 single E14.5 FL Zbtb11hKO and control HSCs were assayed on one 384-well plate using an adapted version of the CelSeq 2 protocol.35,36 Paired-end sequencing was performed on Illumina NextSeq500 with the following run parameters: read1 = 14nt, read2 = 78nt, and phiX = 2%. Expression was quantified by counting the number of unique molecular identifiers mapped to each gene using scPipe (https://www.bioconductor.org/packages/scPipe, Bioconductor). Cells were evenly split between the WT and Zbtb11hKO (n = 93 each). Cell cycle classification was implemented using the cyclone function of the scran R/Bioconductor package. The expression values were normalized and denoised using the scran::denoisePCA function. The output of preprocessing identified 181 cells and 15 069 genes. Seurat was used to perform unsupervised clustering. Clusters corresponding to cell types were identified using the Louvain algorithm in Seurat. Differential expression analyses were performed to ascertain marker genes in each cluster. Marker genes were used to find the top 10 gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathways in each cluster. Cluster-independent differential expression analysis was performed between samples using glmFit from edgeR to compare WT and Zbtb11hKO cells while blocking the estimated cell cycle phase (expanded in supplemental Methods).
RNAseq
Our RNA sequencing (RNAseq) data set comparing differential gene expression between zbtb11C116S and WT neutrophils was generated as previously described.18 It was remined using zebrafishmine.org (AllianceMine, https://www.alliancegenome.org) GO enrichment for genes dysregulated in zbtb11C116S neutrophils compared with those in WT neutrophils, with a false discovery rate < 0.05.
Statistics
Descriptive and analytical statistics were calculated using Prism 9 (GraphPad Software Inc). Unless otherwise stated, the data were obtained from ≥3 experiments. The details are included in the figure legends.
Results
Homozygous deletion of Zbtb11 results in defective FL hematopoiesis
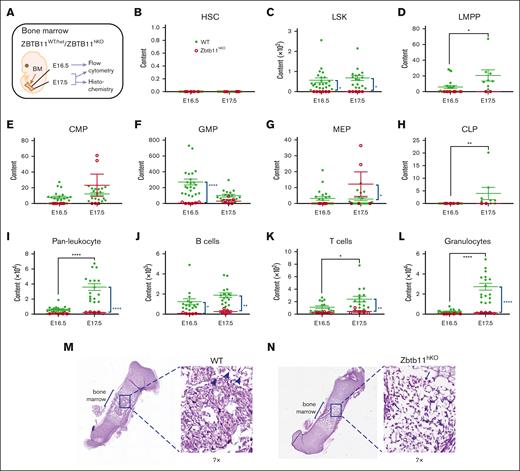
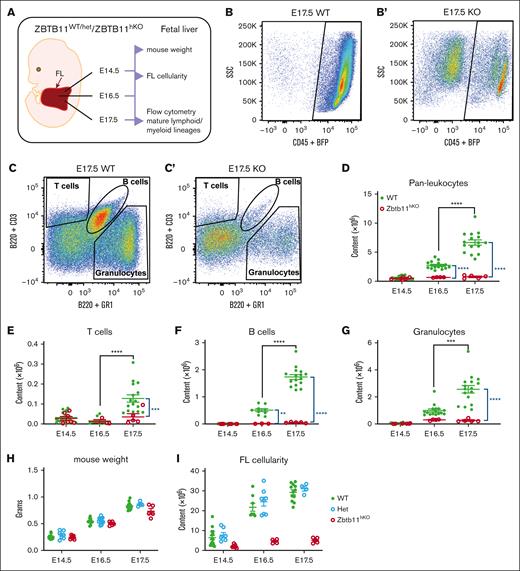
We previously identified Zbtb11 as a transcriptional repressor required for hematopoiesis in zebrafish.18 To determine whether this role is conserved in mammals, Zbtb11fl/fl mice were generated and mated with Vav1-iCre × BFP to disrupt Zbtb11 in the hematopoietic compartment (supplemental Figure 1A). Genomic polymerase chain reaction confirmed the deletion of 976 bp spanning exon 2 of Zbtb11 (supplemental Figure 1B). No Zbtb11fl/flvav-iCre+ (Zbtb11hKO) mice were born (supplemental Figure 1C), and timed mating determined that the absence of Zbtb11 in the hematopoietic compartment resulted in acute lethality by E18.5. From E14.5 to E16.5, gross morphology was largely unaffected in Zbtb11hKO embryos, and the weight remained normal. At E17.5 and E18.5, however, Zbtb11hKO embryos showed significant growth retardation (supplemental Figure 1D,F). Homozygous Zbtb11hKO embryos were immediately distinguishable from heterozygous or Zbtb11-replete siblings based on their pallor from at least E14.5 onwards, indicating severe anemia (supplemental Figure 1F). A schematic description of the analysis is provided in Figure 1A. Despite no differences in embryo weight up to E16.5, FL cellularity significantly decreased from E14.5 to E17.5 in Zbtb11hKO vs in controls (Figure 1I; supplemental Figure 1E). At E16.5 and E17.5, there were significantly fewer Zbtb11hKO FL CD45+ leukocytes (Figure 1B,D). By E17.5, both lymphoid and myeloid lineages were dramatically depleted in the Zbtb11hKO (Figure 1C,E-G). For all Zbtb11hKO populations, the cell number did not significantly increase from E16.5 to E17.5, whereas a robust increase occurred in controls (Figure 1D-G), denoting severely compromised differentiation and/or proliferation potential in FL in the absence of Zbtb11.
