Key Points
Platelet mitochondria undergo a fission step during platelet activation.
The fission of platelet mitochondria correlates with a switch from a predominant mitochondrial to a more glycolytic energy production.
Abstract
Blood platelets undergo several successive motor-driven reorganizations of the cytoskeleton when they are recruited to an injured part of a vessel. These reorganizations take place during the platelet activation phase, the spreading process on the injured vessel or between fibrin fibers of the forming clot, and during clot retraction. All these steps require a lot of energy, especially the retraction of the clot when platelets develop strong forces similar to those of muscle cells. Platelets can produce energy through glycolysis and mitochondrial respiration. However, although resting platelets have only 5 to 8 individual mitochondria, they produce adenosine triphosphate predominantly via oxidative phosphorylation. Activated, spread platelets show an increase in size compared with resting platelets, and the question arises as to where the few mitochondria are located in these larger platelets. Using expansion microscopy, we show that the number of mitochondria per platelet is increased in spread platelets. Live imaging and focused ion beam–scanning electron microscopy suggest that a mitochondrial fission event takes place during platelet activation. Fission is Drp1 dependent because Drp1-deficient platelets have fused mitochondria. In nucleated cells, mitochondrial fission is associated with a shift to a glycolytic phenotype, and using clot retraction assays, we show that platelets have a more glycolytic energy production during clot retraction and that Drp1-deficient platelets show a defect in clot retraction.
Introduction
Blood platelets are cell fragments without a nucleus. They float in the bloodstream as discoid particles controlling vessel integrity. The discoid shape of resting platelets is maintained by a bundle of microtubules forming a ring, called marginal band, in the periphery of the platelets.1 When platelets reach an injured site of the endothelium, they adhere to the exposed extracellular matrix, which leads to their activation. Additional platelets activate in suspension and get trapped in the forming blood clot. Platelet activation leads to a profound shape change, secretion of their granules, aggregation of platelets, and formation of thrombus to ensure hemostasis.2 We have shown previously that the initial shape change during platelet activation from a flat discoid to a spherical state is initiated by microtubule motor proteins and completed by the actin motor myosin. Microtubule motors slide apart the microtubules in the marginal band. This leads to elongation and coiling of the marginal band, promoting the spherical shape of activating platelets. Actin-myosin contraction then compresses the coiled microtubule bundle.3,4 The platelets then spread either on the 2-dimensional subendothelial lining or within the 3-dimensional fibrin clot; both processes depend on further reorganizations of the cytoskeleton. Motor protein actions have again an essential role during late platelet functions that are important for clot retraction. A high amount of energy is required for these motor-driven reorganizations of the cytoskeleton as well as for the polymerization of actin filaments and microtubules. Furthermore, platelets are among the cell types that develop the highest mechanical forces, comparable with those of muscle cells.5,6 Platelet functions are, therefore, dependent on a continuous energy supply, and previous studies have shown that platelets use both glycolysis and mitochondrial respiration for adenosine triphosphate (ATP) production.7
Resting platelets are equipped with 5 to 8 elliptical mitochondria dispersed in the cytoplasm.8-10 Activated platelets, which are spread on the injured vessel or between fibrin fibers show a large increase in size compared with the resting platelets. This increase in surface area is possible because of the membrane reservoir present in the resting platelets, called the open canalicular system, which is evaginated during the spreading process.11 What happens to the few mitochondria present in the resting platelets after their activation and spreading has not been studied so far. Are they redistributed within the larger sized, spread platelets or do they stay clustered in the center? Using expansion microscopy, live imaging, and focused ion beam–scanning electron microscopy (FIB-SEM), we analyzed the impact of the activation and spreading process on platelet mitochondria. We show that mitochondria undergo a fission process during platelet activation, leading to a higher number of smaller mitochondria in the activated, spread platelets than in the resting platelets. This fission step of platelet mitochondria coincides with a switch from a predominantly mitochondrial energy supply (as previously shown)12,13 to a more glycolytic energy production during clot retraction. Mitochondrial fission is dependent on the dynamin related protein 1 (Drp1), and Drp1 knock out (KO) platelets show a defect in clot retraction.
Materials and methods
Reagents
The following reagents were used: neutral formalin (HT5012-1CS, Sigma-Aldrich, Saint-Quentin Fallavier, France), Pluronic F-127 (P2443, Sigma-Aldrich), oligomycin (O4876, Sigma-Aldrich), rotenone (R8875, Sigma-Aldrich), antimycin (A8674, Sigma-Aldrich), 2-deoxy-D-glucose (D8375, Sigma-Aldrich), Mowiol 4 to 88 (475904, Calbiochem), Tubulin Tracker (T34075, Invitrogen), poly-d-lysine (P7280, Sigma-Aldrich), thrombin (T4648, Sigma-Aldrich), mitoTracker Red CMXRos (M7512, Invitrogen), MitoView Fix 640 (Biotium), NHS-Ester DyLight 650 (62265, Thermo Scientific), Mdivi-1 (ab144589, Abcam), y-27632 (688000, Calbiochem), blebbistatin (203391, Calbiochem), and M199 media (Thermo Scientific).
The following antibodies were used: mouse anti–α-tubulin (clone B-5-1-2; T5168; Sigma-Aldrich), mouse anti-integrin αIIb (also called CD41; clone M-148; and sc-7310; Santa-Cruz), rabbit anti-TOMM20 (ab78547; Abcam), Alexa Fluor 488 goat anti–mouse IgG (115-545-003; Jackson), and Cy3 goat anti-rabbit IgG (111-165-003; Jackson).
Preparation of human PRP
Nontherapeutical buffy coats from the French blood bank were diluted with an equal volume of phosphate-buffered saline (PBS), centrifuged for 10 minutes at 400g at room temperature (RT), and the upper phase corresponding to the platelet-rich plasma (PRP) was collected. For some experiments, PRP was prepared from whole blood drawn into sodium-citrate vacutainers with, essentially, the same results. In this case, the blood was not diluted with PBS before centrifugation.
Clot retraction assays
Two different clot retraction assays were used. For classical clot retractions (Figures 5 and 7), the clot was induced in glass tubes (coated with the surfactant pluronic at 2% and washed with PBS before clot induction). PRP was adjusted to a platelet concentration of 1 × 108/mL with PBS and a small number of erythrocytes were added for color contrast. Recalcification of the PRP was not necessary because efficient clot retraction was always observed. Clot formation was initiated by the addition of 10 μL thrombin (2.5 U/mL final concentration) to 400 μL adjusted PRP.
