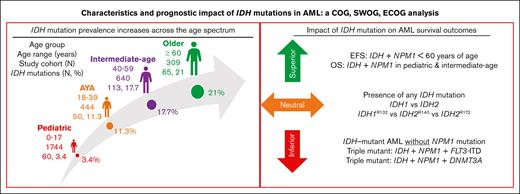
Key Points
IDH mutation did not abrogate the favorable prognostic impact of NPM1 mutation in AML for patients aged <60 years.
Patients with IDH-mutated AML without cooccurring NPM1 or triple-mutated IDH/NPM1/DNMT3A or IDH/NPM1/FLT3-ITD AML had inferior outcomes.
Abstract
Somatic mutations in isocitrate dehydrogenase (IDH) genes occur frequently in adult acute myeloid leukemia (AML) and less commonly in pediatric AML. The objective of this study was to describe the prevalence, mutational profile, and prognostic significance of IDH mutations in AML across age. Our cohort included 3141 patients aged between <1 month and 88 years treated on Children’s Cancer Group/Children’s Oncology Group (n = 1872), Southwest Oncology Group (n = 359), Eastern Cooperative Oncology Group (n = 397) trials, and in Beat AML (n = 333) and The Cancer Genome Atlas (n = 180) genomic characterization cohorts. We retrospectively analyzed patients in 4 age groups (age range, n): pediatric (0-17, 1744), adolescent/young adult (18-39, 444), intermediate-age (40-59, 640), older (≥60, 309). IDH mutations (IDHmut) were identified in 9.2% of the total cohort (n = 288; IDH1 [n = 123, 42.7%]; IDH2 [n = 165, 57.3%]) and were strongly correlated with increased age: 3.4% pediatric vs 21% older, P < .001. Outcomes were similar in IDHmut and IDH-wildtype (IDHWT) AML (event-free survival [EFS]: 35.6% vs 40.0%, P = .368; overall survival [OS]: 50.3% vs 55.4%, P = .196). IDH mutations frequently occurred with NPM1 (47.2%), DNMT3A (29.3%), and FLT3-internal tandem duplication (ITD) (22.4%) mutations. Patients with IDHmut AML with NPM1 mutation (IDHmut/NPM1mut) had significantly improved survival compared with the poor outcomes experienced by patients without (IDHmut/NPM1WT) (EFS: 55.1% vs 17.0%, P < .001; OS: 66.5% vs 35.2%, P < .001). DNTM3A or FLT3-ITD mutations in otherwise favorable IDHmut/NPM1mut AML led to inferior outcomes. Age group analysis demonstrated that IDH mutations did not abrogate the favorable prognostic impact of NPM1mut in patients aged <60 years; older patients had poor outcomes regardless of NPM1 status. These trials were registered at www.clinicaltrials.gov as #NCT00070174, #NCT00372593, #NCT01371981, #NCT00049517, and #NCT00085709.
Introduction
Enhanced genomic and epigenomic profiling of acute myeloid leukemia (AML) has led to identification of recurrent mutations that are prognostic and are candidates for targeted therapy. Somatic mutations in isocitrate dehydrogenase (IDH) genes, IDH1 and IDH2, occur in ∼6% to 16% and ∼8% to 19% of adult patients with AML, respectively.1-5 In pediatric AML, IDH mutations are rare, occurring in <4% of patients.6-11
Mutations in active site arginine residues of IDH1 (R132) and IDH2 (R140, R172) enzymes lead to neomorphic production of the oncometabolite 2-hydroxyglutarate.12-15 Accumulation of 2-hydroxyglutarate alters DNA and histone methylation, which impairs myeloid differentiation and contributes to leukemogenesis.14,16 Inhibitors of IDH-mutant protein including ivosidenib, olutasidenib (IDH1), and enasidenib (IDH2) can be used as single agents in adult patients with IDH-mutant AML, either in the upfront or relapsed/refractory setting.17-20 Clinical trials investigating IDH inhibitors combined with varying intensity chemotherapy are underway for adult patients with newly diagnosed IDH-mutated AML.21-23
Several groups have reported on the prognostic influence of IDH mutations alone and in combination with frequently cooccurring mutations and have found varying results.1-5,24-33 Data on younger patients (aged <30 years) are limited, making it difficult to prognosticate outcomes based on IDH mutation status in these patients.6-11 The aim of the current study is to describe the prevalence, cooccurring mutational profile, and prognostic impact of IDH mutations in a large cohort of patients with AML across the age spectrum. We hypothesize that improved understanding of IDH-mutated AML will allow for optimal integration of targeted agents into risk and age-adapted treatment strategies.
Patients and methods
Characteristics of study cohort
The total patient cohort included 3141 patients with AML ranging in age from <1 month to 88 years. For analysis, patients were divided into 4 age-defined groups: pediatric (0-17 years, n = 1744), adolescent/young adult (AYA; 18-39 years, n = 444), intermediate-age (40-59 years, n = 640), and older (≥60 years, n = 309). The cohort comprised 1872 (59.5%) patients enrolled in Children’s Cancer Group (CCG) or Children’s Oncology Group (COG) trials CCG2961,34 AAML03P1 (NCT00070174),35 AAML0531 (NCT00372593),36 and AAML1031 (NCT01371981)37; 397 (12.6%) patients enrolled on the Eastern Cooperative Oncology Group (ECOG) E1900 trial (NCT00049517)38; 359 (11.4%) patients on the Southwest Oncology Group (SWOG) trial, S0106 (NCT00085709)39; 333 (10.6%) patients included in the Beat AML genomic characterization cohort40; and 180 (5.7%) patients from TCGA AML cohort.41 Details of chemotherapy regimens and randomizations for each treatment protocol and methods for genomic characterization cohorts were previously described.34-41 (supplemental Table 1). Institutional review boards of participating institutions approved clinical protocols. Written informed consent was obtained from study participants in accordance with the Declaration of Helsinki.
Clinical characteristics and treatment outcomes were collected and evaluated per standard practices of respective studies. For our analysis by cytomolecular risk category, we assigned patients to favorable, intermediate, adverse, or indeterminate risk, independent of IDH mutation status. Patients in the ECOG, SWOG, Beat AML, and TCGA cohorts were assigned risk classification based on their designation at the time of the original studies. Earlier CCG/COG studies made very limited use of cytomolecular risk classification; therefore, the current COG risk classification schema was used to classify patients in the CCG/COG cohort for this study (supplemental Table 2).