Homozygous Zbtb11 inactivation in the fetal hematopoietic compartment drastically impairs FL mature lymphoid and myeloid output. (A) Schematic description of the analysis. Flow cytometry analysis of FL at E17.5: dot plots showing the representative incidence of (B/B′) pan leukocytes (CD45+); (C/C′) granulocytes (Gr1+), T cells (CD3+), and B cells (B220+). Scatter plots showing the absolute quantification of total content at E14.5, E16.5, and E17.5: (D) CD45+ leukocytes, (E) T cells, (F) B cells, and (G) granulocytes. Mouse embryo weight (H) and FL cellularity (I) showed no haploinsufficiency effect in heterozygotes. For panels D and G: E14.5 WT, n = 22 and KO, n = 7; E16.5 WT, n = 16 and KO, n = 4; E17.5 WT, n = 16 and KO, n = 5. For panels E and F: E14.5 WT, n = 22 and KO, n = 7; E16.5 WT, n = 9 and KO, n = 3; E17.5 WT, n = 16 and KO, n = 5. For panel H: E14.5 WT, n = 15, Het, n = 7, and KO n = 7; E16.5 WT, n = 10, Het, n = 12, and KO, n = 5; E17.5 WT, n = 12, Het, n = 4, and KO n = 4. For panel I: E14.5 WT, n = 15, Het, n = 7, and KO, n = 7; E16.5 WT, n = 8, Het, n = 8, and KO, n = 4; E17.5 WT, n = 12, Het, n = 4, and KO n = 5. WT/controls; (green); KO, Zbtb11hKO (red); Het, Zbtb11WT/hKO (blue); data ± standard error of the mean. Two-way analysis of variance with Tukey (black) or Šídák (blue) multiple comparisons: ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
Homozygous Zbtb11 inactivation in the fetal hematopoietic compartment drastically impairs FL mature lymphoid and myeloid output. (A) Schematic description of the analysis. Flow cytometry analysis of FL at E17.5: dot plots showing the representative incidence of (B/B′) pan leukocytes (CD45+); (C/C′) granulocytes (Gr1+), T cells (CD3+), and B cells (B220+). Scatter plots showing the absolute quantification of total content at E14.5, E16.5, and E17.5: (D) CD45+ leukocytes, (E) T cells, (F) B cells, and (G) granulocytes. Mouse embryo weight (H) and FL cellularity (I) showed no haploinsufficiency effect in heterozygotes. For panels D and G: E14.5 WT, n = 22 and KO, n = 7; E16.5 WT, n = 16 and KO, n = 4; E17.5 WT, n = 16 and KO, n = 5. For panels E and F: E14.5 WT, n = 22 and KO, n = 7; E16.5 WT, n = 9 and KO, n = 3; E17.5 WT, n = 16 and KO, n = 5. For panel H: E14.5 WT, n = 15, Het, n = 7, and KO n = 7; E16.5 WT, n = 10, Het, n = 12, and KO, n = 5; E17.5 WT, n = 12, Het, n = 4, and KO n = 4. For panel I: E14.5 WT, n = 15, Het, n = 7, and KO, n = 7; E16.5 WT, n = 8, Het, n = 8, and KO, n = 4; E17.5 WT, n = 12, Het, n = 4, and KO n = 5. WT/controls; (green); KO, Zbtb11hKO (red); Het, Zbtb11WT/hKO (blue); data ± standard error of the mean. Two-way analysis of variance with Tukey (black) or Šídák (blue) multiple comparisons: ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001.
With such a strong phenotype in homozygous hematopoietic knockouts, we examined whether there was a Zbtb11 haploinsufficiency phenotype in heterozygotes. No significant difference between the homozygous WT and Zbtb11 heterozygous embryo weight or FL cellularity was apparent (Figure 1H-I). Likewise, across mature lineages, committed progenitors, and multipotent progenitors, cell numbers from embryos heterozygous for the Zbtb11hKO allele were not significantly different from those of WT in the FL or BM (supplemental Figures 2A-K and 3A-K). This demonstrated that hematopoietic compromise resulting from Zbtb11 loss in this model required disruption of both Zbtb11 alleles.
Zbtb11 is required for the generation of committed hematopoietic progenitors during fetal development
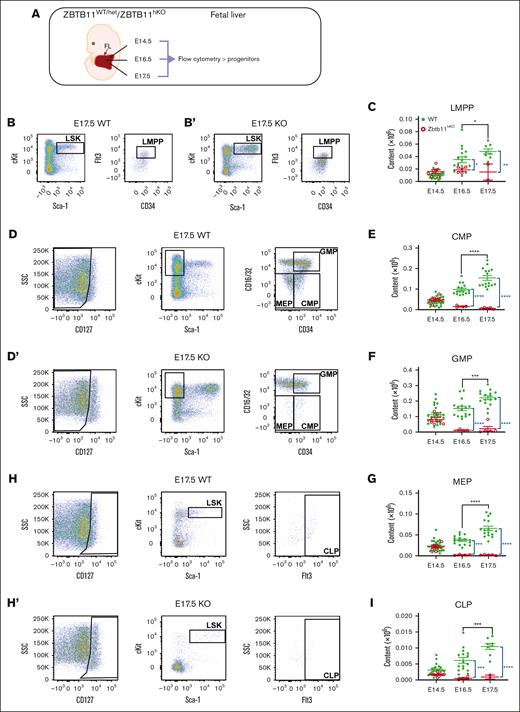
We analyzed the number of lineage-committed progenitors in FL to assess whether Zbtb11 loss preferentially affected the differentiation of specific lineages (Figure 2A). At E14.5, there was no difference in the number of lineage-committed progenitors between Zbtb11hKO and controls (Figure 2E-G,I). In contrast, at E16.5 and E17.5, Zbtb11hKO lymphoid-primed multipotent progenitors, CMPs, granulocyte-monocyte progenitors, myeloerythroid progenitors, and common lymphoid progenitors were all significantly decreased compared with controls (Figure 2B-I). Although all progenitor types significantly increased between E16.5 and E17.5 in controls, there was no increase in Zbtb11hKO progenitors, and CMP progenitor numbers even decreased between E14.5 and E17.5, respectively (P = .030). This is concordant with the reduction in mature differentiated lineages (Figure 1B-I) and is consistent with the requirement for Zbtb11 to support the size of the progenitor cell compartments from which mature cell types are derived by proliferation and differentiation.
In the absence of Zbtb11, FL-committed progenitor numbers fail to increase. (A) Schematic description of the analysis. Flow cytometry analysis dot plots show representative incidence at E17.5: (B/B′) LMPP (LSKCD34+Flt3+); (D/D′) CMP (Lin−CD127−Sca-1−c-Kit+CD34+CD16/32−), GMP (Lin−CD127−Sca-1−c-Kit+CD34+CD16/32+), and MEP (Lin−CD127−Sca-1−c-Kit+CD34−CD16/32−); and (H/H′) CLP (Lin−CD127+Sca-1+cKit+Flt3+). Scatter plots showing the absolute quantification of total content at E14.5, E16.5, and E17.5: (C) LMPP, (E) CMP, (F) GMP, (G) MEP, and (I) CLP. For panels C and I: E14.5 WT, n = 22 and KO n = 7; E16.5 WT, n = 16 and KO n = 4; E17.5 WT, n = 8 and KO n = 2. For panels E-G: E14.5 WT, n = 22 and KO, n = 7; E16.5 WT, n = 16 and KO, n = 4; E17.5 WT, n = 16 and KO, n = 5. WT/controls (green); KO, Zbtb11hKO (red); data ± standard error of the mean. Two-way analysis of variance with Tukey (black) or Šídák (blue) multiple comparisons: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. CLP, common lymphoid progenitor; GMP, granulocyte–macrophage progenitor; LMPP, lymphoid-primed multipotent progenitor; MEP, myeloerythroid progenitor.