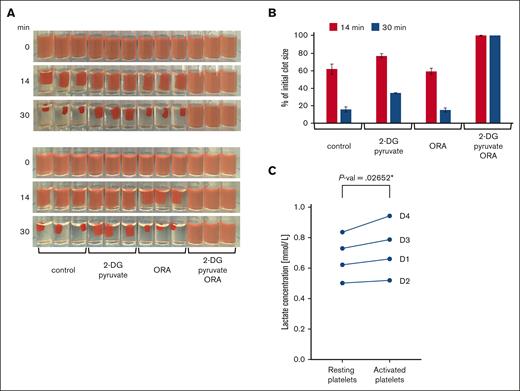
Energy production in platelets during clot retraction. (A) Clot retraction in the presence of glycolysis and oxphos inhibitors. PRP was adjusted to a platelet concentration of 1 × 108/mL with PBS and then preincubated for 20 minutes at RT with vehicle as control or the glycolysis inhibitor 2-deoxy-D-glucose (2-DG, 30 mM, supplemented with 2 mM pyruvate) or a combination of mitochondrial inhibitors, 4 μM oligomycin, 5 μM rotenone, 15 μM antimycin (ORA) as well as with both the glycolysis and oxphos inhibitors (2-DG + pyruvate/ORA). Clot formation was then initiated by addition of thrombin to a final concentration of 2.5 U/mL and clots were kept at RT. Shown are triplicate samples, and photos were taken at different time points during clot retraction as indicated. The retraction assay was repeated twice with the same samples. In the upper panels, clot formation was induced successively from left to right (ie, beginning with control conditions) and in the bottom panels, from right to left (ie, beginning with 2-DG+pyruvate/ORA conditions). This allows to compensate for the delay in the onset of clot induction. Images are representative of 4 independent experiments using platelets from 4 different donors. (B) The extent of clot retraction was quantified as percent of the initial clot volume using the images shown in panel A. Results are expressed as means ± standard deviations. (C) Quantification of lactate release during clot retraction. The concentration of lactate was determined in the serum extruded from triplicates of control clots from 4 different donors (D1-D4) after 30 minutes of retraction and compared with parallel triplicate samples of resting platelets (“Methods”). Data comparison was done using paired 2-tailed t test.
Energy production in platelets during clot retraction. (A) Clot retraction in the presence of glycolysis and oxphos inhibitors. PRP was adjusted to a platelet concentration of 1 × 108/mL with PBS and then preincubated for 20 minutes at RT with vehicle as control or the glycolysis inhibitor 2-deoxy-D-glucose (2-DG, 30 mM, supplemented with 2 mM pyruvate) or a combination of mitochondrial inhibitors, 4 μM oligomycin, 5 μM rotenone, 15 μM antimycin (ORA) as well as with both the glycolysis and oxphos inhibitors (2-DG + pyruvate/ORA). Clot formation was then initiated by addition of thrombin to a final concentration of 2.5 U/mL and clots were kept at RT. Shown are triplicate samples, and photos were taken at different time points during clot retraction as indicated. The retraction assay was repeated twice with the same samples. In the upper panels, clot formation was induced successively from left to right (ie, beginning with control conditions) and in the bottom panels, from right to left (ie, beginning with 2-DG+pyruvate/ORA conditions). This allows to compensate for the delay in the onset of clot induction. Images are representative of 4 independent experiments using platelets from 4 different donors. (B) The extent of clot retraction was quantified as percent of the initial clot volume using the images shown in panel A. Results are expressed as means ± standard deviations. (C) Quantification of lactate release during clot retraction. The concentration of lactate was determined in the serum extruded from triplicates of control clots from 4 different donors (D1-D4) after 30 minutes of retraction and compared with parallel triplicate samples of resting platelets (“Methods”). Data comparison was done using paired 2-tailed t test.
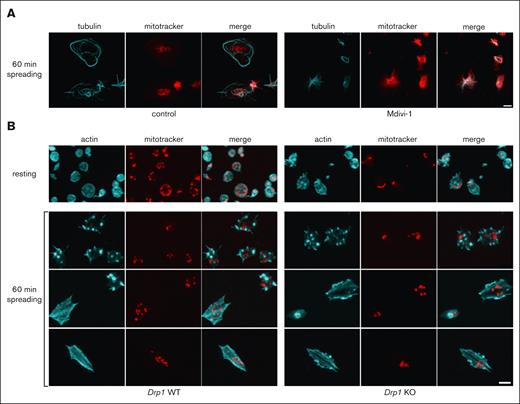
Drp1 is essential for mitochondrial organization in platelets. (A) Resting platelets were stained with mitoTracker (red) and then incubated for 30 minutes at RT with vehicle (control) or with the Drp1 inhibitor, Mdivi-1, at a final concentration of 50 μM. Platelets were then centrifuged onto the glass surface and placed into the incubator for 60 minutes at 37°C. Platelets were stained for tubulin (cyan) and shown are maximal projections of z-stacks. Scale bar, 2.5 μm (B) Resting and spread platelets of Drp1 WT and KO mice were incubated with the MitoView Fix 640, then either fixed immediately (resting) or centrifuged onto fibrinogen coated glass coverslips and incubated for 60 minutes at 37°C. Platelets were stained for actin (cyan) and shown are maximal projections of z-stacks. Scale bar, 2.5 μm
Drp1 is essential for mitochondrial organization in platelets. (A) Resting platelets were stained with mitoTracker (red) and then incubated for 30 minutes at RT with vehicle (control) or with the Drp1 inhibitor, Mdivi-1, at a final concentration of 50 μM. Platelets were then centrifuged onto the glass surface and placed into the incubator for 60 minutes at 37°C. Platelets were stained for tubulin (cyan) and shown are maximal projections of z-stacks. Scale bar, 2.5 μm (B) Resting and spread platelets of Drp1 WT and KO mice were incubated with the MitoView Fix 640, then either fixed immediately (resting) or centrifuged onto fibrinogen coated glass coverslips and incubated for 60 minutes at 37°C. Platelets were stained for actin (cyan) and shown are maximal projections of z-stacks. Scale bar, 2.5 μm
To perform immunofluorescence stainings of individual platelets within a clot, we developed a clot retraction assay around 2 sterile inoculation loops as previously described (Figure 2A-D).14 Briefly, as mentioned earlier, clot formation was induced by the addition of thrombin and immediately transferred to 2 mL eppendorf tubes coated with 2% pluronic and containing the inoculation loops. After 15 or 60 minutes of retraction, clots were fixed for 1 hour with isotonic formalin. Clots were then washed with PBS and incubated overnight at 4°C in 1 mL PBS or 15% sucrose. Clots were further incubated in 1 mL PBS/15% sucrose/7.5% gelatin at 37°C for 4 hours, and then inclusion blocks were formed in the same solution on ice. Blocks were then trimmed and snap frozen for 1 minute in isopentan at −60°C and stored at −80°C until 14 μm sections were cut using a cryostat.