Mutational analysis was performed per each study or subsequent analyses. Cytogenetic analysis, fluorescence in situ hybridization, and reverse transcriptase polymerase chain reaction assays for recurrent cytogenetic lesions were performed as previously described.34-37,39,42 Samples from patients on CCG/COG trials underwent mutational profiling by targeted capture (n = 788), whole-genome (n = 329), and/or transcriptome (n = 1659) sequencing.43 Targeted mutation sequencing was conducted on banked ECOG E1900 samples.44 Samples from SWOG S0106 underwent next-generation sequencing. TCGA employed whole-genome sequencing (n = 50) or whole-exome sequencing (n = 150).41 For Beat AML, whole-exome and RNA sequencing as well as targeted mutation polymerase chain reactions were performed.40 Mutation prevalence was reported as the proportion of patients positive for mutation among patients with available mutation data.
Statistical analysis
IDH and cooccurring mutation prevalence was determined, and overall survival (OS), 5-year event-free survival (EFS), and cumulative incidence of relapse risk (RR) were analyzed across the total cohort and each study-defined age group. EFS and RR data were not available for Beat AML samples, and RR data were not available for TCGA samples, thus Beat AML samples were excluded from all outcome analyses and TCGA samples from RR analyses. OS was measured from date of initial randomization or study entry until death from any cause. EFS was measured from date of randomization or study entry until refractory disease, relapse, or death. Complete remission (CR) was defined as hematopoietic recovery with <5% morphologic leukemic blasts in the bone marrow and no extramedullary disease after induction chemotherapy; response data were not available for the TCGA cohort. RR was measured for all patients who achieved CR from date of CR until first relapse. Observations were censored at date of last contact for patients last known to be alive (OS, EFS, and RR) without report of relapse (EFS and RR). The Kaplan-Meier method was used to estimate survival outcomes.45 The significance of predictor variables was tested with log-rank statistic for OS and EFS and Gray statistic for RR.46 The significance of observed differences in proportions was tested by χ2 test and Fisher exact test when data were sparse. The Mann-Whitney (vs Kruskal-Wallis) test was used to determine the significance between differences in medians. Cox proportional hazard models were used to estimate hazard ratios of OS, EFS for univariable and multivariable analyses.47 Competing risk regression was used to estimate hazard ratios of RR.48 A P value of <.05 was considered statistically significant. Statistical analyses were performed using Statistical Analysis Software (SAS) version 9.4 (SAS Institute Inc., Cary, NC).
Results
Mutation prevalence and characteristics
Among the total cohort, there were 286 patients (9.1%) with IDH-mutant (IDHmut) and 2855 (90.9%) with IDH-wildtype (IDHWT) AML (Table 1). There were 288 IDH mutations identified; 2 patients had cooccurring IDH1 and IDH2 mutations and were excluded from outcomes analyses.
Clinical and disease characteristics of study cohort
| . | IDH1mut . | IDH2mut . | IDH1/2mut∗ . | IDHWT . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| n . | % . | n . | % . | n . | % . | n . | % . | ||
| 123 | 165 | 286 | 2855 | ||||||
| Study cohort | |||||||||
| CCG/COG | 31 | 25.2% | 44 | 26.7% | 74 | 25.9% | 1798 | 63.0% | |
| Beat AML | 24 | 19.5% | 32 | 19.4% | 56 | 19.6% | 277 | 9.7% | |
| ECOG | 24 | 19.5% | 34 | 20.6% | 58 | 20.3% | 339 | 11.9% | |
| SWOG | 25 | 20.3% | 35 | 21.2% | 60 | 21.0% | 299 | 10.5% | |
| TCGA | 19 | 15.4% | 20 | 12.1% | 38 | 13.3% | 142 | 5.0% | |
| Age category (y) | |||||||||
| Pediatric (0-17) | 25 | 20.3% | 35 | 21.2% | 60 | 21.0% | 1684 | 59.1% | |
| AYA (18-39) | 25 | 20.3% | 25 | 15.2% | 49 | 17.1% | 395 | 13.9% | |
| Intermediate-age (40-59) | 48 | 39.0% | 65 | 39.4% | 113 | 39.5% | 527 | 18.5% | |
| Older (≥60) | 25 | 20.3% | 40 | 24.2% | 64 | 22.4% | 245 | 8.6% | |
| P value: IDHmut vs IDHWT | |||||||||
| Age: median (range) | 46.4 (4.3-87) | 51.4 (4.2-83) | 50.7 (4.2-87) | 15.6 (0.01-88) | <.001 | ||||
| IDH1mut | IDH2mut | IDH1/2mut∗ | IDHWT | ||||||
| n | % | n | % | n | % | n | % | ||
| Sex | |||||||||
| Male | 60 | 48.8% | 89 | 53.9% | 147 | 51.4% | 1503 | 52.7% | .683 |
| Female | 63 | 51.2% | 76 | 46.1% | 139 | 48.6% | 1351 | 47.3% | |
| WBC (×103/μL) | |||||||||
| Median (range), n = 3092 | 22.8 (0.6-201.1) | 14.3 (0.8-191.8) | 19.1 (0.6-201.1) | 21.9 (0.2-918.5) | .003 | ||||
| Peripheral blast, % | |||||||||
| Median (range), n = 2961 | 63 (0-98) | 45 (0-97) | 55 (0-98) | 38 (0-100) | .002 | ||||
| Bone marrow blast, % | |||||||||
| Median (range), n = 2582 | 79 (0-99) | 74 (11-100) | 76 (0-100) | 68 (0-100) | <.001 | ||||
| Platelet count (×103/μL) | |||||||||
| Median (range), n = 1799 | 65 (9-650) | 57.5 (8-9300) | 62 (8-9300) | 48 (0.7-7900) | <.001 | ||||
| Cytomolecular risk group | |||||||||
| Favorable | 30 | 26.3% | 42 | 29.2% | 72 | 28.1% | 916 | 33.7% | .071 |
| Intermediate | 61 | 53.5% | 73 | 50.7% | 132 | 51.6% | 968 | 35.6% | <.001 |