In the absence of Zbtb11, FL-committed progenitor numbers fail to increase. (A) Schematic description of the analysis. Flow cytometry analysis dot plots show representative incidence at E17.5: (B/B′) LMPP (LSKCD34+Flt3+); (D/D′) CMP (Lin−CD127−Sca-1−c-Kit+CD34+CD16/32−), GMP (Lin−CD127−Sca-1−c-Kit+CD34+CD16/32+), and MEP (Lin−CD127−Sca-1−c-Kit+CD34−CD16/32−); and (H/H′) CLP (Lin−CD127+Sca-1+cKit+Flt3+). Scatter plots showing the absolute quantification of total content at E14.5, E16.5, and E17.5: (C) LMPP, (E) CMP, (F) GMP, (G) MEP, and (I) CLP. For panels C and I: E14.5 WT, n = 22 and KO n = 7; E16.5 WT, n = 16 and KO n = 4; E17.5 WT, n = 8 and KO n = 2. For panels E-G: E14.5 WT, n = 22 and KO, n = 7; E16.5 WT, n = 16 and KO, n = 4; E17.5 WT, n = 16 and KO, n = 5. WT/controls (green); KO, Zbtb11hKO (red); data ± standard error of the mean. Two-way analysis of variance with Tukey (black) or Šídák (blue) multiple comparisons: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. CLP, common lymphoid progenitor; GMP, granulocyte–macrophage progenitor; LMPP, lymphoid-primed multipotent progenitor; MEP, myeloerythroid progenitor.
Zbtb11-deficient multipotent progenitors and phenotypic HSCs accumulate in the FL during development
Reduced Zbtb11hKO lineage-committed progenitors and mature cells could result from disruption of homeostasis in the HSC compartment. Therefore, we investigated the number of Zbtb11hKO stem cells and multipotent progenitors using flow cytometry (Figure 3A). Surprisingly, and in contrast to committed progenitors and mature cells, there was more than double the number of Zbtb11hKO LSK multipotent progenitors at E14.5, leveling off at E16.5 and E17.5 (Figure 3B-C). Similarly, there were more Zbtb11hKO phenotypic HSCs at E14.5, but HSC overabundance was sustained through E17.5 (Figure 3D-E). These observations provide strong evidence that the initial HSC specification in Zbtb11hKO embryos is intact and that hematopoietic failure is not due to a lack of initial HSC specification but rather a failure of a downstream step incapacitating the development of a fully populated, multilineage hematopoietic system from the abundant number of HSCs that were initially specified. Given the near-100% excision efficiency of vav-iCre in embryonic development,37 the very small but measurable number of lineage-committed progenitors that mostly decrease over time (Figure 2) may potentially arise from primitive HSC-independent progenitors. These may allow for limited lineage commitment but become depleted as development progressively relies on HSC-dependent hematopoiesis in a scenario where HSC function appears to be compromised.
Multipotent Zbtb11hKO progenitors and HSCs are elevated in the FL. (A) Schematic description of the analysis and definition of HSC used in all experiments. Flow cytometry analysis: (B/B′) HSC progenitors (LSK) and (D/D′) HSC (LSKCD150+CD48−) at E14.5, with dot plots showing the representative incidence. Scatter plots showing the absolute quantification at E14.5, E16.5, and E17.5: (C) total LSK (E14.5: Zbtb11hKO = 8.7 × 104 (n = 7); controls = 3.9 × 104 [n = 22]; P = .0011), and (E) total HSC at E14.5 (Zbtb11hKO = 6.1 × 103 [n = 7]; controls = 2.6 × 103 [n = 22]; P = .0072), E16.5 (Zbtb11hKO = 10.4 × 103 [n = 4]; controls = 5.7 × 103 [n = 16]; P = .0062), and E17.5. (Zbtb11hKO =13.7 × 103 [n = 5]; controls = 9.4 × 103 [n = 16]; P = .0061). WT/controls (green); KO, Zbtb11hKO (red); data ± standard error of the mean. Two-way analysis of variance with Šídák multiple comparisons: ∗∗P < .01.
Multipotent Zbtb11hKO progenitors and HSCs are elevated in the FL. (A) Schematic description of the analysis and definition of HSC used in all experiments. Flow cytometry analysis: (B/B′) HSC progenitors (LSK) and (D/D′) HSC (LSKCD150+CD48−) at E14.5, with dot plots showing the representative incidence. Scatter plots showing the absolute quantification at E14.5, E16.5, and E17.5: (C) total LSK (E14.5: Zbtb11hKO = 8.7 × 104 (n = 7); controls = 3.9 × 104 [n = 22]; P = .0011), and (E) total HSC at E14.5 (Zbtb11hKO = 6.1 × 103 [n = 7]; controls = 2.6 × 103 [n = 22]; P = .0072), E16.5 (Zbtb11hKO = 10.4 × 103 [n = 4]; controls = 5.7 × 103 [n = 16]; P = .0062), and E17.5. (Zbtb11hKO =13.7 × 103 [n = 5]; controls = 9.4 × 103 [n = 16]; P = .0061). WT/controls (green); KO, Zbtb11hKO (red); data ± standard error of the mean. Two-way analysis of variance with Šídák multiple comparisons: ∗∗P < .01.