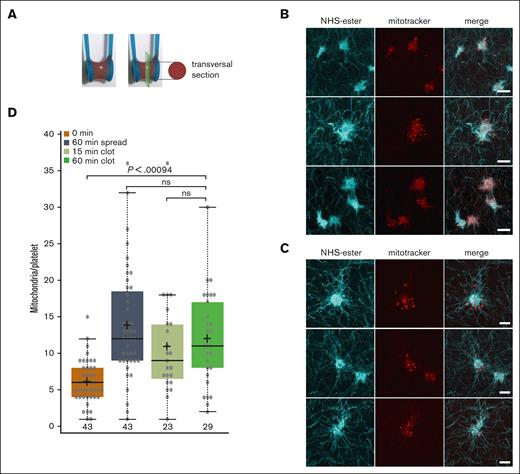
The number of mitochondria in platelets within a clot is similar to that in platelets activated by spreading on a glass surface. (A) Illustration of a clot retraction assay around 2 holders instead of the classical clot retraction assay used in Figures 5 and 7. The strong clot adhesion to both holders allows clot retraction but prevents clot collapse even after prolonged incubation times, thus allowing to visualize individual platelets within the retracted clot as shown in panels B and C. (B) PRP was adjusted to a platelet concentration of 1 × 108/mL with PBS and then preincubated for 30 minutes at RT with mitoTracker (red). Clot formation was then initiated by the addition of thrombin to a final concentration of 2.5 U/mL, and clots were incubated for 15 minutes at 37°C and then fixed, embedded in gelatin, and flash frozen. Transversal clot slices of 14 μm thickness were processed for expansion and then stained using NHS-ester (cyan) to detect the entire platelet dimensions. Shown are maximal intensity projections of confocal image stacks of 3 representative examples, scale bar 10 μm, corresponding to 2.5 μm after correction for expansion. (C) Same conditions as in panel A, but retraction was allowed to proceed for 60 instead of 15 minutes at 37°C. (D) Comparison of the number of mitochondria/platelet in resting and spread platelets (as shown in Figure 1G), with the number of mitochondria per platelet in blood clots after 15 minutes (n = 23; 2 different donors) and 60 minutes (n = 29; 2 different donors) of retraction. Data comparison was performed using 1-way analysis of variance.
The number of mitochondria in platelets within a clot is similar to that in platelets activated by spreading on a glass surface. (A) Illustration of a clot retraction assay around 2 holders instead of the classical clot retraction assay used in Figures 5 and 7. The strong clot adhesion to both holders allows clot retraction but prevents clot collapse even after prolonged incubation times, thus allowing to visualize individual platelets within the retracted clot as shown in panels B and C. (B) PRP was adjusted to a platelet concentration of 1 × 108/mL with PBS and then preincubated for 30 minutes at RT with mitoTracker (red). Clot formation was then initiated by the addition of thrombin to a final concentration of 2.5 U/mL, and clots were incubated for 15 minutes at 37°C and then fixed, embedded in gelatin, and flash frozen. Transversal clot slices of 14 μm thickness were processed for expansion and then stained using NHS-ester (cyan) to detect the entire platelet dimensions. Shown are maximal intensity projections of confocal image stacks of 3 representative examples, scale bar 10 μm, corresponding to 2.5 μm after correction for expansion. (C) Same conditions as in panel A, but retraction was allowed to proceed for 60 instead of 15 minutes at 37°C. (D) Comparison of the number of mitochondria/platelet in resting and spread platelets (as shown in Figure 1G), with the number of mitochondria per platelet in blood clots after 15 minutes (n = 23; 2 different donors) and 60 minutes (n = 29; 2 different donors) of retraction. Data comparison was performed using 1-way analysis of variance.
To determine the effects of the loss of Drp1 on clot retraction, washed platelets from wild type (WT) and Drp1 KO mice were resuspended in Tyrode buffer at 3 × 108/mL. Fibrinogen (800 μg/mL) and CaCl2 (12.5 mM) were added, and clot formation was initiated by adding thrombin (1.7 U/mL). The clots were allowed to retract, and images were taken at indicated time points.
Spreading assay
PRP was diluted in PBS to a concentration of 2.5 × 106 platelets/mL (plasma concentrations were kept constant at 0.3%). To stain mitochondria, 2 μL of 1mM mitoTracker was added to 10 mL of platelet suspension, which was then incubated for 20 minutes at RT in the dark, and 400 μL were then transferred into each well of a 24 well plate containing coverslips (12 mm diameter). The plate was centrifuged for 3 minutes at 600 g at RT to allow synchronized contact of all platelets with the glass surface and placed in the incubator. After 60 minutes of spreading, platelets were fixed with 400 μL isotonic formalin (9 volumes formalin / 1 volume 10 × PBS) for 15 minutes at RT and then kept in PBS at 4°C until further use.
To fix resting platelets (0 minute spreading time point), 400 μL of the same platelet suspension was added to wells of a 24 well plate containing poly-D-lysine-coated coverslips and 800 μL isotonic formalin. After 10 minutes of incubation at RT, plates were centrifuged for 5 minutes, 600g at RT, to attach the fixed, resting platelets to the coated coverslips; after another 5 minutes of incubation at RT, the fixative was replaced by 1 mL of PBS, and the plates were kept at 4°C until further use.
For resting and spread mouse platelets, washed platelets from WT and Drp1 KO mice were resuspended in M199 media. For the resting condition, platelets in M199 media were stained with MitoView Fix 640 for 1 hour at 37°C and then fixed with 2% paraformaldehyde. For the spreading condition, MitoView Fix 640 labeled platelets were allowed to spread for 1 hour at 37°C, fixed with 2% paraformaldehyde, and imaged using spinning disk confocal microscope (Nikon) using 60× oil objective.
Immunofluorescence and microscopy
Fixed platelets were permeabilized with PBS/0.2% Triton X-100 for 15 minutes at RT and then incubated with blocking buffer (3% bovine serum albumin and 10% goat serum in PBS) for 1 hour at RT. Coverslips were then incubated for 2 hours with a primary antibody diluted in blocking buffer and then washed twice with PBS and once with PBS/0.2% Triton X-100 and incubated with a secondary antibody diluted in blocking buffer for 2 hours at RT. After 3 washing steps, coverslips were mounted on glass slides using Mowiol. Image acquisition was performed using a wide-field epi fluorescence microscope (BX41; Olympus), equipped with a Plan 100×/1.25 numerical aperture (NA) oil objective, a camera (DP70; Olympus), and the acquisition software analySIS (Olympus). The software ImageJ was used for image analysis.
Expansion microscopy
Expansion microscopy was essentially performed as described by Gambarotto et al.15 Briefly, fixed platelets on coverslips (diameter, 12 mm) were first immunostained for α-tubulin or the integrin αll subunit and then incubated in PBS/0.7% formaldehyde/1% acrylamide for 5 hours at 37°C. The coverslip was then placed upside down on an ice-cold 35 μL drop of gelation solution (PBS/19% sodium acrylate/10% acrylamide/0.1% bisacrylamide) and placed onto parafilm on ice immediately after the addition of tetramethylenediamine and ammonium persulfate, both to a final concentration of 0.5%. After 5 minutes of incubation on ice, the samples were transferred to 37°C for 1 hour. The coverslip plus gel facing up was then incubated in denaturation buffer (5.7% sodium dodecyl sulfate/0.2 M NaCI/50 mM tris(hydroxymethyl)aminomethane; pH 9) for 15 minutes at RT, with gentle agitation to detach the gel from the coverslip. The gel was then transferred into an eppendorf tube with fresh denaturation buffer and boiled for 30 minutes at 95°C. To expand the gel, it was placed into a large volume of water for expansion, and the water was changed twice before incubation over night at RT in fresh water.