| Adverse | 23 | 20.2% | 29 | 20.1% | 52 | 20.3% | 836 | 30.7% | .001 |
| Unknown | 9 | 21 | 30 | 135 | |||||
| . | IDH1mut . | IDH2mut . | IDH1/2mut∗ . | IDHWT . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| n . | % . | n . | % . | n . | % . | n . | % . | ||
| 123 | 165 | 286 | 2855 | ||||||
| Study cohort | |||||||||
| CCG/COG | 31 | 25.2% | 44 | 26.7% | 74 | 25.9% | 1798 | 63.0% | |
| Beat AML | 24 | 19.5% | 32 | 19.4% | 56 | 19.6% | 277 | 9.7% | |
| ECOG | 24 | 19.5% | 34 | 20.6% | 58 | 20.3% | 339 | 11.9% | |
| SWOG | 25 | 20.3% | 35 | 21.2% | 60 | 21.0% | 299 | 10.5% | |
| TCGA | 19 | 15.4% | 20 | 12.1% | 38 | 13.3% | 142 | 5.0% | |
| Age category (y) | |||||||||
| Pediatric (0-17) | 25 | 20.3% | 35 | 21.2% | 60 | 21.0% | 1684 | 59.1% | |
| AYA (18-39) | 25 | 20.3% | 25 | 15.2% | 49 | 17.1% | 395 | 13.9% | |
| Intermediate-age (40-59) | 48 | 39.0% | 65 | 39.4% | 113 | 39.5% | 527 | 18.5% | |
| Older (≥60) | 25 | 20.3% | 40 | 24.2% | 64 | 22.4% | 245 | 8.6% | |
| P value: IDHmut vs IDHWT | |||||||||
| Age: median (range) | 46.4 (4.3-87) | 51.4 (4.2-83) | 50.7 (4.2-87) | 15.6 (0.01-88) | <.001 | ||||
| IDH1mut | IDH2mut | IDH1/2mut∗ | IDHWT | ||||||
| n | % | n | % | n | % | n | % | ||
| Sex | |||||||||
| Male | 60 | 48.8% | 89 | 53.9% | 147 | 51.4% | 1503 | 52.7% | .683 |
| Female | 63 | 51.2% | 76 | 46.1% | 139 | 48.6% | 1351 | 47.3% | |
| WBC (×103/μL) | |||||||||
| Median (range), n = 3092 | 22.8 (0.6-201.1) | 14.3 (0.8-191.8) | 19.1 (0.6-201.1) | 21.9 (0.2-918.5) | .003 | ||||
| Peripheral blast, % | |||||||||
| Median (range), n = 2961 | 63 (0-98) | 45 (0-97) | 55 (0-98) | 38 (0-100) | .002 | ||||
| Bone marrow blast, % | |||||||||
| Median (range), n = 2582 | 79 (0-99) | 74 (11-100) | 76 (0-100) | 68 (0-100) | <.001 | ||||
| Platelet count (×103/μL) | |||||||||
| Median (range), n = 1799 | 65 (9-650) | 57.5 (8-9300) | 62 (8-9300) | 48 (0.7-7900) | <.001 | ||||
| Cytomolecular risk group | |||||||||
| Favorable | 30 | 26.3% | 42 | 29.2% | 72 | 28.1% | 916 | 33.7% | .071 |
| Intermediate | 61 | 53.5% | 73 | 50.7% | 132 | 51.6% | 968 | 35.6% | <.001 |
| Adverse | 23 | 20.2% | 29 | 20.1% | 52 | 20.3% | 836 | 30.7% | .001 |
| Unknown | 9 | 21 | 30 | 135 | |||||
Bolded values are statisically significant.
There were 123 (42.7%) IDH1 mutations, nearly all point mutations at conserved active site residue R132 (n = 117, 95.1%; R132H [50.4%], R132C [36.8%], R132S [9.4%], and R132G [3.4%]). The remaining IDH1 mutations (n = 6, 4.9%) occurred at alternative residues (V71I [n = 4], F86S [n = 1], and S122N [n = 1]) previously confirmed to be nonfunctional (V71I)49 or outside the active site and were thus excluded from outcomes analyses. There were 165 (57.3%) IDH2 mutations, 164 (99%) located at conserved active site residues R140 (n = 142, 86%; R140Q [95%], R140W [2.1%], R140G [1.4%], and R140L [1.4%]) and R172 (n = 22, 13.3%); 1 frameshift mutation distal to the active site at G145 with unknown functional significance was excluded from outcomes analyses.
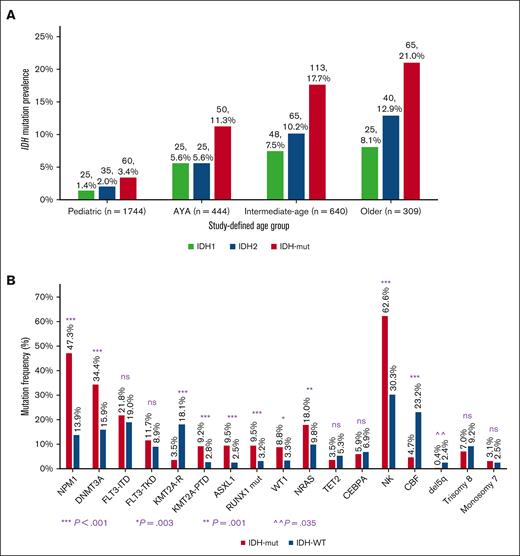
IDH mutation frequency increased significantly with age: pediatric (3.4%, 60 of 1744), AYA (11.3%, 50 of 444), intermediate-age (17.7%, 113 of 640), and older adults (21%, 65 of 309); P < .001. This observed age-dependent mutation prevalence was similar for IDH1 and IDH2 (Figure 1A). IDH mutations were virtually absent (0.3%) in patients aged <5 years.
Profile of IDH mutations in AML according to age and cooccurring mutations. (A) IDH mutation prevalence increases with age shown as IDH1, IDH2, and combined IDH1/IDH2 in study-defined age groups. (B) The cooccurring mutational profile of IDHmut AML varies from that of IDHWT AML. CBF, core-binding factor; KMT2A-R, KMT2A-rearranged; NK, normal karyotype; ns, nonsignificant; PTD, partial tandem duplication; TKD, tyrosine kinase domain.
Profile of IDH mutations in AML according to age and cooccurring mutations. (A) IDH mutation prevalence increases with age shown as IDH1, IDH2, and combined IDH1/IDH2 in study-defined age groups. (B) The cooccurring mutational profile of IDHmut AML varies from that of IDHWT AML. CBF, core-binding factor; KMT2A-R, KMT2A-rearranged; NK, normal karyotype; ns, nonsignificant; PTD, partial tandem duplication; TKD, tyrosine kinase domain.
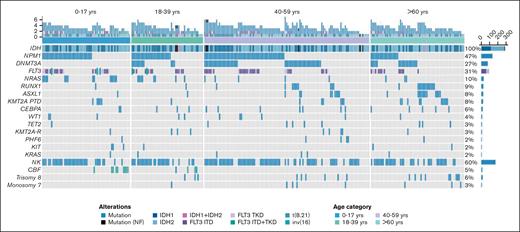
To identify cooccurring mutations, we conducted mutational profiling of IDHmut AML (Figure 2). In the total cohort, NPM1 was the most frequently cooccurring mutation, identified in 47.2% (n = 135) of all patients with IDHmut (IDHmut/NPM1mut), in 53.7% of patients with IDH1mut, and 41.8% of patients with IDH2mut, and was more common in IDHmut than in IDHWT (n = 398) AML (47.2% vs 14.0%; P < .001) (Figure 1B). NPM1 and IDH1 mutations cooccurred with IDH1R132H at 68.8% vs with IDH1R132C at 12.5% (P < .001). NPM1 and IDH2 mutations cooccurred primarily with IDH2R140 (98.6%), and once with IDH2R172K. When analyzed by age group, a greater proportion of younger patients with IDHmut AML had cooccurring NPM1 mutation (56.7% pediatric, 55.1% AYA, 48.7% intermediate-age, and 29.7% older; P = .001).