Fetal BM failure occurs in the absence of Zbtb11
In mice, hematopoietic stem and progenitor cells migrate from the FL to seed the fetal BM, where hematopoiesis establishes gradually from at least E16.5.38-40 Given the consistent elevation of Zbtb11hKO HSC in FL, we investigated whether these HSCs could seed and initiate hematopoiesis in the fetal BM (Figure 4A). Although the establishment of fetal BM hematopoiesis occurred in control embryos as expected, there was near-complete failure of BM hematopoiesis in Zbtb11hKO embryos (Figure 4B-L). In contrast to the FL, analysis of the fetal BM compartment showed that the Zbtb11hKO LSK progenitor pool was absent through E16.5-E17.5 (Figure 4C). Not surprisingly, this lack of Zbtb11hKO LSK cells was accompanied by few, if any, BM-located committed progenitors (Figure 4D-H) and significantly fewer mature lineage cells (Figure 4I-L). Although the number of control hematopoietic cells underwent significant expansion from E16.5 to E17.5, Zbtb11hKO hematopoietic cells did not (Figure 4D,H,I,K,L). Histology of fetal femurs showed disorganization and hypocellular marrow, concordant with the fluorescence-activated cell sorting (FACS) analysis (Figure 4M,N). Collectively, these data indicate that without Zbtb11, the FL to BM hematopoiesis transition does not occur, resulting in fetal BM failure.
The absence of Zbtb11 in the hematopoietic compartment results in fetal BM failure. (A) Schematic description of the analysis. Scatter plots showing absolute quantification of total BM content at E16.5 and E17.5: (B) HSC, (C) LSK, (D) LMPP, (E) CMP, (F) GMP, (G) MEP (H), CLP, (I) pan leukocytes, (J) B cells, (K) T cells, and (L) granulocytes at E16.5 and E17.5. For panels B, C, E, F, G, I, and L: E16.5 WT, n = 22 and KO, n = 6; E17.5 WT, n = 16 description of the KO, n = 5. For panels D and H: E16.5 WT, n = 22 and KO, n = 6; E17.5 WT, n = 8 and KO, n = 2; For panels J and K: E16.5 WT, n = 15 and KO, n = 5; E17.5 WT, n = 16 and KO, n = 5. Hematoxylin and eosin staining of E17.5 femur sections for (M) WT and (N) Zbtb11hKO. Insets are original magnification ×7 of boxed area. Arrowheads indicate enucleated erythroid cells (M), and arrows indicate nucleated erythroid cells (N). WT, wild-type controls (green); KO, Zbtb11hKO (red); E, embryonic day; Data ± standard error of the mean. Two-way analysis of variance with Šídák multiple comparisons: ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001.
The absence of Zbtb11 in the hematopoietic compartment results in fetal BM failure. (A) Schematic description of the analysis. Scatter plots showing absolute quantification of total BM content at E16.5 and E17.5: (B) HSC, (C) LSK, (D) LMPP, (E) CMP, (F) GMP, (G) MEP (H), CLP, (I) pan leukocytes, (J) B cells, (K) T cells, and (L) granulocytes at E16.5 and E17.5. For panels B, C, E, F, G, I, and L: E16.5 WT, n = 22 and KO, n = 6; E17.5 WT, n = 16 description of the KO, n = 5. For panels D and H: E16.5 WT, n = 22 and KO, n = 6; E17.5 WT, n = 8 and KO, n = 2; For panels J and K: E16.5 WT, n = 15 and KO, n = 5; E17.5 WT, n = 16 and KO, n = 5. Hematoxylin and eosin staining of E17.5 femur sections for (M) WT and (N) Zbtb11hKO. Insets are original magnification ×7 of boxed area. Arrowheads indicate enucleated erythroid cells (M), and arrows indicate nucleated erythroid cells (N). WT, wild-type controls (green); KO, Zbtb11hKO (red); E, embryonic day; Data ± standard error of the mean. Two-way analysis of variance with Šídák multiple comparisons: ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001.
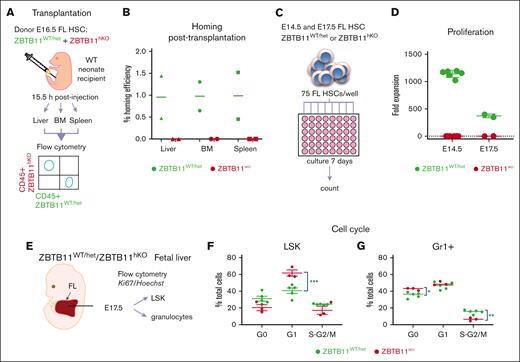
Homing of Zbtb11hKO HSCs is impaired
One reason for fetal BM failure could be defective homing of Zbtb11hKO HSCs. To determine whether Zbtb11hKO HSCs are capable of homing to hematopoietic tissues, a transplantation homing assay was performed. FAC-sorted, differentially labeled Zbtb11hKO and control FL E16.5 HSCs were transplanted into a WT neonate by intraperitoneal injection. After 15.5 hours, hematopoietic organs were assayed for Zbtb11hKO and control donor cells (Figure 5A). Although WT HSCs averaged ∼1% homing efficiency, no Zbtb11hKO donor cells were recoverable from the BM, spleen, or liver, indicating homing impairment of Zbtb11hKO HSCs (Figure 5B). Hence, despite the overabundant Zbtb11hKO HSCs in FL, these HSCs are unable to seed fetal BM even when transplanted to initiate hematopoiesis, contributing to hematopoietic failure in Zbtb11hKO embryos.
