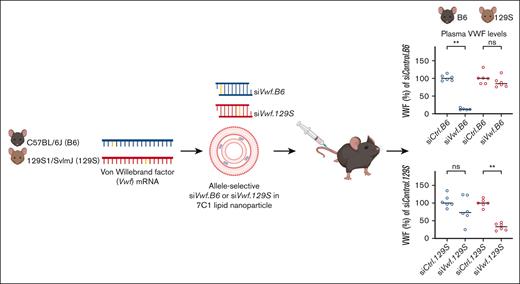
Key Points
Using 7C1 lipid nanoparticles, efficient endothelial delivery of siRNAs targeting VWF can be achieved.
Allele-selective silencing of VWF using siRNAs is feasible based on 1 nucleotide difference within mouse strains.
Abstract
An imbalance in von Willebrand factor (VWF) may either lead to bleeding (von Willebrand disease, VWD) or thrombosis. Both disorders have shortcomings in the currently available treatments. VWF itself could be a potential therapeutic target because of its role in both bleeding and thrombosis. Inhibiting VWF gene expression through allele-selective silencing of VWF with small interfering RNAs (siRNAs) could be a personalized approach to specifically inhibit mutant VWF in VWD or to normalize increased VWF levels in thrombotic disorders without complete VWF knockdown. Therefore, we investigated a method to allele-selectively silence the VWF gene in mice as a therapeutic strategy. Fourteen candidate siRNAs targeting murine Vwf of either the C57BL/6J (B6) or the 129S1/SvImJ (129S) strain were tested in vitro in cells expressing B6- and 129S-Vwf for inhibitory effect and allele-selective potential. Together with a nonselective siVwf, 2 lead candidate siRNAs, siVwf.B6 and siVwf.129S, were further tested in vivo in B6 and 129S mice. Efficient endothelial siRNA delivery was achieved by siRNA encapsulation into 7C1 oligomeric lipid nanoparticles. Treatment with the nonselective siVwf resulted in dose-dependent inhibition of up to 80% of both lung messenger RNA and plasma VWF protein in both mouse strains. In contrast, the allele-selective siVwf.B6 and siVwf.129S were shown to be effective in and selective solely for their corresponding mouse strain. To conclude, we showed efficient endothelial delivery of siRNAs that are highly effective in allele-selective inhibition of Vwf in mice, which constitutes an in vivo proof of principle of allele-selective VWF silencing as a therapeutic approach.
Introduction
Von Willebrand factor (VWF) is a multimeric plasma glycoprotein produced by endothelial cells and megakaryocytes.1 It has a pivotal role in primary hemostasis as it adheres platelets to the exposed subendothelial collagen at sites of vascular damage.2 VWF circulates in inactive form, but upon vascular damage, it binds to the exposed collagen, resulting in its conformational change and activation. This subsequently promotes the binding of VWF to platelets via the platelet glycoprotein Ibα receptor to the A1 domain of VWF.1,3 An imbalance in VWF levels is associated with bleeding and thrombotic disorders: low levels of functional VWF are associated with the bleeding disorder von Willebrand disease (VWD),3 whereas increased levels have been associated with thrombosis.4-7 For both disorders, there are shortcomings in the currently available treatments, such as therapeutic failure and increased bleeding risk.8-11 Given that VWF plays a role in both bleeding and thrombosis, VWF itself may be a potential therapeutic target for both sides of this spectrum.
VWF has been studied as a therapeutic target in the context of thrombotic disorders. Most strategies aim to inhibit VWF binding to platelets or subendothelial collagen by targeting the platelet-binding A1 domain12-16 or the collagen-binding A3 domain.17 A potential disadvantage of inhibiting the function of VWF is the increased risk of bleeding as binding to platelets is blocked and a hemostatic plug can no longer be formed.18 An alternative approach would be to block the translation of VWF messenger RNA (mRNA) into VWF protein, leading to lower but not zero plasma VWF levels. This can be achieved using small interfering RNAs (siRNAs). These short RNA sequences are an established class of US Food and Drug Administration-approved drugs that use the phenomenon of RNA interference to regulate gene expression.19 To avoid complete knockdown of the VWF gene, which would induce a risk of bleeding, the inhibition of VWF could be titrated by adjusting therapeutic dosages of siRNAs, but this will require continuous monitoring. Another approach for preventing complete knockdown of VWF is via allele-selective inhibition of only 1 of the VWF alleles. This can be achieved by designing siRNAs that target a single nucleotide mismatch between both alleles, either a mutation or a heterozygous single nucleotide polymorphism (SNP).20
VWF can also be the therapeutic target in VWD. Many mutations in VWF causing VWD have a dominant-negative effect because of the multimerization of mutant and normal VWF into dysfunctional multimers.21 Inhibiting the expression of the mutant allele will result in the expression of only normal VWF from the untargeted allele. Recently, we have shown the feasibility of this approach. In vitro and ex vivo allele-selective inhibition of mutant VWF led to a correction of the phenotype of the dominant-negative VWF mutation.22,23
Allele-selective VWF inhibition might thus be applied to thrombotic disorders by reducing VWF levels, resulting in a lower risk of (arterial) thrombosis, while at the same time maximizing the knockdown of VWF to roughly 50% to prevent bleeding risk. In contrast, allele-selective inhibition of dominant-negative VWF alleles in VWD may ameliorate the VWD phenotype. As high VWF plasma levels in thrombotic disorders are not caused by specific mutations in the VWF gene5,24 and as VWD is caused by many different mutations,25 in both cases it is not feasible to design siRNAs that target a specific mutation. To achieve inhibition of only 1 of the VWF alleles in thrombosis as well as in VWD, allele-selective siRNAs can be designed to target heterozygous SNPs in VWF that have a high minor allele frequency in humans.23,26
In this proof-of-principle study, we demonstrate that it is feasible to allele-selectively silence VWF in vivo in mice. As mouse inbred strains do not have SNPs in their Vwf genes, we use genetic differences between the Vwf genes of different mouse inbred strains as a proxy for the SNP-targeted approach. We designed strain-selective siRNAs targeting the Vwf gene of C57BL/6J and 129S1/SvImJ mice. We show an in vitro and in vivo proof of principle of siRNA-mediated allele-selective inhibition of murine Vwf, in which we demonstrate that it is possible with siRNAs to discriminate between Vwf genes of 2 different mouse inbred strains. In addition, we show that endothelial targeting of VWF in vivo can be achieved using chemically simple lipid nanoparticles (LNPs) without the use of targeting ligands.27
Methods
siRNA design and synthesis
Eleven genetic differences were found through sequence alignment for the Vwf gene of the 2 mouse inbred strains C57BL/6J (B6; Ensembl ENSMUSG00000001930) and 129S1/SvImJ (129S; Ensembl MGP_129S1SvImJ_G0031299). These mouse strains were chosen based on these differences and comparable plasma VWF levels. In-silico analysis by Axolabs GmbH (Kulmbach, Germany) predicted strain-selective siRNA sets (siVwf.B6 and siVwf.129S) specifically targeting the genetic differences (Figure 1A). In addition, a nonstrain-selective siRNA targeting Vwf of both B6 and 129S mouse strains was designed (siVwf). Scrambled controls (siControl) were designed based on the sequences of the siVwf and the chosen strain-selective siVwf.B6 and siVwf.129S. All siRNAs were chemically modified to improve stability and reduce immunogenicity, with a dT (DNA T) residue at the 5’-end of the antisense strand and a 5’ overhang, a phosphorothioate backbone modification at the 3’-end of both strands, and 2’O-methyl groups on 17 out of 19 residues in both strands.