Mutational analysis of patient cohort. Combined IDH1 and IDH2 mutations with overlapping cytomolecular mutations per age cohort. NF, nonfunctional.
Mutational analysis of patient cohort. Combined IDH1 and IDH2 mutations with overlapping cytomolecular mutations per age cohort. NF, nonfunctional.
DNMT3A mutations (n = 78) were the next most common mutation cooccurring in IDHmut AML and was more prevalent in IDHmut AML than in IDHWT AML (29.3% vs 8.1%; P < .001; Figure 1B). Of the patients with IDHmut/DNMT3Amut AML, 44.8% had triple mutations in IDH, NPM1, and DNMT3A (IDHmut/NPM1mut/DNMT3Amut), mostly in the intermediate-age group (60%), and most were IDH1mut (71.4%). DNTM3A mutation status was assessed in 1281 (68.5%) patients in the CCG/COG trials in the TARGET AML analysis and only 2 overlapping IDH and DNMT3A mutations were identified.
FLT3 internal tandem duplication (ITD) and tyrosine kinase domain mutations occurred with similar frequency in IDHmut and IDHWT AML (ITD: 22.4% vs 19.1%, P = .188; tyrosine kinase domain: 10.9% vs 8.9%, P = .259) (Figure 1B). Of the patients with IDHmut/FLT3-ITD AML, these mutations also cooccurred with NPM1 (IDHmut/NPM1mut/FLT3-ITD) in 63.9%.
Mutations that more frequently occurred in IDHmut vs IDHWT AML included ASXL1 (8.9% vs 2.5%, P < .001), RUNX1 (9.7% vs 3.3%, P < .001), and KMT2A-partial tandem duplication (11.5% vs 4.3%, P < .001) (Figure 1B). The poor risk mutations RUNX1 and ASXL1 were prevalent in intermediate-age and older patients, particularly with IDHmut/NPM1WT (Figure 2). Other comutations were less prevalent in IDHmut than IDHWT AML, namely WT1 (3.8% vs 9.0%, P = .003) and NRAS (11.6% vs 20.0%, P = .001) (Figure 1B).
IDHmut AML was more frequently associated with normal karyotype than with IDHWT AML (62.6% vs 30.3%, P < .001; Figure 1B). Data on cytomolecular risk classification were available in 2976 (94.7%) patients overall and 256 (89.5%) patients with IDHmut AML (Table 1). A greater proportion of IDHmut AML was classified as intermediate risk compared with IDHWT AML in the total cohort (51.6% vs 35.6%, P < .001) and in AYA and intermediate-age groups (AYA: 62.8% vs 36.4%, P = .001; intermediate-age: 74.5% vs 50.5%, P < .001). Among pediatric patients, a greater proportion of those with IDHmut AML had favorable risk classification compared with those with IDHWT AML (71.7% vs 37.4%, P < .001). This was, in large part, because of the combination of NPM1 and IDH mutations (76.7% of pediatric patients with favorable risk IDHmut); cooccurrence with core-binding factor (CBF) mutations accounted for 20.9% of pediatric patients with favorable risk IDHmut because dual CBF and IDH mutations occurred in 15% (9 of 60) of the IDHmut pediatric cohort. There was no difference among cytomolecular risk groups based on IDH mutation status in older patients (IDHmut vs IDHWT, favorable: 16.9% vs 18.2%, P = .820; intermediate: 44.1% vs 46.2%, P = .77; adverse: 39% vs 35.6%, P = .628).
Clinical outcomes
Outcomes analyses were conducted for nearly all patients aged <60 years (pediatric: n = 60/60 [100%], AYA: n = 44/49 [90%], and intermediate-age: n = 92/113 [89]); however, in older patients with IDHmut AML, outcomes analyses were limited to 39% (n = 25/64). There was no significant difference in CR rates between IDHmut and IDHWT AML for the overall cohort (80.4% vs 84.3%; P = .162) or among age groups (supplemental Table 3). Outcome data were available for OS and EFS analysis in 2799 (89.1%) and for RR in 2162 (68.8%) of the total cohort. In the overall cohort, patients with IDHmut or IDHWT AML experienced similar OS (50.3%; 95% confidence interval [CI], 42.8-57.2 vs 55.4%; 95% CI, 53.3-57.3; P = .196) and EFS (35.6% [95% CI, 29-42.3] vs 40% [95% CI, 38.1-42]; P = .368). Further analysis by age group demonstrated no difference in OS or EFS between IDHmut and IDHWT AML (Table 2). There was no significant OS or EFS difference when comparing IDHWT and IDHmut by IDH isoform (IDH1 vs IDH2) or mutation subtype (IDH1R132 vs IDH2R140 vs IDH2R172K) overall (Figure 3) or in any age group (Table 2). Among the cohort for RR analysis, there was no RR difference based on IDH mutation status (IDHmut: 47.3% [95% CI, 38.7-55.5] vs IDHWT: 45.5% [95% CI, 43.3-47.7]; P = .858; supplemental Table 4). When the cohort was analyzed by cytomolecular risk group, presence of IDH mutation did not significantly modify outcomes (OS for IDHmut vs IDHWT: favorable: 78.3% [95% CI, 63.1-87.8] vs 79.8% [95% CI, 76.8-82.5]; P = .705; intermediate: 44.1% [95% CI, 33.7-54] vs 52.5% [95% CI, 49.1-55.9]; P = .25; adverse: 36.8% [95% CI, 18.8-55.0] vs 36.2% [95% CI, 32.6-39.8]; P = .448). Those with IDHmut/normal karyotype had better outcomes compared with those with IDHmut with abnormal karyotype (EFS: 46.5% vs 22.9%, P = .002; OS: 58.4% vs 41.5%, P = .007).