Clot sections (14 μm thick) were prepared using a cryostat and recovered on superfrost plus slides. The sections were delimitated using a pap-pen and then incubated as mentioned earlier, except the drop of gelation solution was added to the slide kept on ice, and a coverslip was put on top. Before the incubation in denaturation solution, the coverslip was removed from the slide, the gel was trimmed to retain only the part of the gel above the clot section, and the gel above the gelatin inclusion part was removed. Denaturation and expansion of the gel was continued as described earlier. After the expansion, a part of the gel was shrinked in PBS and then incubated in 100 mM sodium bicarbonate (pH 8.3) and 20 μg/mL N-hydroxysuccinimide (NHS)-Ester DyLight 650 for 90 minutes at RT with agitation. Samples were then washed 3× 20 minutes with PBS/0.1% tween-20. As a nonspecific protein stain, the NHS-ester staining was used to reveal the fibrin fibers and the platelets within the clot.
For image acquisition of expanded samples, a confocal microscope (Nikon A1R+MP) equipped with a homemade adaptive optics corrector was used with a 40× long distance water immersion objective. The geometric optical aberrations were corrected both in excitation and detection light paths in the open loop mode with a deformable mirror (AlpAO DM97-15) inserted between the confocal scanning head and the microscope inverted stand (Nikon Ti2E). Local aberrations were sensed using the metrics of molecular brightness, derived from fluorescence fluctuations, and were iteratively corrected as individual Zernike modes. This configuration allowed to compensate for the refractive index mismatch, specimen tilt, and eventual heterogeneities, in depth, of the expanded samples and to bring back the confocal resolution to its diffraction limit in the vicinity of the imaged platelet.
Live cell imaging
For live imaging, platelets were diluted in PBS to 1.6 × 107/mL. Then 1 μL microtubule-tracker reagent (500 nM final) and 1 uL mitoTracker (200 nM final) were added to 1 mL of platelet suspension. After 30 to 60 min of incubation at RT, the suspension was diluted with an equal volume of PBS, and 500 μL was pipetted into 4-well slides (Lab-Tek; Thermo Fisher Scientific). Platelets were imaged at RT and activated by the addition of drops (∼36 μL per drop) of a diluted thrombin solution (0.125 U/mL PBS) until activation was observed (Figure 3A, final thrombin concentration is indicated in the figure legend).
3D confocal video microscopy was performed in single-photon counting mode with a confocal microscope (LSM 710 ConfoCor 3; Zeiss), 63×/1.4 NA Plan Apochromat objective, long pass 505-nm emission filter, and single-photon avalanche photodiode detector (ConfoCor 3), as described previously.3 The 488 nm excitation power at the objective output was only 30 nW (0.005% acousto-optic tunable filter transmission), ensuring no fluorescence photobleaching throughout the whole time-lapse acquisition and no laser-induced platelet activation, as compared with the nonirradiated zones at the end of the experiment. The pinhole was closed to 1 arbitrary unit, and the voxel size was set to 66 × 66 × 500 nm with a pixel dwell time of 6.3 microseconds. A z-stack of 6 planes spaced by 0.5 μm were imaged.
FIB-SEM
Platelets were fixed with 2.5% glutaraldehyde in 0.1 mol/L sodium cacodylate buffer containing 20 mg/mL sucrose (305 mOsm; pH 7.3) directly in the aggregation cuvette after addition of collagen (1.25 μg/ml) at the time points corresponding to shape change. The fixed samples were then prepared for FIB-SEM as described previously.9 Briefly, platelets were embedded in epon after a contrast-enhancing step, consisting of incubating the cells in 1.5% potassium ferrocyanide and 1% osmium tetroxide in 0.1 M sodium cacodylate buffer. The blocks were mounted on SEM stubs, coated with platinum/palladium, and examined under a Helios NanoLab dual beam microscope (Field Electron and Ion Company). Samples were milled with the FIB (30 kV; 2.5 nA) at a thickness of 20 nm per section and images were acquired at 2048 × 1768 in backscattering mode (3 kV; 1.4 nA). Three-dimensional models were computed using Amira software (version 5.4, Visualization Sciences Group). The plasma membrane, granulomere, and mitochondria were manually traced to generate a 3D representation of the intracellular membrane organization. The volumes of the 3D reconstructed objects, represented in triangular surface meshes, were determined with Amira software (n = 3 whole platelets).
Lactate release during clot retraction
PRP was adjusted to a platelet concentration of 1 × 108/mL with PBS, and a small number of erythrocytes were added for color contrast. Clot formation was initiated by the addition of thrombin to a final concentration of 2.5 U/mL. After 30 minutes of retraction at RT, the extruded serum was recovered and used for lactate quantification.
Parallel samples, to which no thrombin was added, so no clot was formed, were also incubated for 30 minutes at RT and then centrifuged (1 min; 12000 g; RT) to pellet the resting platelets. Thrombin was then added to the supernatant (platelet-free plasma) to induce coagulation, samples were then centrifuged (5 min; 16000 g; RT) to pellet the fibrin fibers of the unretracted formed clot, and the serum was recovered for lactate determination.
Lactic acid quantification was carried out by nuclear magnetic resonance spectroscopy on a Bruker 600 Avance Neo IVDr spectrometer with the Bruker B.I.Quant-PS 2.0 module and in accordance with associated standard operating protocols for plasma and serum (B.I.Methods) sample preparation and measurement.
Generation of Drp1 KO mice
All animal experiments complied with the regulatory standards of the University of Utah. Megakaryocyte/platelet-specific Drp1–null mice (KO) were generated by crossing mice harboring flox (Drp1 fl/fl) sites flanking exons 3 to 5 of the gene that codes for Drp1 (Dnm1l) with mice harboring Cre recombinase driven by the MK Pf4 promoter (Pf4-Cre). All experiments were performed using age- and sex-matched adult littermates aged 4 to 6 months. A detailed characterization of the Drp1 KO phenotype of megakaryocytes and platelets will be published elsewhere.
Results
Distribution of mitochondria in resting and spread platelets
Resting platelets have an average diameter of 2 to 4 μm and a thickness of 0.5 μm. The activation and spreading of platelets can be mimicked in vitro by letting platelets attach to and spread on a glass surface. The mean area covered by a projection of a resting, disk-shaped platelet is 7 μm2, whereas the average area covered by platelets spread on a glass surface is ∼30 μm2, and even areas up to 40 to 70 μm2 were frequently observed.16 The question, thus, arose as to where the 5 to 8 mitochondria present in resting platelets were localized within spread platelets. Were they concentrated in the center of the spread platelet, evenly distributed over the whole platelet, or localized at sites of high energy demands?