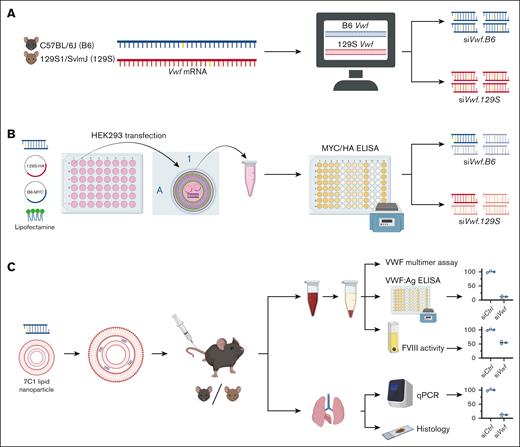
Schematic overview of the stepwise experimental approach to achieve siRNA-mediated allele-selective silencing of VWF; from in silico, to in vitro, and in vivo studies. (A) Using an alignment on the cDNA sequences of mouse inbred strains C57BL/6J (B6) and 129S1/SvImJ (129S), genetic differences between the strains’ Vwf gene were determined. Based on these genetic differences, an in-silico prediction analysis was performed to identify candidate strain-selective siRNAs. (B) Next, the candidate siRNAs were tested in vitro on activity and strain-selectivity. HEK293 cells were transiently co-transfected with a candidate siRNA and plasmids permitting expression of HA-tagged 129S-Vwf and MYC-tagged B6-Vwf using Lipofectamine 2000 as transfection reagent. VWF-MYC or VWF-HA protein was measured using an enzyme-linked immunosorbent assay (ELISA). siRNAs that were active and strain-selective in vitro were tested further in vivo on allele-selectivity. (C) For in vivo endothelial delivery, the lead candidate siRNAs were encapsulated in 7C1 LNPs. B6 or 129S mice were intravenously injected with nanoparticle-encapsulated siRNAs. Blood was drawn from the inferior vena cava, and plasma VWF protein levels were measured using ELISA. Lungs were harvested for measuring Vwf mRNA using quantitative polymerase chain reaction (qPCR) or VWF protein localization using immunofluorescent staining. This figure was created with Biorender.com.
Schematic overview of the stepwise experimental approach to achieve siRNA-mediated allele-selective silencing of VWF; from in silico, to in vitro, and in vivo studies. (A) Using an alignment on the cDNA sequences of mouse inbred strains C57BL/6J (B6) and 129S1/SvImJ (129S), genetic differences between the strains’ Vwf gene were determined. Based on these genetic differences, an in-silico prediction analysis was performed to identify candidate strain-selective siRNAs. (B) Next, the candidate siRNAs were tested in vitro on activity and strain-selectivity. HEK293 cells were transiently co-transfected with a candidate siRNA and plasmids permitting expression of HA-tagged 129S-Vwf and MYC-tagged B6-Vwf using Lipofectamine 2000 as transfection reagent. VWF-MYC or VWF-HA protein was measured using an enzyme-linked immunosorbent assay (ELISA). siRNAs that were active and strain-selective in vitro were tested further in vivo on allele-selectivity. (C) For in vivo endothelial delivery, the lead candidate siRNAs were encapsulated in 7C1 LNPs. B6 or 129S mice were intravenously injected with nanoparticle-encapsulated siRNAs. Blood was drawn from the inferior vena cava, and plasma VWF protein levels were measured using ELISA. Lungs were harvested for measuring Vwf mRNA using quantitative polymerase chain reaction (qPCR) or VWF protein localization using immunofluorescent staining. This figure was created with Biorender.com.
Plasmid expression vectors for B6- and 129S-Vwf and transfection
Recombinant pcDNA3.1/Zeo(+) containing full-length B6-Vwf complementary DNA (cDNA) (pcDNA3.1/Zeo(+)mVwf.B6) was used and modified. The 11 genetic differences between the B6 and 129S strains were introduced into pcDNA3.1/Zeo(+)mVwf.B6 using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA), resulting in pcDNA3.1/Zeo(+)mVwf.129S. To distinguish between the 2 Vwf alleles at the protein level, c-Myc (MYC) or hemagglutinin (HA) peptide tags were inserted, resulting in mVwf.B6-MYC and mVwf.129S-HA, respectively. Mutagenesis primers were designed with NEBaseChanger (New England Biolabs). Plasmids were purified with the PureYield Plasmid Maxiprep system (Promega, Madison, WI), and plasmid sequences were verified by Sanger sequencing (Leiden Genome Technology Center). Plasmid transfection was in human embryonic kidney 293 (HEK293) cells (ATCC, Rockville, MD) at 5 ng/mL, with or without siRNAs in different concentrations as described previously23 (Figure 1B).
Animal procedures
Male and female C57BL/6J (B6) mice (strain #000664, The Jackson Laboratory, Bar Harbor, ME) were obtained from Charles River, France, while 129S1/SvImJ (129S) mice were bred inhouse using breeder pairs from The Jackson Laboratory (strain #002448). Five- to 8-week-old mice received a single tail vein injection with 0.5, 1, or 1.5 mg siRNA per kg bodyweight (Figure 1C). For endothelial delivery, siRNAs were encapsulated in 7C1 oligomeric LNPs as described.27,28 After 72 hours, 96 hours, or 10 days of injection, mice were anesthetized by an intraperitoneal injection of a mixture of ketamine (100 mg/kg; Ketalin, aa-vet, Biddinghuizen, The Netherlands), xylazine (10 mg/kg; Sedamun 20 mg/mL, Dechra, Bladel, The Netherlands), and atropine-sulfate (0.1 mg/kg; Teva, Haarlem, The Netherlands). Citrated blood was collected from the inferior vena cava. Plasma was obtained by centrifugation at 12 000g (10 minutes) at room temperature (RT) and stored at −80°C. Euthanasia is followed by disruption of the aorta and inferior vena cava. Left lung lobules were harvested, snap frozen, and stored at −80°C, whereas right lung lobules were fixed with 10% formalin (24 hours), followed by storage in 70% ethanol. During experiments, mice were housed in conventional cages with a 12:12 hour light-dark cycle and ad libitum access to food and water. The experimental setup was in accordance with the institutional ethical guidelines. Sample sizes were based on non-parametric power calculations using G∗Power version 3.1.9.4.29 Randomization was done using RandoMice software,30 and the primary researcher remained blinded until data analysis.