Comparison of survival outcomes of IDHmut vs IDHWT AML including by IDH isoform or mutation subtype in total cohort and by age group
| OS (n, 5-y estimated OS, 95% CI) . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | IDHWT . | IDHmut . | P value . | IDH1mut . | IDH2mut . | P value . | IDH1R132 . | IDH2R140 . | IDH2R172K . | P value∗ . |
| Total | 2578, 55.4%, (53.3-57.3) | 221, 50.3%, (42.8-57.2) | .196 | 91, 50.6%, (38.9-61.2) | 130, 50.1%, (40.4-59.1) | .778 | 91, 50.6%, (38.9-61.2) | 110, 50.4%, (39.8-60.0) | 19, 45.6%, (20.0-68.1) | .939 |
| Pediatric | 1681, 63.2%, (60.8-65.6) | 60, 73.4%, (59.7-83.1) | .269 | 25, 78.7%, (56.1-90.6) | 35, 69.2%, (50.0-82.3) | .471 | 25, 78.7%, (56.1-90.6) | 34, 68.2%, (48.6-81.7) | 1, 100%, | .610 |
| AYA | 358, 52.0%, (46.3-57.3) | 44, 57.3%, (38.1-72.5) | .225 | 21, 58.3%, (27.7-79.7) | 23, 56.6%, (31.5-75.5) | .548 | 21, 58.3%, (27.7-79.7) | 17, 52.9%, (27.6-73.0) | 5, 66.7%, (5.4-94.5) | .312 |
| Intermediate age | 446, 34.4%, (29.7-39.1) | 92, 40.6%, (29.3-51.5) | .187 | 38, 33.8%, (18.2-50.1) | 54, 46.4%, (31.2-60.3) | .425 | 38, 33.8%, (18.2-50.1) | 43, 50.7%, (33.1-65.8) | 11, 32.7%, (8.3-60.6) | .479 |
| Older | 91, 19.8%, (12.3-28.7) | 25, 9.2%, (0.9-30.2) | .5 | 7, 14.3%, (0.7-46.5) | 18, 0% | .751 | 7, 14.3%, (0.7-46.5) | 16, 0% | 2, 50.0%, (0.6-91) | .692 |
| OS (n, 5-y estimated OS, 95% CI) . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | IDHWT . | IDHmut . | P value . | IDH1mut . | IDH2mut . | P value . | IDH1R132 . | IDH2R140 . | IDH2R172K . | P value∗ . |
| Total | 2578, 55.4%, (53.3-57.3) | 221, 50.3%, (42.8-57.2) | .196 | 91, 50.6%, (38.9-61.2) | 130, 50.1%, (40.4-59.1) | .778 | 91, 50.6%, (38.9-61.2) | 110, 50.4%, (39.8-60.0) | 19, 45.6%, (20.0-68.1) | .939 |
| Pediatric | 1681, 63.2%, (60.8-65.6) | 60, 73.4%, (59.7-83.1) | .269 | 25, 78.7%, (56.1-90.6) | 35, 69.2%, (50.0-82.3) | .471 | 25, 78.7%, (56.1-90.6) | 34, 68.2%, (48.6-81.7) | 1, 100%, | .610 |
| AYA | 358, 52.0%, (46.3-57.3) | 44, 57.3%, (38.1-72.5) | .225 | 21, 58.3%, (27.7-79.7) | 23, 56.6%, (31.5-75.5) | .548 | 21, 58.3%, (27.7-79.7) | 17, 52.9%, (27.6-73.0) | 5, 66.7%, (5.4-94.5) | .312 |
| Intermediate age | 446, 34.4%, (29.7-39.1) | 92, 40.6%, (29.3-51.5) | .187 | 38, 33.8%, (18.2-50.1) | 54, 46.4%, (31.2-60.3) | .425 | 38, 33.8%, (18.2-50.1) | 43, 50.7%, (33.1-65.8) | 11, 32.7%, (8.3-60.6) | .479 |
| Older | 91, 19.8%, (12.3-28.7) | 25, 9.2%, (0.9-30.2) | .5 | 7, 14.3%, (0.7-46.5) | 18, 0% | .751 | 7, 14.3%, (0.7-46.5) | 16, 0% | 2, 50.0%, (0.6-91) | .692 |
| EFS (n, 5-y estimate EFS, 95% CI) . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | IDHWT . | IDHmut . | P value . | IDH1mut . | IDH2mut . | P value . | IDH1R132 . | IDH2R140 . | IDH2R172K . | P value∗ . |
| Total | 2578, 40.0%, (38.1-42) | 221, 35.6%, (29.0-42.3) | .368 | 91, 35.8%, (25.7-46.0) | 130, 35.4%, (26.8-44.2) | .920 | 91, 35.8%, (25.7-46.0) | 110, 36.5%, (27.2-45.8) | 19, 12.4%, (5.8-50.1) | .892 |
| Pediatric | 1681, 45.6%, (43.2-48.0) | 60, 49.5%, (35.8-61.7) | .484 | 25, 57.6%, (35.4-74.6) | 35, 43.3%, (26.0-59.5) | .279 | 25, 57.6%, (35.4-74.6) | 34, 44.6%, (26.8-61.0) | 1, 0% | .342 |
| AYA | 358, 38.2%, (33.1-43.3) | 44, 52.6%, (36.3-66.6) | .061 | 21, 61.1%, (36.9-78.4) | 23, 46.4%, (25.0-65.4) | .473 | 21, 61.1%, (36.9-78.4) | 17, 40.3%, (17.6-62.2) | 5, 60.0%, (12.6-88.2) | .437 |
| Intermediate age | 446, 26.1%, (22-30.5) | 92, 24.2%, (15.3-34.3) | .980 | 38, 12.2%, (3.6-26.4) | 54, 33.4%, (20.5-46.9) | .086 | 38, 12.2%, (3.6-26.4) | 43, 39.3%, (24.5-53.8) | 11, 13.6%, (1.0-2.6) | .129 |
| Older | 91, 11.5%, (5.8-19.2) | 25, 13.7%, (3.6-30.6) | .293 | 7, 14.3%, (0.7-46.5) | 18, 13.3%, (2.3-34.0) | .636 | 7, 14.3%, (0.7-46.5) | 16, 7.8%, (0.5-29.1) | 2, 50.00%, (0.6-91.0) | .559 |
| EFS (n, 5-y estimate EFS, 95% CI) . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | IDHWT . | IDHmut . | P value . | IDH1mut . | IDH2mut . | P value . | IDH1R132 . | IDH2R140 . | IDH2R172K . | P value∗ . |
| Total | 2578, 40.0%, (38.1-42) | 221, 35.6%, (29.0-42.3) | .368 | 91, 35.8%, (25.7-46.0) | 130, 35.4%, (26.8-44.2) | .920 | 91, 35.8%, (25.7-46.0) | 110, 36.5%, (27.2-45.8) | 19, 12.4%, (5.8-50.1) | .892 |
| Pediatric | 1681, 45.6%, (43.2-48.0) | 60, 49.5%, (35.8-61.7) | .484 | 25, 57.6%, (35.4-74.6) | 35, 43.3%, (26.0-59.5) | .279 | 25, 57.6%, (35.4-74.6) | 34, 44.6%, (26.8-61.0) | 1, 0% | .342 |
| AYA | 358, 38.2%, (33.1-43.3) | 44, 52.6%, (36.3-66.6) | .061 | 21, 61.1%, (36.9-78.4) | 23, 46.4%, (25.0-65.4) | .473 | 21, 61.1%, (36.9-78.4) | 17, 40.3%, (17.6-62.2) | 5, 60.0%, (12.6-88.2) | .437 |
| Intermediate age | 446, 26.1%, (22-30.5) | 92, 24.2%, (15.3-34.3) | .980 | 38, 12.2%, (3.6-26.4) | 54, 33.4%, (20.5-46.9) | .086 | 38, 12.2%, (3.6-26.4) | 43, 39.3%, (24.5-53.8) | 11, 13.6%, (1.0-2.6) | .129 |
| Older | 91, 11.5%, (5.8-19.2) | 25, 13.7%, (3.6-30.6) | .293 | 7, 14.3%, (0.7-46.5) | 18, 13.3%, (2.3-34.0) | .636 | 7, 14.3%, (0.7-46.5) | 16, 7.8%, (0.5-29.1) | 2, 50.00%, (0.6-91.0) | .559 |
Five-year estimates of OS and EFS, 95% CI. P values calculated by log-rank test.