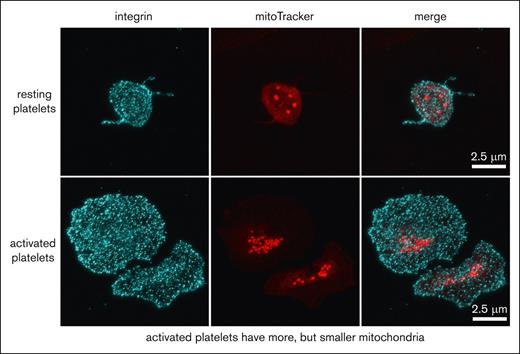
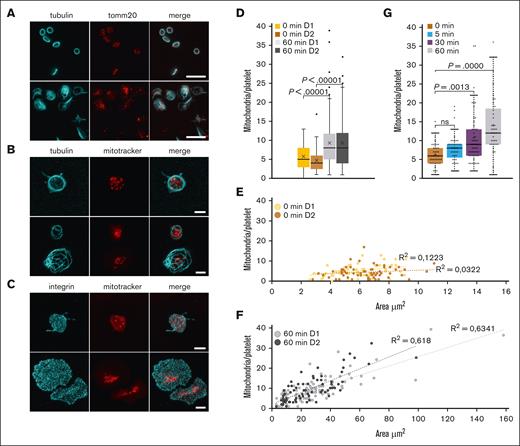
To compare the localization of mitochondria in the resting and spread platelets, we stained platelets for TOMM20, a specific translocase of the mitochondrial outer membrane. We observed a dot-like staining, and, as expected, in the resting platelets 5 to 8 dots were visible, which corresponded to the number of mitochondria in the resting platelets. Surprisingly, however, more TOMM20 spots were visible in the spread platelets (Figure 1A). The TOMM20 staining could result in >1 patch per mitochondrion, which could artificially increase the count of mitochondria per platelet. We, therefore, decided to confirm these observations by using a mitoTracker reagent as an alternative method to label mitochondria. Another concern was that the resolution limit of wide-field and confocal fluorescence microscopy is 200 to 500 nm. The diameter of mitochondria in resting platelets is also ∼200 to 500 nm and, thus, around the limit of resolution. To circumvent this problem, we performed expansion microscopy, which leads to an isometric fourfold increase of the sample size in all 3 dimensions, indirectly increasing the resolution of microscope acquisitions.17 Resting platelets were preincubated with a cell permeant mitoTracker, which accumulates in active mitochondria. This property presents an additional advantage because only functional mitochondria will be taken into account. Platelets were then either fixed or allowed to spread on a glass surface before fixation. The fixed platelets were stained for tubulin (Figure 1B) or the integrin subunit αIIb (Figure 1C) to detect the surface area occupied by the platelets. Samples were then either used directly for image acquisition (not shown) and quantification of mitochondria (Figure 1D-F) or processed for expansion before image acquisition and quantification (Figure 1B,C,G). We determined an average number of 5 mitochondria for resting platelets, which is in good agreement with results of previous studies.8,18,19 In platelets spread for 60 minutes, the average number of mitochondria was found to be 9 (Figure 1D). These results confirmed the initial observation of the TOMM20 staining, which suggested a higher number of mitochondria in the spread platelets than in the resting platelets. Furthermore, mitochondria appear to be smaller in the spread platelets than in the resting platelets and are not uniformly distributed but rather concentrated in the center of the spread platelet (Figure 1B-C).
The number of mitochondria per platelet differs between resting and spread platelets. (A) Detection of mitochondria using TOMM20 immunostainings (red). Microtubules were stained using a monoclonal antibody against α-tubulin (cyan). Epifluorescence images of resting (top) and spread platelets (bottom) are shown; scale bar, 10 μm. (B) Detection of mitochondria in platelets incubated for 20 minutes with a mitoTracker (red) before allowing them to spread for 60 minutes on a glass surface and then fixing them (bottom). Resting platelets were fixed in suspension and centrifuged onto PDL-coated coverslips (top). Platelets were then stained for α-tubulin (cyan) and processed for expansion. Maximum intensity projections of confocal image stacks are shown; scale bar, 10 μm, corresponding to 2.5 μm after correction for expansion. (C) Detection of mitochondria in platelets incubated for 20 min with a mitoTracker (red) before allowing them to spread for 60 min on a glass surface and then fixing them (bottom). Resting platelets were fixed in suspension and centrifuged onto PDL-coated coverslips (top). Platelets were then stained for the integrin subunit αIIb (cyan) and processed for expansion. Maximum intensity projections of confocal image stacks are shown; scale bar, 10 μm, corresponding to 2.5 μm after correction for expansion. (D) Quantification of the number of mitochondria in resting platelets after an incubation at 4°C to depolymerize microtubules and in platelets spread for 60 minutes at 37°C on a glass surface using maximal projections of confocal image stacks of unexpanded samples; results for 2 representative donors (D1 and D2) are shown. Data comparison was performed using the 2-tailed Mann-Whitney test. (E) Graph illustrating the number of mitochondria per resting platelet with respect to platelet size (2 representative donors used in panel D). (F) Graph illustrating the number of mitochondria per spread platelet with respect to platelet size (2 representative donors used in panel D). (G) Time course of platelet spreading on a glass surface and quantification of the number of mitochondria per platelet using maximal projections of confocal image stacks of expanded samples. Data comparison was performed using 1-way analysis of variance.
The number of mitochondria per platelet differs between resting and spread platelets. (A) Detection of mitochondria using TOMM20 immunostainings (red). Microtubules were stained using a monoclonal antibody against α-tubulin (cyan). Epifluorescence images of resting (top) and spread platelets (bottom) are shown; scale bar, 10 μm. (B) Detection of mitochondria in platelets incubated for 20 minutes with a mitoTracker (red) before allowing them to spread for 60 minutes on a glass surface and then fixing them (bottom). Resting platelets were fixed in suspension and centrifuged onto PDL-coated coverslips (top). Platelets were then stained for α-tubulin (cyan) and processed for expansion. Maximum intensity projections of confocal image stacks are shown; scale bar, 10 μm, corresponding to 2.5 μm after correction for expansion. (C) Detection of mitochondria in platelets incubated for 20 min with a mitoTracker (red) before allowing them to spread for 60 min on a glass surface and then fixing them (bottom). Resting platelets were fixed in suspension and centrifuged onto PDL-coated coverslips (top). Platelets were then stained for the integrin subunit αIIb (cyan) and processed for expansion. Maximum intensity projections of confocal image stacks are shown; scale bar, 10 μm, corresponding to 2.5 μm after correction for expansion. (D) Quantification of the number of mitochondria in resting platelets after an incubation at 4°C to depolymerize microtubules and in platelets spread for 60 minutes at 37°C on a glass surface using maximal projections of confocal image stacks of unexpanded samples; results for 2 representative donors (D1 and D2) are shown. Data comparison was performed using the 2-tailed Mann-Whitney test. (E) Graph illustrating the number of mitochondria per resting platelet with respect to platelet size (2 representative donors used in panel D). (F) Graph illustrating the number of mitochondria per spread platelet with respect to platelet size (2 representative donors used in panel D). (G) Time course of platelet spreading on a glass surface and quantification of the number of mitochondria per platelet using maximal projections of confocal image stacks of expanded samples. Data comparison was performed using 1-way analysis of variance.
To investigate whether there is a correlation between platelet size and the number of mitochondria per platelet, we measured the area occupied by individual platelets. Discoid resting platelets either lie as flat disks on the glass surface or are captured as upright standing disks and all intermediate situations can also be observed (Figure 1A, top, tubulin staining), which does not allow for obtaining a correct size of all resting platelets because of the lateral-axial resolution anisotropy of the optical microscopy. We, therefore, used a preincubation step at 4°C for resting platelets, which leads to depolymerization of the marginal microtubule bundle and consequently to a spherical shape, and the projection of confocal image stacks gives a good size indication for each platelet in the population. For spread platelets, we observed a strong correlation between platelet size and the number of mitochondria per platelet (Figure 1F); this was not the case for resting platelets (Figure 1E). The spreading time course shows that the longer the platelets can spread on the glass surface, the higher is the number of mitochondria (Figure 1G). Of note, the number of mitochondria in activated platelets spread for 60 minutes is about twice the number of mitochondria in resting platelets (median of 12 and 6, respectively). The spreading time course has been quantified using expansion microscopy and the better separation of individual mitochondria using this technique might explain the higher number of mitochondria at 60 minutes of spreading than in the boxplots for 60 minutes shown in Figure 1D.