Quantification of lung Vwf mRNA
Lung RNA isolation using RNA STAT-60 (Tel-Test Inc, Friendswood, TX) and complementary DNA synthesis were performed as described.31,32 An additional DNAse step was included (Turbo DNA-free kit, Invitrogen, Carlsbad, CA, USA). Lung Vwf transcript levels were determined by a quantitative polymerase chain reaction. The glyceraldehyde-3-phosphate dehydrogenase (Gapdh) gene (forward primer: ACTCCCACTCTTCCACCTTC; reverse: CACCACCCTGTTGCTGTAG) was used as the endogenous reference gene and analyzed within the same quantitative polymerase chain reaction run as the Vwf gene (forward: GCCTCAAGCAGAGCACAAAC, reverse: TCCTGCAGGCACAGGTAAAG) and F8 gene (forward: CTTCACCTCCAGGGAAGGACTA, reverse: TCCACTTGCAACCATTGTTTTG). Data were analyzed using the comparative Ct method and normalized against the median transcript level of control siRNA-treated animals.33
Quantification of mouse VWF protein levels in conditioned medium and plasma
VWF:MYC and VWF:HA proteins were measured in conditioned medium and VWF:Ag in plasma. Ninety-six–wells enzyme-linked immunosorbent assay plates (ELISA, Greiner, Frickenhausen, Germany) were coated overnight (4°C) with polyclonal rabbit anti-VWF immunoglobulin G (IgG) antibody (A0082-4.1 g/L, DAKO, Glostrup, Denmark) diluted 1:5000 in coating buffer (100 mM NaHCO3, 500 mM NaCl, pH 9.0) for VWF:Ag and VWF:MYC, and with polyclonal sheep anti-VWF IgG (ab11713-1 mg/mL, Abcam, Cambridge, United Kingdom; 1:1000 in coating buffer) for VWF:HA. Recombinant mVWF-WT, mVWF-MYC, and mVWF-HA served as references and were normalized to VWF:Ag levels of wild-type B6 normal mouse plasma (NMP). Samples and detection antibodies were diluted in phosphate-buffered saline (PBS) with 0.1% Tween-20 (Sigma-Aldrich, Saint Louis, MO) or, for VWF:HA, in PBS with 3% bovine serum albumin (BSA). Diluted samples were incubated for 2 hours RT. For VWF:HA, before sample incubation, wells were blocked with PBS with 3% BSA for 30 minutes. Wells were incubated with detection antibodies, rabbit anti-human VWF-IgG coupled to horseradish peroxidase (HRP) (P0226-1.3 g/L, DAKO; 1:1000) for VWF:Ag and rabbit anti-MYC-IgG-HRP (A00173-0.5 mg/mL, Genscript, Piscataway, NJ; 1:4000) for VWF:MYC, for 2 hours RT. Incubation was with rabbit anti–HA-IgG (C29F4-66 μg/mL, Cell Signaling, Leiden, The Netherlands; 1:1500) for VWF:HA for 1 hour RT, followed by 1-hour incubation with goat anti-rabbit IgG-HRP (172-1019, Bio-Rad, Veenendaal, The Netherlands; 1:8000). HRP detection was with O-phenylenediamine dihydrochloride (Sigma-Aldrich).
VWF Multimer analysis
Preparation and quantification of platelet lysates
Whole blood was diluted 1:4 in wash buffer (36 mM C₆H₈O₇, 103 mM NaCl, 5 mM KCl, 5 mM EDTA, 5.6 mM glucose, pH 6.5), centrifuged at 150g to obtain platelet-rich plasma, and centrifuged at 800g to pellet platelets. Platelet pellets were washed twice in wash buffer with 1% 1 mM prostaglandin E1 (P8908, Sigma-Aldrich), followed by lysis in lysis buffer (1% [v/v] Triton X-100, 10% [v/v] glycerol, 50 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, pH 7.4; containing 1:100 Complete Protease Inhibitor Cocktail [Sigma-Aldrich]). Measurement was with a modified VWF:Ag ELISA. Adjustments included sample incubation overnight at 37°C and detection with biotinylated rabbit anti-VWF (1.5 hours) diluted in PBS with 1% BSA and streptavidin poly-HRP (21140-0.5 mg/mL, Thermo Fisher Scientific, Leiden, The Netherlands; 45 minutes), RT. Values were normalized to platelet counts measured in whole blood using a Sysmex automated cell counter.
Immunofluorescent imaging of VWF
Formalin-fixed paraffin-embedded lungs were sectioned (5 μm), deparaffinized, and rehydrated. Antigen retrieval was with tris(hydroxymethyl)aminomethane-ethylenediaminetetraacetic acid (Tris-EDTA) buffer, pH 9.0. Blocking was with PBS with 3% BSA (2 hours, RT), followed by incubation with the primary antibody rabbit anti-mouse VWF (A0082-4.1 g/L, DAKO; 1:1000 in blocking buffer) (4 hours, 4°C). For detection (30 minutes, RT), goat anti-rabbit IgG AF568 (A11-11-2 mg/mL Invitrogen; 1:750 in blocking buffer) was used. Nuclear counterstaining was with Hoechst 33258 (H3569-10 mg/mL, Thermo Fisher Scientific) and mounting with ProLong Gold (Life Technologies, Bleiswijk, The Netherlands). Fluorescent images were made using the ZEISS Axio Scan.Z1 slide scanner and processed with ZEN blue 3.6 software.
Quantification of plasma FVIII activity
Plasma coagulation factor VIII (FVIII) was measured using a 2-stage modified activated partial thromboplastin time (APTT) clotting assay.35 Samples were diluted 1:5 in Owren-Koller buffer (Stago, Leiden, The Netherlands) and mixed with equal volume (25 μL) human factor VIII–deficient plasma (STA-ImmunoDef VIII; Stago) and 50 μL APTT reagents (Triniclot Automated APTT; Stago). After a 3-minute incubation at 37°C (STA Start clotting machine; Stago) and recalcification with 50 μL prewarmed 25 mM CaCl2 (Stago), clotting time was recorded. NMP was used as a reference for calculating FVIII concentration.