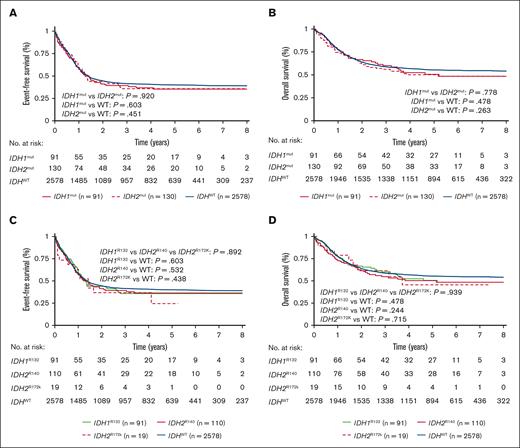
Survival outcomes of IDHmut AML in the total study cohort based on IDH isoform and mutation subtype. There was no difference between isoform IDH1 vs IDH2 for (A) EFS or (B) OS. Similarly, there was no difference in outcomes according to mutation subtype IDH1R132 vs IDH2R140 vs IDH2R172 for (C) EFS or (D) OS. P values calculated by log-rank test.
Survival outcomes of IDHmut AML in the total study cohort based on IDH isoform and mutation subtype. There was no difference between isoform IDH1 vs IDH2 for (A) EFS or (B) OS. Similarly, there was no difference in outcomes according to mutation subtype IDH1R132 vs IDH2R140 vs IDH2R172 for (C) EFS or (D) OS. P values calculated by log-rank test.
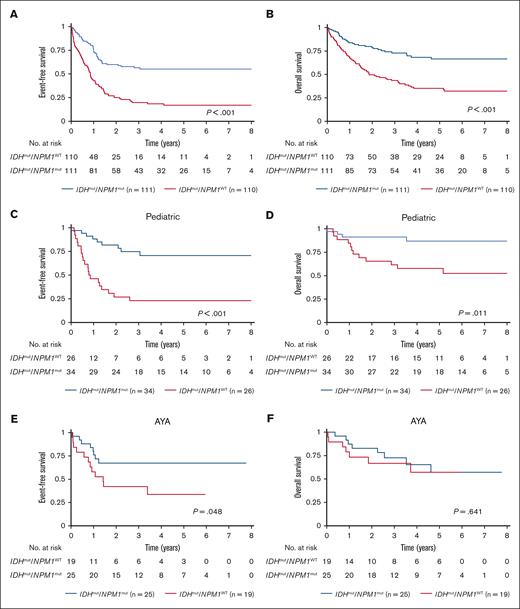
Nearly half of all patients with IDHmut AML had cooccurring NPM1 mutation (NPM1mut; 47.2%), thus we evaluated outcomes of IDHmut AML based on presence of NPM1 mutation. In the overall cohort, patients with dual mutant IDHmut/NPM1mut had significantly better EFS and OS compared with the particularly poor outcome of those with IDHmut/NPM1WT (EFS: 55.1% [95% CI, 44.9-64.2] vs 17% [95% CI, 10.4-25.1]; P < .001; Figure 4A; OS: 66.5% [95% CI, 55.4-75.4] vs 35.2% [95% CI, 25.9-44.6]; P < .001; Figure 4B). RR was significantly lower for IDHmut/NPM1mut vs IDHmut/NPM1WT AML (35.3% [95% CI, 25.2-45.6] vs 66% [95% CI, 51.3-77.3]; P < .001).
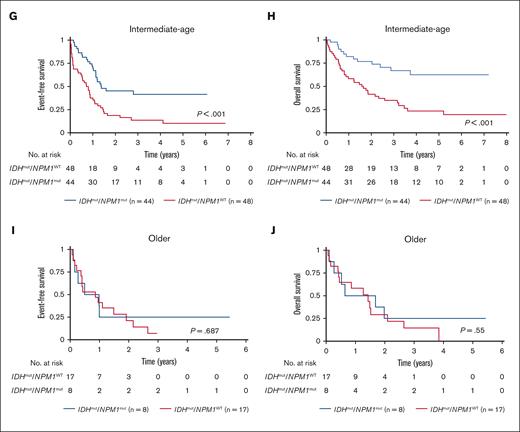
Cooccurrence of NPM1 and IDH mutations is associated with improved survival outcomes in the total study cohort. Survival outcomes based on NPM1 and IDH mutation status in AML (A) EFS and (B) OS for the total study cohort; EFS and OS in each age group: pediatric (C-D), AYA (E-F), intermediate-age (G-H), and older patients (I-J). P values calculated by log-rank test.
Cooccurrence of NPM1 and IDH mutations is associated with improved survival outcomes in the total study cohort. Survival outcomes based on NPM1 and IDH mutation status in AML (A) EFS and (B) OS for the total study cohort; EFS and OS in each age group: pediatric (C-D), AYA (E-F), intermediate-age (G-H), and older patients (I-J). P values calculated by log-rank test.