Distribution of mitochondria within platelets in a clot
Under physiological conditions, activated platelets end up in a forming blood clot, and the fibrin fibers are pulled together by the platelets to retract the clot. Activated platelets trapped in a forming clot are spread between the fibrin fibers in 3 dimensions, extending filopodia and lamellipodia. Thus, in a clot, there is not a large 2D area covered by a spread platelet but rather a large volume of complex shape. The question is whether under these conditions, one can also observe a higher number of mitochondria within activated platelets in the clot and whether they also stay grouped in the center of the platelet or are distributed through the whole platelets, including the extremities of their plasma membrane extensions. To investigate the number, size, and distribution of mitochondria within individual platelets in a clot, we had to use a clot retraction assay that prevents complete clot collapse. We, thus, used an assay that we described previously14 and is illustrated in Figure 2A. PRP was preincubated with mitoTracker before clots were induced to form between 2 holders by the addition of thrombin. Clots were then fixed at indicated time points of retraction, and clot slices were analyzed using expansion microscopy. Expanded samples were also stained using an NHS-ester, a nonspecific protein staining reagent, to detect fibrin fibers and platelets. Similar to the platelets spread on a 2D surface, the number of mitochondria per platelet was higher (median of 11 after 60 min of retraction; Figure 2D) and mitochondria were smaller than in resting platelets. Most mitochondria were quite evenly distributed in the center, but several were located in extended regions of the platelets (Figure 2B-C).
A constriction step of mitochondria can be observed during platelet activation by live cell video microscopy and FIB-SEM
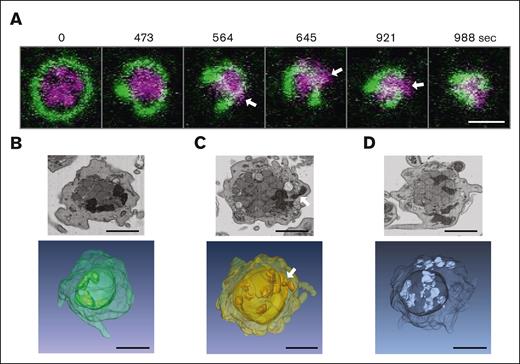
In nucleated cells, mitochondria have been described to be highly dynamic organelles that adapt to energetic needs by fission and fusion events, regulating their number and cellular distribution.20 It has been shown that the fission process can be induced by mechanical forces exerted on the mitochondrial membranes.21 In platelets, mitochondrial fission or fusion events have not been described so far. Our results showing a higher number of mitochondria in the activated platelets than the resting platelets suggest that mitochondria might undergo a fission process after or during platelet activation. To test this hypothesis, we followed the activation process by video microscopy. Resting platelets in suspension were incubated with a microtubule-tracker and a mitoTracker reagent. Using fast scan/low intensity confocal live cell imaging, we could follow in space and time the reorganization of microtubules and mitochondria during platelet activation induced by thrombin (Figure 3A; supplemental Video 1). During early time points, the time-lapse video shows that mitochondria get compacted in the center of the activating platelet as in a cage owing to the coiling of the marginal band and its compression by the surrounding acto-myosin cortex as described previously.3 At later time points, mitochondria seem to get squeezed through the encircled central space (Figure 3A, arrows). This strongly suggests that mechanical forces are applied on the mitochondria during the activation process, which may trigger mitochondrial fission, leading to the appearance of additional, smaller mitochondria in the periphery of activating platelets.
Compression and squeezing of mitochondria during platelet activation observed by live cell video microscopy and FIB-SEM images. (A) Live cell video microscopy of microtubule-tracker (green) and mitoTracker (magenta) stained human platelets during activation with thrombin (final concentration 0.034 U/mL). Still images of the supplemental Video 1 are shown at different time points as indicated. White arrows indicate mitochondria that get squeezed through the coiled marginal band; scale bar, 2 μm. (B-D) Single FIB-SEM image planes of platelets after activation with 1.25 μg/mL collagen (top) and below 3D reconstructions of FIB-SEM image stacks of platelets at activation states similar to those in the top panels. The white arrow indicates a mitochondrion that gets squeezed through the encircled center; scale bars, 1 μm.
Compression and squeezing of mitochondria during platelet activation observed by live cell video microscopy and FIB-SEM images. (A) Live cell video microscopy of microtubule-tracker (green) and mitoTracker (magenta) stained human platelets during activation with thrombin (final concentration 0.034 U/mL). Still images of the supplemental Video 1 are shown at different time points as indicated. White arrows indicate mitochondria that get squeezed through the coiled marginal band; scale bar, 2 μm. (B-D) Single FIB-SEM image planes of platelets after activation with 1.25 μg/mL collagen (top) and below 3D reconstructions of FIB-SEM image stacks of platelets at activation states similar to those in the top panels. The white arrow indicates a mitochondrion that gets squeezed through the encircled center; scale bars, 1 μm.
We wondered whether it would be possible to use FIB-SEM to observe situations reflecting this squeezing of mitochondria through the enclosed inner space during platelet activation. Indeed, careful examination of individual planes of FIB-SEM images as well as 3D reconstructions of FIB-SEM stacks of platelets fixed at the onset of collagen-induced activation shows platelets in which mitochondria are either compressed in the center (Figure 3B), squeezed through the encircled space (Figure 3C), or have escaped to peripheral locations (Figure 3D).
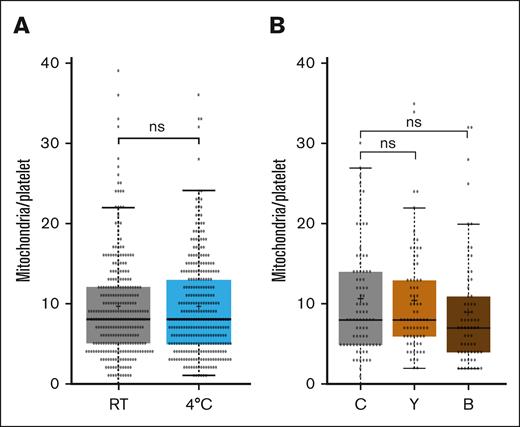
We then investigated whether the coiling process of the marginal band microtubules during platelet activation could play a role and facilitate the fission process. However, depolymerization of microtubules in resting platelets by incubation at 4°C before the activation and spreading process did not change the number of mitochondria in the spread platelets (Figure 4A). Similar to nucleated cells, it could probably be the acto-myosin–mediated compression that is providing the mechanical force to facilitate the fission process.22 However, again, the incubation with inhibitors of acto-myosin contraction did not allow for the clear confirmation of a role of the cytoskeleton (Figure 4B).