Statistical analysis
Graphic illustrations were generated, and statistical analysis was performed using GraphPad Prism 9.3.1 (GraphPad Software, La Jolla, CA). All in vitro data are represented as mean ± standard deviation, and all in vivo data are represented as median with range. For in vivo data, Mann-Whitney U or Kruskal-Wallis tests were used for testing between 2 or 3 experimental groups, respectively. P ≤ 0.05 was considered statistically significant.
Results
In-silico design and in vitro selection of mouse strain-selective siRNAs targeting Vwf
From 139 possible siRNA sequences per mouse strain, 7 candidate strain-selective siRNAs were selected for both the B6 and 129S strains that were predicted to be (i) active inhibitors of Vwf, (ii) specific, meaning <25 off-targets matched with 2 mismatches predicted for antisense strand and siRNA sequence is not identical to (conserved) microRNA seed regions in mice and (preferably) other species, and iii) strain-selective for Vwf of either the B6 or 129S mouse (Table 1). A nonselective siVwf was chosen based on the first 2 criteria and full complementarity to both B6- and 129S-Vwf mRNA.
Overview of selected candidate siRNAs with their target nucleotide(s)
| ID . | Target . | . | siRNA sequence . |
|---|---|---|---|
| siVwf | B6/129S | c.6348-c.6365 | 5'-dTAUGUUUUCACCaACGUACgsg-3' |
| siVwf.B6-1 | B6 | c.848G>A; c.852T>C | 5'-dTCGUUUACACCGcUGUUCCusc-3' |
| siVwf.B6-2 | B6 | c.1010T>C | 5'-dTGUACACAAAAUcUUCUCAcsa-3' |
| siVwf.B6-3 | B6 | c.1010T>C | 5'-dTCGUACACAAAAuCUUCUCasc-3' |
| siVwf.B6-4 | B6 | c.1010T>C | 5'-dTACGUACACAAAaUCUUCUcsa-3' |
| siVwf.B6-5 | B6 | c.2745T>C; c.2754A>G | 5'-dTGUACUCUGCACcCUGAUGgsa-3' |
| siVwf.B6-6 | B6 | c.2996G>A | 5'-dTUCACCGAGGGAuAGCUGCasa-3' |
| siVwf.B6-7 | B6 | c.2996G>A | 5'-dTUUCACCGAGGGaUAGCUGcsa-3' |
| siVwf.129S-1 | 129S | c.1089G>T; c.1090A>C | 5'-dTGACCUUCCUGGgCGCAGGusu-3' |
| siVwf.129S-2 | 129S | c.1089G>T; c.1090A>C | 5'-dTAGACCUUCCUGgGCGCAGgsu-3' |
| siVwf.129S-3 | 129S | c.2739C>T | 5'-dTAAACAGGGACAcCGCUCCasa-3' |
| siVwf.129S-4 | 129S | c.2745T>C; c.2754A>G | 5'-dTUACUCCGCACCcUGGUGGasa-3' |
| siVwf.129S-5 | 129S | c.2745T>C; c.2754A>G | 5'-dTGUACUCCGCACcCUGGUGgsa-3' |
| siVwf.129S-6 | 129S | c.2996G>A | 5'-dTGUUGCAACCCUcAUUUCCcsa-3' |
| siVwf.129S-7 | 129S | c.2996G>A | 5'-dTUUCACCGAGGGaUAGUUGcsa-3' |
| siControl | - | scrambled siVwf | 5'-dTGAUAUUUCCCUcAGUAACgsg-3' |
| siControl.B6 | - | scrambled siVwf.B6 | 5'-dTAUAUCCAAUUAcFCCUAAcsa-3' |
| siControl.129S | - | scrambled siVwf.129S | 5'-dTCGACAUCGAACgCGCACAasa-3' |
| ID . | Target . | . | siRNA sequence . |
|---|---|---|---|
| siVwf | B6/129S | c.6348-c.6365 | 5'-dTAUGUUUUCACCaACGUACgsg-3' |
| siVwf.B6-1 | B6 | c.848G>A; c.852T>C | 5'-dTCGUUUACACCGcUGUUCCusc-3' |
| siVwf.B6-2 | B6 | c.1010T>C | 5'-dTGUACACAAAAUcUUCUCAcsa-3' |
| siVwf.B6-3 | B6 | c.1010T>C | 5'-dTCGUACACAAAAuCUUCUCasc-3' |
| siVwf.B6-4 | B6 | c.1010T>C | 5'-dTACGUACACAAAaUCUUCUcsa-3' |
| siVwf.B6-5 | B6 | c.2745T>C; c.2754A>G | 5'-dTGUACUCUGCACcCUGAUGgsa-3' |
| siVwf.B6-6 | B6 | c.2996G>A | 5'-dTUCACCGAGGGAuAGCUGCasa-3' |
| siVwf.B6-7 | B6 | c.2996G>A | 5'-dTUUCACCGAGGGaUAGCUGcsa-3' |
| siVwf.129S-1 | 129S | c.1089G>T; c.1090A>C | 5'-dTGACCUUCCUGGgCGCAGGusu-3' |
| siVwf.129S-2 | 129S | c.1089G>T; c.1090A>C | 5'-dTAGACCUUCCUGgGCGCAGgsu-3' |
| siVwf.129S-3 | 129S | c.2739C>T | 5'-dTAAACAGGGACAcCGCUCCasa-3' |
| siVwf.129S-4 | 129S | c.2745T>C; c.2754A>G | 5'-dTUACUCCGCACCcUGGUGGasa-3' |
| siVwf.129S-5 | 129S | c.2745T>C; c.2754A>G | 5'-dTGUACUCCGCACcCUGGUGgsa-3' |
| siVwf.129S-6 | 129S | c.2996G>A | 5'-dTGUUGCAACCCUcAUUUCCcsa-3' |
| siVwf.129S-7 | 129S | c.2996G>A | 5'-dTUUCACCGAGGGaUAGUUGcsa-3' |
| siControl | - | scrambled siVwf | 5'-dTGAUAUUUCCCUcAGUAACgsg-3' |
| siControl.B6 | - | scrambled siVwf.B6 | 5'-dTAUAUCCAAUUAcFCCUAAcsa-3' |
| siControl.129S | - | scrambled siVwf.129S | 5'-dTCGACAUCGAACgCGCACAasa-3' |
Note: For both mouse strains, 7 strain-selective siRNAs were selected with the target nucleotide present in the B6 strain represented in bold and the corresponding nucleotide present in the 129S strain represented in roman font. siRNA sequences shown are of the antisense strands with the underscored nucleotides indicating the position of the nucleotide targets. Most siRNAs have only 1 nucleotide mismatch as target, whereas some have 2 mismatches. For the chosen in vivo candidate siRNAs, a corresponding scrambled siRNA was designed to act as a control. dT = DNA residue; G, C, A, U = RNA residue; g, c, a, u = 2’-O-methyl modified residue; s = phosphorothioate backbone modification.