Functional failure of Zbtb11hKO HSC: failure to home after transplantation, failure to proliferate in vitro, and impairment of cell cycle progression. (A) Schematic description of the transplantation and homing analysis. (B) Homing efficiency of E16.5 Zbtb11hKO and WT FL HSCs transplanted into WT neonate. (C) Schematic description of the in vitro proliferation assay in which 75 immunophenotyped Zbtb11hKO or WT FL HSCs were seeded per well and cultured in growth medium. Cells were counted in each well on day 7 and divided by 75 to determine the fold expansion. (D) Flow cytometry quantification of in vitro proliferation assays: on day 7, E14.5 HSC culture resulted in mean = 133 Zbtb11hKO cells and 85 078 control cells, respectively; the average fold expansion was 1.78 and 1142.7 over initial seeding of 75 cells. (E) Schematic description of the cell cycle analysis. Cell cycle progression was assessed using the Ki67/nuclear DNA (Hoechst) ratio for (F) LSK and (G) granulocytes (Gr1+) at E17.5. For panel B, n = 2 independent experiments. For panel D: E14.5 WT, n = 6 and KO, n = 6; E17.5 WT, n = 2 and KO, n = 2. For panels F and G: WT, n = 5 and KO, n = 3; WT/controls (green); KO, Zbtb11hKO (red); data ± standard error of the mean. Two-way analysis of variance with Šídák multiple comparisons: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
Functional failure of Zbtb11hKO HSC: failure to home after transplantation, failure to proliferate in vitro, and impairment of cell cycle progression. (A) Schematic description of the transplantation and homing analysis. (B) Homing efficiency of E16.5 Zbtb11hKO and WT FL HSCs transplanted into WT neonate. (C) Schematic description of the in vitro proliferation assay in which 75 immunophenotyped Zbtb11hKO or WT FL HSCs were seeded per well and cultured in growth medium. Cells were counted in each well on day 7 and divided by 75 to determine the fold expansion. (D) Flow cytometry quantification of in vitro proliferation assays: on day 7, E14.5 HSC culture resulted in mean = 133 Zbtb11hKO cells and 85 078 control cells, respectively; the average fold expansion was 1.78 and 1142.7 over initial seeding of 75 cells. (E) Schematic description of the cell cycle analysis. Cell cycle progression was assessed using the Ki67/nuclear DNA (Hoechst) ratio for (F) LSK and (G) granulocytes (Gr1+) at E17.5. For panel B, n = 2 independent experiments. For panel D: E14.5 WT, n = 6 and KO, n = 6; E17.5 WT, n = 2 and KO, n = 2. For panels F and G: WT, n = 5 and KO, n = 3; WT/controls (green); KO, Zbtb11hKO (red); data ± standard error of the mean. Two-way analysis of variance with Šídák multiple comparisons: ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
Proliferation and cell cycle progression impairment in Zbtb11-deficient embryonic hematopoietic cells
Excessive proliferation of HSCs can result in their subsequent loss.41,42 Given the increased number of Zbtb11hKO HSC, we evaluated HSC proliferation directly by a 7-day in vitro culture assay seeded with 75 E14.5 or E17.5 FAC-sorted HSCs (Figure 5C). At this stage, HSC are still highly proliferative.3,43 The hematopoietic cell output of HSCs, as measured based on the number of cells per well, was increased >1000-fold in the WT cultures but not in the Zbtb11hKO cultures, which were less than doubled at E14.5 (Figure 5D; average = 133 per well). Cell death could not account for the failure of Zbtb11hKO cell numbers to expand because there were slightly more cells at the end of the assay than at the beginning (75 per well). However, at E17.5, 5 out of 6 wells seeded with Zbtb11hKO HSCs had no viable cells on day 7, suggesting that cell survival was compromised and that cell death largely contributed to the proliferation failure of these older HSCs.
Cell cycle progression in HSCs is very tightly regulated, and adult HSCs can remain quiescent for long periods.44 Because Zbtb11 loss completely arrests cell cycle progression in zebrafish,18 we examined these processes in murine Zbtb11hKO hematopoietic cells (Figure 5E). At E17.5, a greater proportion of Zbtb11hKO FL LSK multipotent progenitors accumulated in G1 than in controls (Figure 5F). Of the cells capable of differentiating along the granulocyte lineage, a significantly higher proportion of Zbtb11hKO cells were quiescent (G0), accompanied by a reduced proportion progressing to the S/M phase (Figure 5G). This is consistent with our previous zebrafish 5-ethynyl 2′-deoxyuridine incorporation data showing cell cycle arrest before the S phase18 and demonstrates that Zbtb11 is specifically required for cell cycle progression in the mouse hematopoietic compartment. Because cell death did not increase in E14.5 Zbtb11hKO HSCs, the lack of in vitro proliferation was likely due to checkpoint restriction. At E17.5, checkpoint restriction may initially impair proliferation (G1 accumulation; Figure 5F) but becomes more severe, causing cells to exit the cell cycle (G0 accumulation and decreased division; Figure 5G) and ultimately undergo cell death, as observed in HSCs in the in vitro proliferation assay at E17.5. This suggests that the overabundance of Zbtb11hKO FL HSCs may arise from increased specification rather than aberrant self-renewal.
In the absence of Zbtb11, phenotypic HSC lack an erythroid-primed subpopulation, and niche and OXPHOS gene signatures are downregulated
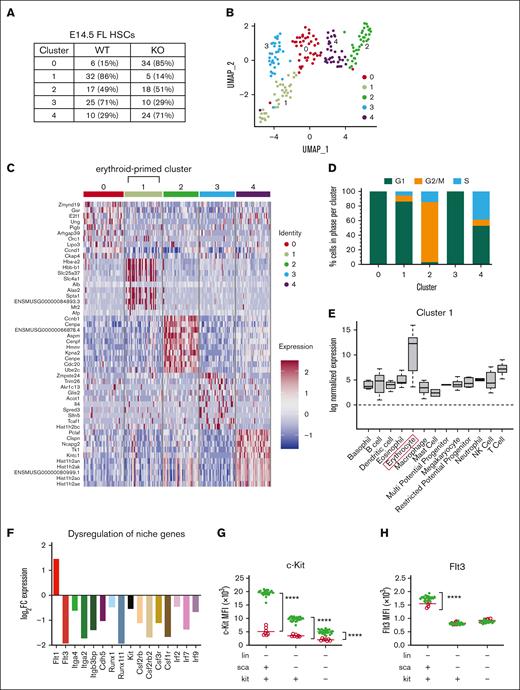
Unsupervised clustering of E14.5 Zbtb11hKO and control FL HSC single-cell RNAseq (scRNAseq) generated k = 5 clusters: 2 in which Zbtb11hKO HSCs were underrepresented (1 and 3), 2 in which Zbtb11hKO HSCs were overrepresented (0 and 4) and 1 cluster (2) in which Zbtb11hKO and control HSCs were balanced (Figure 6A-B). Cells were classified into the cell cycle phase based on gene expression data ratios,45 and most cells were in G1 (WT = 71.1%, KO = 68.1%). Although there was no significant difference in the proportion of cells assigned to each cell cycle phase based on the genotype (G2/M: WT = 18.9% and KO = 19.8%; S: WT = 10.0% and KO = 12.1%), the proportion of cells within each cluster was separable based on the cell cycle (Figure 6D). Differential gene expression between clusters revealed upregulation of erythroid genes in cluster 1, suggesting a subpopulation of erythroid-primed HSCs (Figure 6C). When the expression of marker genes for each cluster was evaluated across individual hematopoietic lineages defined in the hemosphere,46 marked enrichment for erythroid lineage signature genes occurred only in cluster 1 (Figure 6E; supplemental Figure 4A-D). Furthermore, GO analysis showed enrichment for erythropoiesis in cluster 1 (supplemental Figure 4E), revealing a subpopulation in this cluster poised for differentiation along the erythroid lineage.47 Because cluster 1 mainly comprised control HSCs (Figure 6A), this uncovered a potential genetic signature underlying the profound early anemia of Zbtb11hKO embryos. Indeed, the E17.5 Zbtb11hKO BM showed only nucleated erythroid cells, indicating a delay or arrest in erythrocyte development in the absence of Zbtb11 (Figure 4N). Because Zbtb11hKO HSCs were abundantly specified but hematopoiesis failed, we examined whether there was a disruption in genes associated with the HSC niche. Figure 6F shows the downregulation of several families of genes crucial for the correct trafficking of HSCs to the FL, including receptor tyrosine kinases (Flt3), adhesion molecules (integrins and cadherin 5), cytokine receptors (colony stimulating factor receptors), and transcription factors (Runx1 [RUNX family transcription factor 1, also known as Acute Myeloid Leukemia 1 (AML1)] and RUNX1 partner transcriptional co-repressor 1 [Runx1T].; interferon regulatory factors). Furthermore, c-Kit and Flt3 cell surface proteins were correspondingly decreased in LSK populations (Figure 6G,H).