The fission of mitochondria during platelet activation/spreading does not depend on microtubules or acto-myosin contraction. (A) Comparison of the number of mitochondria in platelets spread for 60 minutes on a glass surface under different assay conditions. Resting platelets were stained with mitoTracker and then incubated for 30 minutes either at RT (intact microtubules) or at 4°C (depolymerized microtubules). Platelets were then centrifuged onto glass surface in the cold before being placed at 37°C for 60 minutes. The graph illustrates the number of mitochondria in platelets combined from 4 different donors. Data comparison was performed using the 2-tailed Mann-Whitney test. (B-C) Comparison of the number of mitochondria in platelets spread for 60 minutes on a glass surface under different assay conditions. Resting platelets were stained with mitoTracker and then incubated for 30 minutes at RT with vehicle (C), 10 μM y-27632 (Y) or 50 μM blebbistatin (B). Platelets were then centrifuged onto the glass surface and placed at 37°C for 60 minutes. The graph illustrates the number of mitochondria in platelets combined from 2 different donors. Data comparison was performed using the 2-tailed Mann-Whitney test.
The fission of mitochondria during platelet activation/spreading does not depend on microtubules or acto-myosin contraction. (A) Comparison of the number of mitochondria in platelets spread for 60 minutes on a glass surface under different assay conditions. Resting platelets were stained with mitoTracker and then incubated for 30 minutes either at RT (intact microtubules) or at 4°C (depolymerized microtubules). Platelets were then centrifuged onto glass surface in the cold before being placed at 37°C for 60 minutes. The graph illustrates the number of mitochondria in platelets combined from 4 different donors. Data comparison was performed using the 2-tailed Mann-Whitney test. (B-C) Comparison of the number of mitochondria in platelets spread for 60 minutes on a glass surface under different assay conditions. Resting platelets were stained with mitoTracker and then incubated for 30 minutes at RT with vehicle (C), 10 μM y-27632 (Y) or 50 μM blebbistatin (B). Platelets were then centrifuged onto the glass surface and placed at 37°C for 60 minutes. The graph illustrates the number of mitochondria in platelets combined from 2 different donors. Data comparison was performed using the 2-tailed Mann-Whitney test.
Platelets switch from a predominantly oxidative metabolism at resting state to a more glycolytic metabolism during clot retraction
Previous studies have shown that resting platelets have a high mitochondrial activity compared with other blood cell types.12,13 Although they can use both glycolysis and oxidative phosphorylation for energy production, they predominantly produce ATP via mitochondria. In nucleated cells, mitochondrial fission is associated with a shift to a more glycolytic phenotype. We, therefore, wondered whether the observed fission of mitochondria during platelet activation/spreading could coincide with the induction of a more glycolytic ATP production during late phases of platelet function. To investigate this hypothesis, we used a classical clot retraction assay. PRP was preincubated with inhibitors of glycolysis or oxidative phosphorylation before the addition of thrombin to induce platelet activation and fibrin clot formation. Surprisingly, the inhibition of oxidative phosphorylation has no influence on clot retraction, whereas clot retraction is retarded after the inhibition of glycolysis, and the inhibition of both ATP production pathways completely prevents clot retraction (Figure 5A,B).
These results demonstrate that both ATP supply pathways can compensate for each other. Moreover, at this stage of platelet function, glycolysis becomes more important than mitochondrial respiration, indicating a switch from a predominantly mitochondrial energy production at resting state13 to a more glycolytic energy production during clot retraction. To test this possibility, we compared lactate release rates of resting platelets and that of activated platelets during clot contraction. Our results show that more lactate is released by activated than resting platelets (Figure 5C). These findings are in agreement with earlier observations demonstrating an increase in aerobic glycolysis after thrombin-induced platelet activation.23,24
Mitochondrial fission in platelets depends on the Drp1
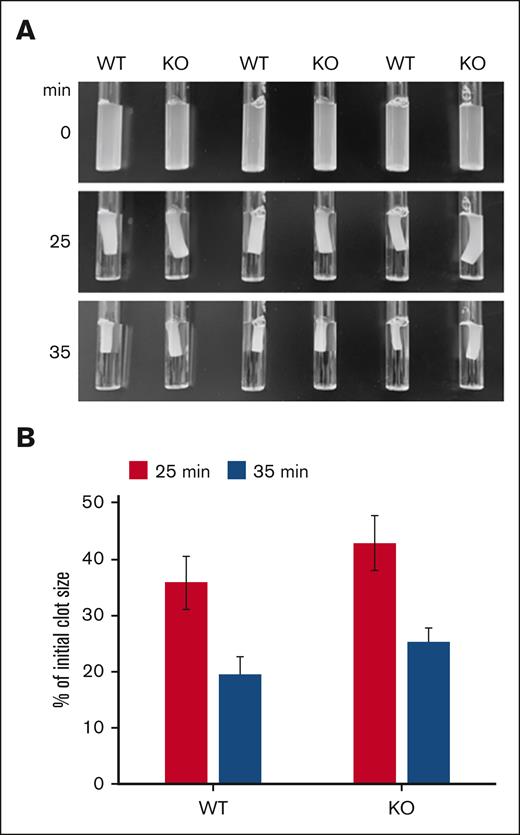
To obtain a mechanistic insight into the fission event of mitochondria in platelets, we decided to test whether Drp1 could be implicated in this process similar to that in nucleated cells.25 We used the Drp1 inhibitor Mdivi-1, which is known to inhibit mitochondrial fission in nucleated cells.26,27 When platelets were incubated with Mdivi-1 before allowing them to spread on a glass surface, it was difficult to distinguish individual mitochondria in the spread platelets (Figure 6A), indicating that mitochondria have a more fused morphology. Because the specifity of Mdivi-1 has been questioned,28 we decided to test the role of Drp1 in the fission process using a genetic approach. Platelets were isolated from WT and conditional Drp1 KO mice and incubated with a mitoTracker reagent (MitoView Fix 640). Although mitochondria of resting WT platelets showed the characteristic organization of punctate mitochondria, more fused mitochondria were observed in the resting platelets of Drp1 KO mice (Figure 6B). In the fully spread WT platelets, a higher number of mitochondria than those in the resting platelets was frequently observed (average of 10 vs 2-6, respectively), similar to human platelets. In platelets that were not completely spread and had an actin organization in the form of nodules,29 a similar number of mitochondria to the resting platelets could be seen. In contrast, even in the fully spread Drp1 KO platelets, the number of mitochondria was lower than in WT platelets and sometimes fused. We then wondered whether this phenotype could have an influence on clot retraction. As shown in Figure 7A-B, there is a reproducible retardation of clot retraction in KO vs WT clots.
Drp1-deficient platelets show a delay in clot retraction. (A) Washed platelets from WT and Drp1-KO mice were resuspended in Tyrode buffer at 3×108/mL. Fibrinogen (800 μg/mL) and CaCl2 (12.5 mM) were added, and clot formation was initiated by adding thrombin (1.7 U/mL). The clots were allowed to retract, and images were taken at indicated time points. (B) The extent of clot retraction was quantified as percent of the initial clot volume using the images shown in panel B. Results are expressed as means ± standard deviations (P = .03078 at 35 minutes). Shown is a representative experiment repeated 3 times.