Similarly, when analyzed by age group, patients aged <60 years with IDHmut/NPM1mut had superior EFS compared with those with IDHmut/NPM1WT (pediatric: 70.6% [95% CI, 50.7-83.7] vs 23.1% [95% CI, 9.4-40.3], P < .001; AYA: 67.3% [95% CI, 45.0-82.2] vs 33.7% [95% CI, 13-56], P = .048; intermediate-age: 41.5% [95% CI, 26.0-56.4] vs 10.3% [95% CI, 3.3-22.0], P < .001; Figure 4C,E,G). OS was significantly better for IDHmut/NPM1mut vs IDHmut/NPM1WT AML in the pediatric (86.8% [95% CI, 68-95] vs 57.7% [95% CI, 36.8-73.9], P = .011; Figure 4D) and intermediate-age groups (62.5% [95% CI, 43.5-76.8] vs 23.4% [95% CI, 11.9-37.2], P < .001; Figure 4H); however, this effect was not observed in OS in the AYA group (57.2% [95% CI, 30.3-76.9] vs 57.1% [95% CI, 28.7-77.8], P = .641; Figure 4F). Among older patients with IDHmut AML, outcomes were poor irrespective of cooccurring NPM1 mutation IDHmut/NPM1mut vs IDHmut/NPM1WT, EFS: 25% [95% CI, 3.7-55.8] vs 7.1% [95% CI, 0.5-27.0]; P = .687 and OS: 25% [95% CI, 3.7-55.8] vs 0%; P = .55; Figure 4I-J.
When comparing outcomes of IDHmut AML by mutation subtypes, IDH1R132 and IDH2R140, the favorable prognostic impact of cooccurring NPM1 was retained (IDH1R132/NPM1mut vs IDH1R132/NPM1WT: OS, 65.1% [95% CI, 49.5-77] vs 26.1% [95% CI, 12.2-42.4]; P < .001; IDH2R140/NPM1mut vs IDH2R140/NPM1WT: OS, 68.4% [95% CI, 51.8-80.4] vs 36.3% [95% CI, 23.7-49]; P < .001). IDH2R172/NPM1mut occurred only once, thus precluding outcome analysis.
To further evaluate the effect of mutations in IDH in combination with NPM1, we analyzed outcomes for NPM1mut AML with or without IDH mutation. We found that EFS and OS were comparable between dual mutant NPM1mut/IDHmut vs NPM1mut/IDHWT in pediatric, AYA, and older patients (supplemental Table 5). In the intermediate-age cohort, NPM1mut/IDHmut patients had improved OS compared with NPM1mut/IDHWT (62.5% [95% CI, 43.5-76.8] vs 38.2% [95% CI, 29.3-47.0]; P = .009), but EFS differences did not achieve significance for NPM1mut/IDHmut vs NPM1mut/IDHWT (41.5% [95% CI, 26.0-56.4] vs 27.4% [95% CI, 19.7-35.6]; P = .089) (supplemental Figure 1). In the overall cohort, to which intermediate age patients contributed significantly, patients with NPM1mut/IDHmut compared with NPM1mut/IDHWT showed improved OS at 66.5% (95% CI, 55.4-75.5) vs 54.1% (95% CI, 48.2-59.6); P = .017 (supplemental Figure 1) and improved EFS, albeit just short of statistical significance at 55.1% (95% CI, 44.9-64.2) vs 44.3% (95% CI, 38.6-49.9); P = .056.
We considered the prognostic impact of IDH mutation in favorable risk AML without mutated NPM1. With the nontrivial overlap of CBF in IDHmut AML in younger patients (11.2%, n = 12 of 107 pediatric/AYA patients with IDHmut AML), we evaluated the outcomes of CBF AML in pediatric and AYA patients treated on COG trials (n = 11). The favorable prognosis of CBF AML was modulated by the addition of IDH mutation; cooccurrence of IDH mutation (CBF/IDHmut) was associated with inferior outcomes when compared with wildtype IDH (CBF/IDHWT); OS: 54.6% (95% CI, 22.9-78) vs 81.5% (95% CI, 77.6-84.7; P = .03) and EFS: 27.3% (95% CI, 6.5-53.9) vs 62.4% (95% CI, 57.9-66.6; P = .001).
We analyzed the prognostic impact of DNMT3A in the presence of cooccurring IDH and NPM1 mutations in the intermediate-age cohort in which this mutation combination was most prevalent. Intermediate-age patients with IDHmut/NPM1mut/DNMT3Amut AML had inferior outcomes compared with those with IDHmut/NPM1mut/DNMT3AWT; EFS: 19% (95% CI, 5.9-37.7) vs 64% (95% CI, 38.1-81.3; P < .001); and OS: 47.7% (95% CI, 24.5-67.8) vs 76.5% (95% CI, 40.8-92.3; P = .019) and higher RR (87.6% [95% CI, 49.3-97.1] vs 30.4% [95% CI, 11.9-51.3]; P < .001) (supplemental Figure 2).
Comparison of outcomes for IDHmut AML based on the presence of cooccurring FLT3-ITD mutation demonstrated no difference based on presence of FLT3-ITD (OS: IDHmut/FLT3-ITD vs IDHmut/FLT3-non-ITD; 44.4% [95% CI, 22.6-55.9] vs 51.6% [95% CI, 43.2-59.3]; P = .409) (supplemental Figure 3). We evaluated the impact of FLT3-ITD in combination with both IDH and NPM1 mutations in the overall cohort and found that IDHmut/NPM1mut/FLT3-ITD compared with IDHmut/NPM1mut/FLT3-non-ITD was associated with inferior OS (45.8% [95% CI, 24.6-64.7] vs 74.3% [95% CI, 61.4-83.4]; P = .018), whereas no significant difference in EFS was seen (45.7% [95% CI, 26.7-62.9] vs 58.5% [95% CI, 46.7-69.1]; P = .274, supplemental Figure 3). This difference in OS was largely driven by intermediate-age patients because those with IDHmut/NPM1mut/FLT3-ITD experienced inferior OS compared with those with IDHmut/NPM1mut/FLT3-non-ITD (33.3% [95% CI, 6.3-64.6] vs 73.2% [95% CI, 51.4-86.4]; P = .021). Pediatric and AYA patients with IDHmut/NPM1mut/FLT3-ITD AML experienced inferior OS compared with those with IDHmut/NPM1mut/FLT3-non-ITD, although these differences did not reach statistical significance. In a multivariable analysis, there was no statistical difference for EFS, OS, and RR by IDH mutation status when adjusting for age group, cooperative group, risk group, white blood cell count, blast percentage, NPM1 mutation, or FLT3-ITD mutation (supplemental Table 6).