scRNAseq analysis of WT and Zbtb11hKO HSCs reveals underrepresentation of Zbtb11hKO HSCs in the cluster characterized by an erythroid signature and dysregulation of niche gene expression. (A) Distribution of cells based on the number, proportion (%), and genotype for each cluster. Clusters 1 and 3 are disproportionately comprised of WT cells, whereas clusters 0 and 4 are disproportionately comprised of KO cells. (B) Uniform Manifold Approximation and Projection plot of unsupervised clustering identified 5 subpopulations of E14.5 FL HSCs (LSK/SLAM). (C) Heatmap of normalized log-expression values for the top 10 cluster-specific marker genes. Column colors represent the cluster identity to which each cell is assigned. (D) Distribution of cell cycle phase within each cluster. (E) Enrichment of log normalized gene expression of top 10 upregulated genes in each cluster was evaluated across individual hematopoietic lineages defined using the Haemopedia Mouse RNAseq data set, which contains 129 samples classified into 13 cell lineages and 57 cell types.46 Expression of marker genes for cluster 1 shows enrichment for erythrocyte lineage (boxed) genes. (F) Differential expression of hematopoietic niche genes in unclustered E14.5 FL Zbtb11hKO HSCs compared with WT HSCs. Y-axis shows the log2FC (fold change) gene expression level, with a false discovery rate < 0.05. Mean fluorescence intensity (MFI) of c-Kit (G) and Flt3 (H) protein in Zbtb11hKO and WT control progenitors at E14.5. WT/controls (green); KO, (red); fluorescence intensity of the protein is shown along the y-axis; data ± standard error of the mean. Two-way analysis of variance with Šídák multiple comparisons: ∗∗∗∗P < .0001.
scRNAseq analysis of WT and Zbtb11hKO HSCs reveals underrepresentation of Zbtb11hKO HSCs in the cluster characterized by an erythroid signature and dysregulation of niche gene expression. (A) Distribution of cells based on the number, proportion (%), and genotype for each cluster. Clusters 1 and 3 are disproportionately comprised of WT cells, whereas clusters 0 and 4 are disproportionately comprised of KO cells. (B) Uniform Manifold Approximation and Projection plot of unsupervised clustering identified 5 subpopulations of E14.5 FL HSCs (LSK/SLAM). (C) Heatmap of normalized log-expression values for the top 10 cluster-specific marker genes. Column colors represent the cluster identity to which each cell is assigned. (D) Distribution of cell cycle phase within each cluster. (E) Enrichment of log normalized gene expression of top 10 upregulated genes in each cluster was evaluated across individual hematopoietic lineages defined using the Haemopedia Mouse RNAseq data set, which contains 129 samples classified into 13 cell lineages and 57 cell types.46 Expression of marker genes for cluster 1 shows enrichment for erythrocyte lineage (boxed) genes. (F) Differential expression of hematopoietic niche genes in unclustered E14.5 FL Zbtb11hKO HSCs compared with WT HSCs. Y-axis shows the log2FC (fold change) gene expression level, with a false discovery rate < 0.05. Mean fluorescence intensity (MFI) of c-Kit (G) and Flt3 (H) protein in Zbtb11hKO and WT control progenitors at E14.5. WT/controls (green); KO, (red); fluorescence intensity of the protein is shown along the y-axis; data ± standard error of the mean. Two-way analysis of variance with Šídák multiple comparisons: ∗∗∗∗P < .0001.
Clusters were analyzed for GO terms and KEGG pathway enrichment (supplemental Figure 5A-E). The cells in cluster 0 were predominantly Zbtb11hKO HSCs. Interestingly, one of the most overrepresented pathways in this cluster was necroptosis (supplemental Figure 5A), a caspase-independent genetically programmed form of cell death.48 Cluster 3 cells were all in G1 phase, and GO analysis showed enrichment of terms associated with nucleoside triphosphate metabolism, adenosine triphosphate synthesis and metabolism, and respiratory transport chain, supported by KEGG pathway analysis, in which OXPHOS was in the top 10 terms (supplemental Figure 5D). This likely reflects cells in G1 undergoing preparation for DNA synthesis. Both clusters (1 and 3), which were predominantly composed of control HSCs, showed enrichment of gene signatures for OXPHOS and metabolic pathways (supplemental Figure 5B,D). These gene signatures were not enriched in clusters predominantly lacking functional Zbtb11, suggesting that these pathways may be defective in Zbtb11hKO HSCs. Interestingly, cluster 4, consisting predominantly of Zbtb11hKO HSCs, showed enrichment of GO terms for DNA repair, replication, cellular response to DNA damage stimulus, and cellular response to stress (supplemental Figure 5E). This was supported by the disproportionate enrichment of 5 KEGG pathways involved in DNA repair: base excision, nucleotide excision, mismatch repair, homologous recombination, and the Fanconi anemia pathway as well as DNA replication and pyrimidine metabolism. These analyses revealed susceptibility to DNA damage and potential replicative stress in this subpopulation of Zbtb11hKO HSCs.