Drp1-deficient platelets show a delay in clot retraction. (A) Washed platelets from WT and Drp1-KO mice were resuspended in Tyrode buffer at 3×108/mL. Fibrinogen (800 μg/mL) and CaCl2 (12.5 mM) were added, and clot formation was initiated by adding thrombin (1.7 U/mL). The clots were allowed to retract, and images were taken at indicated time points. (B) The extent of clot retraction was quantified as percent of the initial clot volume using the images shown in panel B. Results are expressed as means ± standard deviations (P = .03078 at 35 minutes). Shown is a representative experiment repeated 3 times.
Discussion
In platelets, motor mediated reorganization/contraction processes take place during the initial activation,3 the spreading,30 and during the sequential elongation and shortening of filopodia responsible for the retraction of blood clots.31 Especially during the process of clot retraction, platelets are able to develop strong mechanical forces similar to muscle cells.5,6 Thus, platelets need high amounts of energy for cytoskeletal reorganizations. How platelets manage to produce a sufficient amount of energy has been the subject of several studies, and our results confirm and extend previous observations.7,13,24,32-35 We show that platelets use both glycolysis and mitochondrial respiration for ATP production during clot retraction and that they are highly flexible, able to switch between these processes, thereby adapting to the environmental conditions. To our knowledge, we also demonstrate, for the first time, that the number of mitochondria per platelet increases after the activation process, whereas their size is reduced. Furthermore, for the spread but not the resting platelets, there is a strong correlation between the number of mitochondria and the size of the spread platelets, suggesting that the more mitochondria there are, the better the distribution of mitochondrial ATP supply36 and, thus, the more extensive the spreading process. Alternatively, it is possible that an extensive spreading process leads to a better separation of the divided mitochondria through transport along the cytoskeletal elements and, thus, the more the platelet is spread, the higher is the number of smaller individual mitochondria that can be observed.
The fission of the small number of mitochondria of the resting platelets during the activation and spreading process most probably relies on a mechanical stress-induced preconstriction step on the mitochondrial membrane.21 The sensing of the mitochondrial membrane curvature at the constricted site can then facilitate the recruitment of Drp1,37,38 which we have shown to be implicated in mitochondrial fission in platelets. We tested whether the coiling process of the marginal band microtubules during platelet activation could facilitate the fission process. However, platelets that were activated and spread after microtubule depolymerization still have a higher number of mitochondria than the resting platelets. This is in agreement with an earlier study in yeast demonstrating that the association of mitochondria with microtubules inhibits the fission process and that the number of mitochondria is inversely correlated with the microtubule length.39 The fact that upon platelet activation, the peripheral microtubule bundle first coils, the microtubules are then depolymerized,3 and mitochondrial fission occurs, is in agreement with the coupling of microtubule dynamics and mitochondrial fission frequencies observed in yeast. Inhibition of acto-myosin contraction during the activation process has also no pronounced effect on the fission process, which may suggest that multiple forces are necessary to induce the preconstriction of the mitochondrial membrane or that no special site on the small dot-like mitochondria has to be defined for the recruitment of Drp1.
What could be the reason and/or the consequences of mitochondrial fission leading to a higher number of smaller mitochondria in activated platelets? If mitochondrial functions, including ATP production but also, calcium homeostasis or apoptosis induction40 play an important role in the larger sized activated platelets, it seems fundamental that mitochondria are divided so that they can be more evenly distributed throughout the platelet. However, we did not observe a uniform distribution of mitochondria in platelets spread in 2 dimensions on a glass surface (Figure 1B-C). This might be due to the unphysiological activation conditions on glass, and, indeed, mitochondria are more evenly distributed in activated platelets within a clot (Figure 2B-C), an experimental condition, which is close to the physiological situation of platelets in a thrombus.
Another consequence might be that mitochondrial fission imposes a switch to a more glycolytic energy supply pathway. Indeed, in nucleated cells, it has been shown that mitochondrial fusion increases ATP production through oxidative phosphorylation, whereas fission leads to a more glycolytic phenotype.25,41 The change to a glycolytic phenotype could be particularly useful for platelets in a clot where oxygen availability should decline rapidly but ATP production needs to be maintained for the highly energetic process of clot retraction. Our experiment of clot retraction after inhibition of glycolysis or oxidative phosphorylation supports this hypothesis because clot retraction is more affected after the inhibition of glycolysis than after the inhibition of mitochondrial respiration, indicating a more important role of glycolysis at this stage of platelet function. Moreover, we show that more lactate is released by the activated platelets during clot retraction than by the resting platelets, further supporting a metabolic switch after platelet activation. Furthermore, platelets contain high amounts of glycogen, comparable with those in muscle cells, to fuel glycolysis throughout the whole process of clot retraction.42 The fact that Drp1-deficient platelets show a reduced capacity to retract blood clots could indicate that the switch to a more glycolytic phenotype is less efficient when mitochondrial fission does not take place. However, other causes such as reduced granule release could also account for the impaired clot retraction because Drp1 inhibition has been shown to impact granule exocytosis.43 Detailed analysis of Drp1 KO platelets will be necessary to determine the extent to which markers of platelet activation (increased cytosolic Ca+2, αIIbβ3 integrin activation, P-selectin expression, and phosphatidylserine exposure) depend on the mitochondrial fission event.
Future studies may reveal that a higher number of smaller mitochondria might also be an advantage for other processes in which platelets are known to be implicated, including immune response, infection, and inflammation and how to regulate the fission process in pathological situations.44
Acknowledgments
The authors thank Marine Laporte for her advice concerning expansion microscopy and Monika Dolega and Sandrine Fraboulet for critical reading of the manuscript.
This work was supported by Fondation pour la Recherche Médicale grant DEI20151234416 and University Grenoble Alpes grants AGIR-POLE FRAG15CS08, GM103806, and HL144957 (J.W.R.). Nuclear magnetic resonance analyses were carried out by the Grenoble Metabolomics and Lipidomics for Health platform, supported by Installations de Recherche et d'Innovation Centrées Entreprises funding from the Auvergne-Rhône-Alpes region.
Authorship
Contribution: A.G. developed the microscope with adaptive optics corrections and participated in manuscript writing; J.M. acquired images; A.E. and J.-Y.R. performed FIB-SEM experiments and image rendering; C.J. and A.-S.R. performed experiments; F.A., M.P., and E.D. helped with different image acquisition approaches and analyses; J.V. and B.E.-H. performed lactic acid quantifications; S.J. and J.W.R. generated Drp1 mice and associated data; L.L. participated in manuscript writing; K.S. initiated and planned the study, performed experiments, interpreted the results, and wrote the manuscript; and all authors read the manuscript and agreed to its content.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karin Sadoul, Research Center UGA/Inserm U 1209/CNRS 5309, Cytoskeletal Dynamics and Nuclear Functions, Site Albert Bonniot-Allée des Alpes, Institute for Advanced Biosciences, 38700 La Tronche, France; e-mail: karin.sadoul@univ-grenoble-alpes.fr.
References
Author notes
∗A.G. and S.J. contributed equally to this study.
Data are available on request from the corresponding author, Karin Sadoul (karin.sadoul@univ-grenoble-alpes.fr).
The full-text version of this article contains a data supplement.