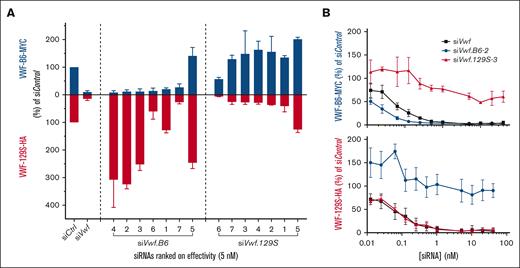
The 14 candidate siRNAs were screened for in vitro activity and selectivity in a concentration range of 5 to 20 nM in HEK293 cells transiently expressing Vwf from both B6 and 129S (Figure 2A; supplemental Figure 1). The nonselective siVwf showed strong inhibition of both Vwf from B6 (86%) and 129S (81%) at 5 nM as compared with siControl-treated cells. Of the 7 siRNA candidates predicted to target B6-Vwf, only 3 (siVwf.B6-2, -3, -4) were very potent inhibitors (>85% reduction on VWF-MYC) as well as strain-selective (no reduction on VWF-HA); 1 siRNA was strain-selective but less strong (siVwf.B6-1; 75% inhibition); 2 siRNAs (siVwf.B6-6, -7) inhibited VWF of both strains; and 1 siRNA (siVwf.B6-5) did not inhibit at all. Of the 7 candidates predicted to target 129S-Vwf, only 3 (siVwf.129S-3, -4, -7) were good inhibitors as well as strain-selective (>70% inhibition on VWF-HA); 2 (siVwf.129S-1, -2) were strain-selective but medium inhibitors (>55% inhibition); 1 siRNA inhibited VWF of both strains (siVwf.129S-6) and 1 siRNA did not inhibit at all (siVwf.129S-5). Remarkably, candidates with 2 genetic differences included in their nucleotide sequence were not more strain-selective than the siRNAs with 1 nucleotide difference. The overexpression of the nontargeted plasmid is assumed to not be a true overexpression but rather due to oversaturation in the case of the coexpressed plasmids vs the allowance of extra expression by 1 plasmid through inhibition of another.
In vitro selection of strain-selective siRNAs. (A) HEK293 cells were transfected with 5 nM candidate siVwf, siVwf.B6, siVwf.129S, or scrambled siControl together with plasmids expressing VWF-B6-MYC (blue bars) and VWF-129S-HA (red bars). Seventy-two hours after transfection, VWF-MYC and VWF-HA were measured in conditioned medium. For both B6 and 129S, siRNAs are ranked based on highest inhibitory effect in their corresponding strain relative to siControl. (B) HEK293 cells transfected with VWF-B6-MYC and VWF-129S-HA were cotransfected with siVwf (black squares), siVwf.B6-2 (blue circles), or siVwf.129S-3 (red triangles) at a concentration range of 0.0125 to 40 nM and MYC (VWF-B6, upper panel) and HA (VWF-129S, lower panel) were determined. Data are represented as the mean ± standard deviation (SD) of an n = 6.
In vitro selection of strain-selective siRNAs. (A) HEK293 cells were transfected with 5 nM candidate siVwf, siVwf.B6, siVwf.129S, or scrambled siControl together with plasmids expressing VWF-B6-MYC (blue bars) and VWF-129S-HA (red bars). Seventy-two hours after transfection, VWF-MYC and VWF-HA were measured in conditioned medium. For both B6 and 129S, siRNAs are ranked based on highest inhibitory effect in their corresponding strain relative to siControl. (B) HEK293 cells transfected with VWF-B6-MYC and VWF-129S-HA were cotransfected with siVwf (black squares), siVwf.B6-2 (blue circles), or siVwf.129S-3 (red triangles) at a concentration range of 0.0125 to 40 nM and MYC (VWF-B6, upper panel) and HA (VWF-129S, lower panel) were determined. Data are represented as the mean ± standard deviation (SD) of an n = 6.
siVwf.B6-2 and siVwf.129S-3 were further tested in a larger concentration range (0.0125-40 nM siRNA) as these were strain-selective as well as the most potent inhibitors of their corresponding strain at 5, 10, and 20 nM siRNA (Figure 2B). In addition, we selected these siRNAs based on a 1-nucleotide difference as this is more similar to the ultimate clinical SNP-based targeting approach. siVwf, siVwf.B6-2, and siVwf.129S-3 showed to be potent inhibitors (>70%) of their corresponding strain(s) even at lower dosages (<1 nM).
Feasibility of endothelial VWF inhibition in vivo
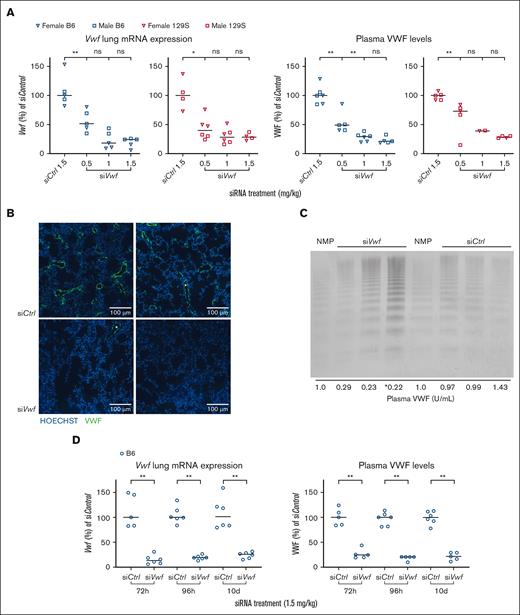
The nonselective siVwf was encapsulated in 7C1 LNPs, which has previously delivered siRNAs to endothelial cells in mice27 and non-human primates.28 After a single intravenous injection of nanoparticle-encapsulated siVwf both male and female B6 and 129S animals showed a dose-dependent reduction in plasma VWF (median inhibition 78.7% for the 1.5 mg/kg dose in B6 and 71.5% in 129S mice). Similar effects at 1.5 mg/kg dose were measured at mRNA level with 75.6% reduction in lungs of B6 mice and 71.9% in lungs of 129S mice (Figure 3A). siControl did not affect lung Vwf mRNA or plasma VWF at 1.5 mg/kg dose. Comparable, strong VWF reduction was observed for both male and female mice, therefore sexes were combined in subsequent experimental groups.