Discussion
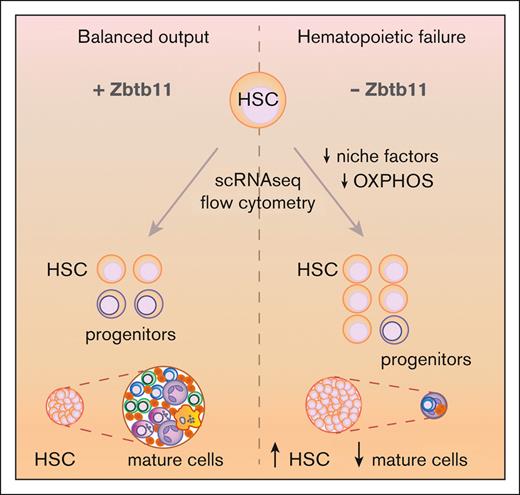
We have shown that Zbtb11 is a key regulator of the HSC function required for self-renewal and differentiation. Zbtb11 ablation during murine hematopoietic development generated overabundant HSCs in FL, yet profound BM failure and embryonic lethality followed. The intricacies of this severe disturbance in HSC function warrant further investigation because the requirement for Zbtb11 and its regulatory networks present an attractive avenue for therapeutic manipulation in HSC-directed therapies. Currently, it is unknown whether the overabundance of HSCs in Zbtb11-depleted hematopoietic compartments results from increased specification or increased cycling before E14.5, leading to HSC exhaustion by E14.5. Failure of transplanted Zbtb11hKO HSCs to home to major hematopoietic organs could arise from HSC death or insensitivity to homing signaling pathways. In vitro studies suggest that E14.5 Zbtb11hKO HSCs are quiescent and that Zbtb11hKO HSCs mostly do not survive by E17.5; however, it is possible that E16.5 Zbtb11hKO HSCs survive in vivo for a short duration in the homing study. Another important question is whether Zbtb11 affects the capacity of HSCs to exit FL, given the accumulation of Zbtb11hKO HSCs in FL but a shift to significantly fewer Zbtb11hKO lineage-committed progenitors over time. The inability of transplanted E16.5 Zbtb11hKO HSCs to home to hematopoietic organs in a WT recipient precludes additional classical transplantation studies to dissect HSC function in this model. In future studies, inducible models of Zbtb11 deletion in HSCs and specific mature hematopoietic lineages will determine the precise onset of Zbtb11 requirement, function of Zbtb11 in adult HSCs, and whether the impact observed in mature lineages in Zbtb11hKO is due to defective HSCs and/or a cell-intrinsic Zbtb11 requirement in some or all of these lineages.
Increased specification of HSCs followed by complete BM failure is an uncommon phenotype. Ablation of geminin is one example in which HSCs increase but at a cost of proliferation and differentiation of lineage-committed progenitors, postulated to occur through the epigenetic regulation of multipotency vs lineage specification genetic programs.49 We previously showed that Zbtb11 is under the regulatory control of myeloid transcription factors Pu.1, Gfi1a, Gfi1b, and C/ebpα.18 Hematopoiesis-specific transcriptional binding partners remain to be identified for Zbtb11, and determining these would provide further mechanistic insight along with interrogating Zbtb11-mediated alterations in the hematopoietic gene regulatory landscape.
The crucial role played by mitochondrial reactive oxygen species (ROS) in regulating HSC fate and its therapeutic application is currently a topic of much interest.50,51 Changes in OXPHOS and its byproduct, ROS, determine the balance between HSC self-renewal and differentiation, with a switch to OXPHOS needed to support the metabolic demands of differentiating HSCs.52-56 Both scRNAseq of mouse HSCs and our bulk RNAseq of Zbtb11-deficient zebrafish neutrophils showed significantly affected OXPHOS and mitochondrial pathways (supplemental Figure 5A-B,D-F),18 concordant with reports in Zbtb11–/– mouse embryonic stem cells.27,57 A recent study showed that Zbtb11 is required for mouse embryonic stem cell pluripotency by maintaining the repression of poised prodifferentiation genes.27 However, Zbtb11hKO HSCs, which are overabundant yet intrinsically incompetent to undergo differentiation and proliferation, appear to have lost multipotency or the function of key cellular components, such as respiratory complex I and mitoribosome,57 required for the execution of differentiation programs. Cluster 4 showed an increase in genes responsible for DNA damage repair, suggesting these HSCs are likely under replicative stress, consistent with their accumulation in the S phase (Figure 6D; supplemental Figure 5E). This ties in with the dysregulation of OXPHOS/ROS and suboptimal metabolic requirements for replication, which may exacerbate the failure of Zbtb11hKO HSCs to progress through the intra-S-phase checkpoint.58 Notably, Zbtb11hKO and mitochondrial phosphatase Ptpmt1–/– HSCs share a similar phenotype.55 Measuring ROS, adenosine triphosphate, and oxygen consumption in Zbtb11hKO HSCs and the remaining lineage-committed and mature cells could elucidate Zbtb11-mediated defects in mitochondrial function as a source of failed hematopoiesis. Together, these observations highlight the potential role of Zbtb11 in regulating mitochondrial activity in HSC.