Feasibility of endothelial Vwf inhibition in vivo in mice using siVwf encapsulated in 7C1 oligomeric LNPs. (A) The nonselective siVwf was encapsulated in 7C1 LNPs and injected at a dose of 0.5 to 1.5 siRNA mg/kg in male (squares) and female (inverse triangles) B6 (blue symbols) and 129S (red symbols) mice. Lung Vwf mRNA (left 2 panels) and plasma VWF (right 2 panels) levels were determined and normalized to the scrambled siControl. (B) Images of immunofluorescent staining of 2 representative areas of the lungs of siControl- (left) and siVwf-treated (right) B6 mice. Images include nuclei staining (Hoechst) in blue and VWF in green. Scale bar is representative for 100 μm. (C) Representative VWF multimeric pattern of B6 mice treated with siVwf or siControl. Analysis was performed on platelet-free plasma, and NMP was used as a reference for normal multimer patterns. Samples loaded on the gel were diluted to a final sample concentration of 0.05 U/mL, except for ∗, where the sample concentration was accidentally 0.10 U/mL. At the bottom of this panel, the plasma VWF levels are indicated for the individual mice that were used for this VWF multimeric pattern analysis. (D) Duration of siRNA–mediated endothelial Vwf inhibition in B6 mice treated with siVwf (1.5 mg/kg) and at 72 hours, 96 hours, and 10 days after injection sacrificed for analysis of lung Vwf mRNA (left panel) and plasma VWF protein (right panel). n = 6 mice per group; however, 1 siControl-treated animal was removed because of a failed siRNA injection, incidentally a blood sample was removed because of unwanted clotting activation (as established visually). Values are presented as median with range. P ≥ 0.05, denotes not significant (ns); ∗P ≤ 0.05; ∗∗P ≤ 0.01.
Feasibility of endothelial Vwf inhibition in vivo in mice using siVwf encapsulated in 7C1 oligomeric LNPs. (A) The nonselective siVwf was encapsulated in 7C1 LNPs and injected at a dose of 0.5 to 1.5 siRNA mg/kg in male (squares) and female (inverse triangles) B6 (blue symbols) and 129S (red symbols) mice. Lung Vwf mRNA (left 2 panels) and plasma VWF (right 2 panels) levels were determined and normalized to the scrambled siControl. (B) Images of immunofluorescent staining of 2 representative areas of the lungs of siControl- (left) and siVwf-treated (right) B6 mice. Images include nuclei staining (Hoechst) in blue and VWF in green. Scale bar is representative for 100 μm. (C) Representative VWF multimeric pattern of B6 mice treated with siVwf or siControl. Analysis was performed on platelet-free plasma, and NMP was used as a reference for normal multimer patterns. Samples loaded on the gel were diluted to a final sample concentration of 0.05 U/mL, except for ∗, where the sample concentration was accidentally 0.10 U/mL. At the bottom of this panel, the plasma VWF levels are indicated for the individual mice that were used for this VWF multimeric pattern analysis. (D) Duration of siRNA–mediated endothelial Vwf inhibition in B6 mice treated with siVwf (1.5 mg/kg) and at 72 hours, 96 hours, and 10 days after injection sacrificed for analysis of lung Vwf mRNA (left panel) and plasma VWF protein (right panel). n = 6 mice per group; however, 1 siControl-treated animal was removed because of a failed siRNA injection, incidentally a blood sample was removed because of unwanted clotting activation (as established visually). Values are presented as median with range. P ≥ 0.05, denotes not significant (ns); ∗P ≤ 0.05; ∗∗P ≤ 0.01.
Immunofluorescent imaging of the lungs revealed, for both siControl- and siVwf-treated mice, VWF staining in the cells of the lining of structures morphologically identified as lung blood vessels (Figure 3B). Images presented are from murine lungs that were sectioned, stained, and processed in the same series and imaged with identical settings. Despite a qualitative analysis, VWF-specific fluorescence in the lung vessels of siVwf-treated mice was clearly lower than in siControl-treated mice. In siVwf mice, some lung regions did not express any VWF at all (Figure 3B, right panel). Overall, these findings are in line with the observed strong reduction of lung Vwf mRNA after siVwf treatment (Figure 3A). Qualitative visualization of the VWF multimers showed that the strong reduction found on both the mRNA and plasma levels did not affect the VWF multimer pattern (Figure 3C; supplemental Figure 2). These results indicate that upon siRNA-mediated Vwf silencing, VWF is only quantitatively reduced and multimeric structure remains unaffected.
Further detailing the time point of maximal siRNA-mediated endothelial Vwf inhibition showed that lung Vwf mRNA as well as plasma VWF were reduced to a comparable level 72 and 96 hours after injection; however, at 96 hours there was less variability among mice (Figure 3D). Therefore, for subsequent experiments, 96 hours after injection was used as the optimal time point for sacrifice.
Regarding duration of effect, our previous studies on siIcam2 indicated that the inhibitory effect can last for a prolonged period, and up to 21 days.27 Here, 10 days was chosen as the end point of Vwf inhibition, as this period is sufficient for future experiments. Interestingly, 10 days after siRNA injection, the reducing effect of siVwf on plasma VWF and lung Vwf mRNA was comparable to that 72 and 96 hours after injection (Figure 3D).
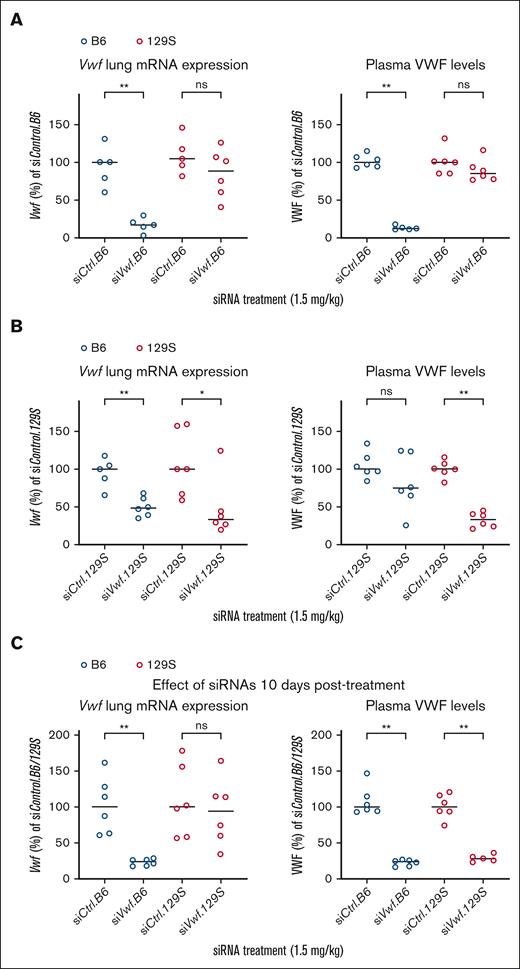
Allele-selectivity of strain-selective siRNAs in vivo
B6 and 129S mice were injected with either siVwf.B6(-2) or siVwf.129S(-3) with the corresponding scrambled siRNA serving as control (siControl.B6 or siControl.129S; Table 1). siVwf.B6 in B6 mice reduced lung Vwf mRNA levels with a median of 83.0% and a corresponding 87.7% reduction for plasma VWF. When siVwf.B6 was injected in mice of the noncorresponding 129S strain, this siRNA had a minimal inhibitory effect (inhibition of lung Vwf mRNA by 15.4% and plasma VWF by 14.9% (Figure 4A)). Largely comparable results were observed for siVwf.129S on plasma VWF levels. Injected into 129S mice, a considerable reduction of 67.1% in plasma VWF was observed, whereas in the noncorresponding B6 strain, nonselective inhibition by siVwf.129S was limited to 26.5% (Figure 4B). At the lung Vwf mRNA level, inhibition of the corresponding strain was comparable to plasma (66.6%). However, allele-selectivity was less outspoken at mRNA levels, as nonselective median inhibition of Vwf was 51.5% in the noncorresponding B6 mice.
Allele-selective inhibition of Vwf in vivo in B6 and 129S mice treated with siVwf.B6 or siVwf.129S. (A) Lung mRNA expression and plasma levels of VWF in B6 (blue) and 129S (red) mice treated with siVwf.B6. Plasma and lungs were collected 96 hours after injection with siVwf.B6. (B) Lung mRNA expression and plasma levels of VWF in B6 (blue) and 129S (red) mice treated with siVwf.129S. Plasma and lungs were collected 96 hours postinjection with siVwf.129S. (C) Lung mRNA expression and plasma levels of VWF of siVwf.B6-treated B6 (blue) mice and siVwf.129S-treated 129S (red) mice. Mice were sacrificed 10 days after receiving their respective siRNA treatments to indicate the effect of a single siRNA injection after a prolonged time period. n = 6 mice per group, however, incidentally a blood sample was removed because of unwanted clotting activation (as established visually). Values are presented as median with range and is compared with the median of the corresponding scrambled siControl.B6 or siControl.129S-treated mice. P ≥ 0.05, ns; ∗P ≤ 0.05; ∗∗P ≤ 0.01.
Allele-selective inhibition of Vwf in vivo in B6 and 129S mice treated with siVwf.B6 or siVwf.129S. (A) Lung mRNA expression and plasma levels of VWF in B6 (blue) and 129S (red) mice treated with siVwf.B6. Plasma and lungs were collected 96 hours after injection with siVwf.B6. (B) Lung mRNA expression and plasma levels of VWF in B6 (blue) and 129S (red) mice treated with siVwf.129S. Plasma and lungs were collected 96 hours postinjection with siVwf.129S. (C) Lung mRNA expression and plasma levels of VWF of siVwf.B6-treated B6 (blue) mice and siVwf.129S-treated 129S (red) mice. Mice were sacrificed 10 days after receiving their respective siRNA treatments to indicate the effect of a single siRNA injection after a prolonged time period. n = 6 mice per group, however, incidentally a blood sample was removed because of unwanted clotting activation (as established visually). Values are presented as median with range and is compared with the median of the corresponding scrambled siControl.B6 or siControl.129S-treated mice. P ≥ 0.05, ns; ∗P ≤ 0.05; ∗∗P ≤ 0.01.
At 10 days after injection (Figure 4C), the inhibitory effect on mRNA level was still present for the siVwf.B6, although it started to fade for the siVwf.129S. However, at plasma level, both siVwf.B6- and siVwf.129S induce strong (>72%) and persistent inhibition 10 days after injection.
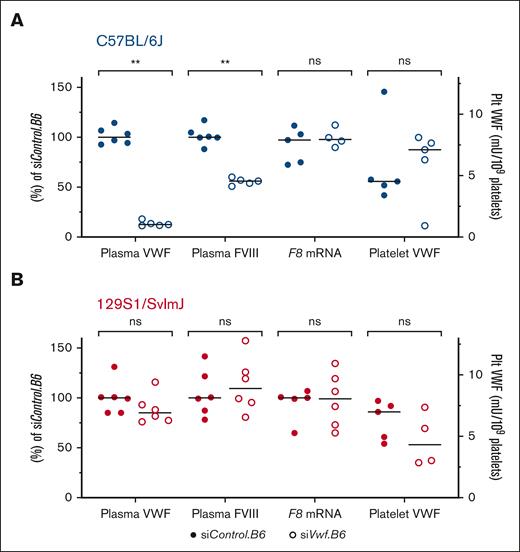
Effect of endothelial Vwf inhibition on FVIII and platelet VWF
A subsequent effect of low plasma VWF levels is decreased plasma FVIII levels. To investigate whether this also occurs after siRNA-mediated Vwf inhibition, FVIII activity was measured in the plasma of siVwf.B6-treated animals. In comparison to the median 87.7% reduction in plasma VWF, FVIII activity was reduced with a median of 44.0% in siVwf.B6-treated B6 mice (Figure 5A). In the noncorresponding 129S strain (Figure 5B), there was no reduction in FVIII activity, which is in accordance with the nonreduced plasma VWF levels. To investigate whether reduced plasma FVIII in the B6 mice is due to the lower plasma VWF and not to reduced endothelial F8 gene transcription, F8 lung mRNA levels were measured. For both mouse strains, treatment with siVwf.B6 did not affect F8 mRNA levels (Figure 5). In addition to the secondary effects of Vwf silencing on FVIII, the effect on platelet VWF was also investigated. The platelets were not expected to be targeted because of the endothelial-specific nanoparticles.27 Platelet VWF levels varied among individual siVwf.B6-treated B6 and 129S mice, with siRNA treatment not affecting platelet VWF, suggesting that the siRNAs did not target platelets or megakaryocytes.
Secondary effects of allele–selective Vwf silencing. (A) The effect on plasma FVIII and platelet (Plt) VWF was assessed in plasma of siVwf.B6-treated B6 mice, and F8 transcript levels were measured on mRNA isolated from lungs of siVwf.B6-treated B6 mice. (B) The effect on plasma FVIII and Plt VWF was assessed in plasma of siVwf.B6-treated 129S mice, and F8 transcript levels were measured on mRNA isolated from lungs of siVwf.B6-treated 129S mice. The effect of plasma VWF, plasma FVIII, and F8 lung mRNA was all relative to the median of their respective siControl.B6 values (left y-axis), whereas for Plt VWF data were normalized as mU per 109 platelets (right y-axis). Individual values are presented with median. P ≥ 0.05, ns; ∗∗P ≤ 0.01. Note: plasma VWF data are the same as shown in Figure 4A and were used as reference for the other parameters of interest for clarity.