During murine development, the first wave of primitive hematopoiesis occurs in the yolk sac between E7.5 and E9.5 to sustain the early embryonic tissues. Around E9, a definitive (late fetal/adult) population of erythroid-myeloid progenitors capable of forming a broader range of lineages also emerges in the yolk sac.59 Zbtb11 expression has been detected in the brain or central nervous system and several other organs during the relevant stages of HSC specification.60 Importantly, Zbtb11 is expressed in the yolk sac (8.9), aorta-gonads-mesonephros (7.4), placenta (8.8), and FL (E12.5 = 8.5; E13.5 = 8.2; E14.5 = 7.4; mean expression of replicate samples, StemSite61). Our studies demonstrate that when Zbtb11 is deleted in the hematopoietic compartment, there is a loss of an erythroid subpopulation, evidenced by predominantly WT erythroid signature-expressing HSCs in cluster 1, early FL anemia, and the presence of only immature nucleated erythroid cells at E17.5 in the BM. Therefore, it is interesting to speculate whether ZBTB11 acts in the FL microenvironment to support the differentiation of yolk sac-derived erythroid-myeloid progenitors, which are thought to be the source for the establishment of the adult hematopoietic system in this niche. Vav-Cre expression initiates at approximately E11.5;62 therefore, earlier hematopoietic development in the yolk sac, including specification of erythroid-myeloid progenitors, will not be affected by Zbtb11 loss in this vav-iCre deletion-dependent model. However, at E14.5, while HSCs were within the FL niche, Zbtb11 loss in HSCs resulted in dysregulation of key genes involved in hematopoietic differentiation and proliferation and cell-cell interactions, including c-Kit and Flt3. In HSCs, c-Kit acts to preserve self-renewal,63,64 with c-Kit density inversely correlated with stemness.64,65 A fourfold decrease in the mean fluorescence intensity of c-Kit cell surface protein expression in Zbtb11hKO LSK cells at E14.5 suggested that c-Kit signaling disruption could contribute to the inhibition of Zbtb11hKO stem and progenitor differentiation (Figure 6G). Consistent with this, unclustered scRNAseq showed that c-Kit was underexpressed in Zbtb11hKO HSCs (Figure 6F), whereas kit ligand expression was normal. Flt3 is also crucial for hematopoietic development and mutations in this gene are commonly associated with acute myeloid leukemia.66-68 The downregulation of both c-Kit and Flt3 in Zbtb11hKO progenitor cells and multiple other factors that support HSC differentiation and expansion (Figure 6F-H) highlight the role of Zbtb11 in maintaining a gene regulatory landscape permitting HSCs to functionally interact with their microenvironment.
In conclusion, these data demonstrate an absolute requirement for Zbtb11 to sustain murine fetal hematopoiesis after HSCs are specified. This requirement for Zbtb11 acts through cell-essential mechanisms affecting HSC homing, proliferation, and mitochondrial function and the regulatory gene networks required for productive hematopoietic output from the hematopoietic niche.
Acknowledgments
The authors thank the FlowCore and Monash Animal Research Platform for their equipment and technical support. The authors thank Jeanette Rientjes from Gene Recombineering (Monash) and Arianna Nenci from the Monash Gene Technology Facility, Jessica Hatwell-Humble for mouse husbandry and technical expertise, Brenda Williams and Madeline Fulton for technical expertise, Chad Heazlewood for histology, and Tracey Baldwin from Single Cell Open Research Endeavour (The Walter and Eliza Hall Institute of Medical Research) for technical assistance.
This work was supported by the National Health and Medical Research Council (1070687) (G.J.L. and M.C.K.), Cancer Council Victoria (1047660) (G.J.L. and M.C.K.), National Institutes of Health (R01 HL079545) (G.J.L.), and the La Trobe Institute for Molecular Science, VIC Australia (M.C.K.). The Australian Regenerative Medicine Institute is supported by funds from the State Government of Victoria and the Australian Federal Government.
Authorship
Contribution: M.C.K., S.K.N., and G.J.L. conceptualized the study; H.C., B.C., S.K.N., and M.C.K. conducted the experiments; M.C.K., H.C., B.C., S.K.N., P.H., S.H.N., and A.S. performed the formal analysis; D.A.-Z. and S.H.N. provided resources for the scRNAseq; M.C.K., S.K.N., and G.J.L. supervised the study; M.C.K. and G.J.L. wrote the original draft of the manuscript; M.C.K., S.K.N., and G.J.L. acquired funding; and all authors contributed to the review and editing process.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Graham J. Lieschke, Australian Regenerative Medicine Institute, Level 1, Building 75 Monash University, Wellington Road, Clayton, VIC 3800, Australia; e-mail: graham.lieschke@monash.edu; and M. Cristina Keightley, La Trobe Rural Health School, La Trobe University, PO Box 199, Bendigo, VIC 3552, Australia; e-mail: c.keightley@latrobe.edu.au.
References
Author notes
∗S.K.N., M.C.K., and G.J.L. contributed equally to this study.
The scRNA-seq mouse HSC data have been deposited in the Gene Expression Omnibus database (accession number GSE240459).
RNAseq data of WT and zbtb11-deficient zebrafish neutrophils have been deposited in the Gene Expression Omnibus database (accession number GSE239949).
The authors declare that all remaining data supporting the findings of this study are available within the article and its supplemental Files, or on request from the corresponding authors, Graham J. Lieschke (graham.lieschke@monash.edu) and Cristina Keightley (c.keightley@latrobe.edu.au).
The full-text version of this article contains a data supplement.

![Multipotent Zbtb11hKO progenitors and HSCs are elevated in the FL. (A) Schematic description of the analysis and definition of HSC used in all experiments. Flow cytometry analysis: (B/B′) HSC progenitors (LSK) and (D/D′) HSC (LSKCD150+CD48−) at E14.5, with dot plots showing the representative incidence. Scatter plots showing the absolute quantification at E14.5, E16.5, and E17.5: (C) total LSK (E14.5: Zbtb11hKO = 8.7 × 104 (n = 7); controls = 3.9 × 104 [n = 22]; P = .0011), and (E) total HSC at E14.5 (Zbtb11hKO = 6.1 × 103 [n = 7]; controls = 2.6 × 103 [n = 22]; P = .0072), E16.5 (Zbtb11hKO = 10.4 × 103 [n = 4]; controls = 5.7 × 103 [n = 16]; P = .0062), and E17.5. (Zbtb11hKO =13.7 × 103 [n = 5]; controls = 9.4 × 103 [n = 16]; P = .0061). WT/controls (green); KO, Zbtb11hKO (red); data ± standard error of the mean. Two-way analysis of variance with Šídák multiple comparisons: ∗∗P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/7/21/10.1182_bloodadvances.2022009580/4/m_blooda_adv-2022-009580-gr3.jpeg?Expires=1769829711&Signature=Yoz2-e5LX7hifKloWvvzE3vxqqhfBqHFqrhzzcFE3~RCg8~iiu8M07HRSeXLpzy4sQnGCQ9tWLEAkmFjQGCQTu7QBLjVbbda4nXBChkej1V1sBB2xvcWtHs7-g8uFe3P9XNufhwfUlNXF3PS2VJ2MmXTqMIqHJZ9k7u2GV63KYxdpiBoA4ffHOURLjz8oC-f8rz8nM7iR0UVLYsUKE4drRH4CP-~OobVJ0dPXi2u5W15i3bWpgICOgKvw5tJ28cdch2GxAThMPQ5BwdZKH4MEga2L6pY7SGJrOf6-bH-IXbApftnCSglMxR3wC3d1pXOLNg-l0cacC0xpMg47A4dww__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)