Secondary effects of allele–selective Vwf silencing. (A) The effect on plasma FVIII and platelet (Plt) VWF was assessed in plasma of siVwf.B6-treated B6 mice, and F8 transcript levels were measured on mRNA isolated from lungs of siVwf.B6-treated B6 mice. (B) The effect on plasma FVIII and Plt VWF was assessed in plasma of siVwf.B6-treated 129S mice, and F8 transcript levels were measured on mRNA isolated from lungs of siVwf.B6-treated 129S mice. The effect of plasma VWF, plasma FVIII, and F8 lung mRNA was all relative to the median of their respective siControl.B6 values (left y-axis), whereas for Plt VWF data were normalized as mU per 109 platelets (right y-axis). Individual values are presented with median. P ≥ 0.05, ns; ∗∗P ≤ 0.01. Note: plasma VWF data are the same as shown in Figure 4A and were used as reference for the other parameters of interest for clarity.
Discussion
Here, we have shown that it is feasible to silence endogenous endothelial Vwf allele-selectively and effectively in mice. In addition, VWF was reduced for at least 10 days. This approach is a proof of principle of therapeutic strategies for both bleeding and thrombotic disorders as plasma VWF is a determinant in VWD and (arterial) thrombosis.
Based on previous data by Magner et al,36 it was expected that siRNAs targeting a combination of 2 nucleotide differences would be superior to those targeting a single nucleotide difference. However, both lead candidate siRNAs target a single nucleotide difference and were surprisingly effective (up to 99% in vitro and 90% in vivo at low concentration or dose). siRNAs showing relatively weak effects, like siVwf.B6-5, siVwf.129S-5, and siVwf.129S-6 could be due to the presence of a so-called G:U wobble base pair (Table 1). This interaction between the G and U bases plays essential roles in the structure and amino acid acceptor identity of RNAs but for siRNAs, a G:U wobble can lead to a decreased specificity or activity.37,38 However, depending on the position of the G:U wobble, siRNA activity is not necessarily decreased, which could explain the relatively good inhibitory and allele-selective effects of siVwf.129S-4 and siVwf.129S-7.38
Our siRNA approach resulted in an up to 90% reduction of plasma VWF in vivo, in line with our in vitro data. Previously, we showed with siIcam2 maximal endothelial knockdown of 96% in vivo,27 which is somewhat deeper than for the Vwf siRNAs in this study. A stronger Vwf inhibition might be achieved by further chemically modifying the siRNA. Full knockdown of the targeted allele at plasma level is unlikely as the 7C1 LNP delivers the siRNAs to the endothelial compartment and leaves VWF production in the megakaryocytes unaffected, which may contribute to up to 5% of circulating VWF in plasma.39 Although we did not assess VWF production by the megakaryocytes, we did observe no effect on platelet VWF, suggesting that the 7C1 LNP, as expected, did not deliver the siRNAs to platelets or megakaryocytes. For this proof of principle study, the difference in inhibitory effect of the chosen lead candidate siRNAs on the corresponding vs noncorresponding mouse strain is significant enough to show the feasibility of effective and allele-selective Vwf silencing.
In-silico prediction yielded siRNAs with a minimal chance of off-target effects, but this was not further assessed in the in vitro or in vivo studies. Mice did not show any adverse reactions to the siRNA treatment. We considered further assessment beyond the scope of the present proof of principle study. Moreover, these siRNAs were specifically designed to target murine Vwf and were obviously not intended to serve our ultimate goal, that is, in a clinical setting. Future studies using potential clinically therapeutic siRNAs targeting human VWF should consider the assessment of off-target effects and potential chemical modifications to optimize siRNA effectivity, allele-selectivity, and duration of effect.
In our previous in vitro studies on the correction of dominant-negative VWD phenotypes, we allele-selectively inhibited mutated VWF based on siRNAs that target SNP-based differences between VWF alleles.22,23 In this in vivo study, we used the genetic differences between Vwf of 2 mouse inbred strains targeted by siRNAs as a proxy for the human SNPs. For future clinical application of this approach, siRNAs should be designed for human VWF, specifically targeting common SNPs.23 An advantage of SNP-targeting instead of mutation-specific targeting is the possibility to apply this approach with the same small set of siRNAs to both VWD, which is caused by many different mutations, and (arterial) thrombosis, where there are no VWF mutations at play. Further analysis of the phenotypic effects of this siRNA-based approach can be performed using the murine-specific siRNAs that we describe here. This can be done in experimental bleeding and vascular injury models in mice, in nonhumanized type 2B knock-in mice for VWD,40 or in atherothrombosis-prone mice,41 as long as these mice have either a B6 or 129S background or a B6x129S background to assess allele-selectivity.
To conclude, we show efficient in vivo endothelial delivery of siRNAs with a specific delivery vehicle, resulting in highly effective and allele-selective Vwf inhibition in mice, which is, to our knowledge, the first in vivo proof of principle of allele-selective VWF silencing as a therapeutic target. Although this approach with murine-specific siRNAs cannot directly be used for clinical purposes, the siRNAs can be applied in future studies on the potential therapeutic effect of allele-selective silencing of Vwf in experimental murine models for bleeding and thrombosis.
Acknowledgments
The authors thank Rob F.P. van den Akker, Rayna J. S. Anijs, Ka Lei Cheung, and Sebastiaan N. J. Laan for assisting during the animal experiments.
This study was supported by the Dutch Thrombosis Foundation (Trombosestichting Nederland grant #2018-01).
Authorship
Contribution: Y.K.J., A.d.J., B.J.M.v.V., and J.C.J.E. designed the study; E.S.E., K.P., and J.E.D. prepared and contributed vital reagents; Y.K.J., R.J.D., J.C.W., and K.D.v.d.G. performed in vitro experiments; Y.K.J., N.A.L., and B.J.M.v.V. performed in vivo experiments; Y.K.J., B.J.M.v.V., and J.C.J.E. analyzed and interpreted the data and wrote the manuscript; and all authors contributed to revisions of the manuscript.
Conflict-of-interest disclosure: J.C.J.E. declares financial support from CSL Behring. The remaining authors declare no competing financial interests.
Correspondence: Jeroen C. J. Eikenboom, Department of Internal Medicine, Division of Thrombosis and Hemostasis, Einthoven Laboratory for Vascular and Regenerative Medicine, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, The Netherlands; e-mail: h.c.j.eikenboom@lumc.nl.
References
Author notes
Data are available on request from the corresponding author, Jeroen C. J. Eikenboom (h.c.j.eikenboom@lumc.nl).
The full-text version of this article contains a data supplement.