Discussion
In this study we investigated the prevalence, cooccurring mutational profile, and prognostic significance of IDH mutations in AML across the age spectrum, using a large cohort of 3141 patients. We demonstrated an age-associated prevalence of IDH1 and IDH2 mutations in AML and showed that IDH mutations increased in frequency from 3.4% in pediatric patients to 21% in those older than 60 years. Our findings are concordant with prior reports of mutation prevalence.4,7,9,50 Furthermore, we identified cooccurring mutational patterns that were associated with prognostic outcomes. Although limited by small numbers, patients aged ≥60 years had poor outcomes regardless of mutational status. In patients younger than 60 years, those with dual IDHmut/NPM1mut AML had better EFS compared with those with IDHmut/NPM1WT AML; furthermore, IDH mutation did not abolish the favorable prognostic impact of NPM1 mutation. Patients of all ages with IDHmut AML who lacked a cooperating NPM1 mutation, experienced unfavorable outcomes. We observed that the favorable outcomes of IDHmut/NPM1mut AML were abrogated by cooccurrence of DNMT3A or FLT3-ITD.
Our age-expansive cohort showed that younger patients with IDHmut AML had superior survival outcomes compared with older patients. This can be attributed to many factors including enrichment of favorable risk mutations and paucity of adverse risk mutations in the younger age cohorts. We found that favorable risk NPM1 mutations were least prevalent in patients aged >60 years with IDHmut AML, occurring in less than one-third of older patients, in contrast to at least half of patients aged <60 years. In contrast, adverse risk mutations including RUNX1 and ASXL1 were far more prevalent in intermediate-age and older adults with IDHmut AML. The improved pediatric outcomes may also reflect better tolerance of more intensive therapeutic regimens, irrespective of IDH mutational profile.
When we compared patients with IDHmut AML with those with IDHWT AML, we found no difference in response to induction therapy or survival outcome in our total cohort or in any age group. Among our large cohort, there was no difference in OS or EFS between IDH1mut and IDH2mut AML, consistent with recent analysis from a large German cohort.51 Furthermore, we demonstrated no outcome differences among IDH1R132, IDH2R140, and IDH2R172K for the overall study cohort or in any age group.
IDH and NPM1 mutations frequently co-occur, but there has been lack of consensus on the prognostic significance of IDH mutations in conjunction with NPM1.5,32,44 Recent reports from several large study cohorts have provided clarity about the favorable prognostic influence of cooccurring NPM1 in IDHmut AML in adult patients, most of whom were treated with intensive chemotherapy.51-53 In our study, after separating patients with IDHmut based on NPM1 status, we observed that almost all patients with IDHmut/NPM1WTAML had dismal EFS, regardless of age, IDH isoform, or mutation subtype. This is consistent with previous reports in smaller cohorts of adult patients for IDHmut/NPM1WTAML, including when analyzed by subtype.3,24,44,53,54 We found that patients aged <60 years with IDHmut/NPM1mut AML had significantly improved EFS compared with patients with IDHmut/NPM1WT. Of note, despite significantly worse EFS, the AYA cohort with IDHmut/NPM1WT AML had overlapping OS with those with IDHmut/NPM1mut AML, which suggests salvage strategies were effective in this group; however, the patient cohort was limited in number and by heterogenous treatment across several studies, thereby limiting our ability to better understand why dismal outcomes were salvageable in this age cohort. Although limited by smaller numbers of NPM1 mutations compared with the younger age groups, patients aged >60 years in this study had poor survival outcomes regardless of IDH and NPM1 mutation status, thus there were no discernible differences based on mutational profile.
Furthermore, in our study we found that IDH mutation did not abrogate the favorable prognostic impact of NPM1 mutation in patients aged <60 years. We also observed that in the intermediate-age cohort, patients with NPM1mut AML had improved outcomes in the presence of cooccurring IDH mutation compared with those with NPM1mut/IDHWT AML. Further analysis of this finding was limited by the small numbers of patients with dual IDHmut/NPM1mut AML, especially when divided by age; however, the favorable outcome in these patients is of interest for further study. In a multivariate analysis, our findings showed that cooccurrence with NPM1 had a more powerful impact on prognosis than IDH mutation alone and suggests that risk stratification per IDH should incorporate NPM1 mutational status.
Our findings demonstrated that in patients aged <60 years with IDHmut/NPM1mut AML, the frequently cooccurring DNMT3A and FLT3-ITD mutations could further inform prognostication. We demonstrated that patients with IDHmut/NPM1mut/DNMT3Amut AML had inferior EFS and OS compared with those with IDHmut/NPM1mut AML, suggesting that DNMT3A mutation abrogated the positive prognostic effect of NPM1. Our findings align with previous findings of inferior survival for this mutation combination.53,55 The addition of FLT3-ITD mutation in IDHmut/NPM1mut AML led to inferior OS in the total cohort. This decline in OS for IDHmut/NPM1mut/FLT3-ITD AML reached statistical significance in intermediate-age patients with a similar pattern in pediatric and AYA groups. Thus, FLT3-ITD mutation negated the favorable impact of NPM1 mutation in IDHmut AML. With the availability of FLT3 and IDH inhibitors, patients with IDHmut/NPM1mut/FLT3-ITD AML may be ideal candidates for prospective clinical investigation to incorporate these therapeutic strategies.56
Our study included the largest cohort of pediatric and AYA patients to undergo IDH mutation analysis. With the inclusion of patients from several large pediatric trials, the distribution of ages for IDHWT AML was significantly skewed to younger age, and we conducted analyses by total cohort and age group to account for this whenever possible. Because of limited available data, we reported outcomes for only 25 patients aged >60 years; thus, these conclusions should be considered descriptive, particularly for subanalyses by mutation profile. We were unable to evaluate outcomes in the older patient (≥75 years) age group because only 2 patients had outcomes data available. The retrospective nature of our study was an inherent limitation and comparative outcomes must be interpreted with caution. Our analyses were confounded by nonuniform treatment of patients enrolled in studies conducted by various cooperative groups in different diagnostic and treatment eras. Given the time period represented in our study, we were unable to assess the impact of modern-day targeted therapies including IDH inhibitors. Certain analyses were limited by small patient numbers; for instance, we could not determine the impact of low-intensity approaches for older patients or higher-intensity treatments including hematopoietic stem cell transplant (HSCT) for younger adults. The role of HSCT on treatment outcomes could not be determined either because the clinical trials in our cohort did not capture data on HSCT, or too few patients with IDHmut AML underwent transplantation in first CR. Future studies should pay special attention to these subsets to better understand differential outcomes with targeted therapies and treatment approaches of varying intensity.
This expansive study of IDHmut AML across the age spectrum identified specific mutation combinations with inferior outcomes including patients with IDHmut/NPM1WT AML and IDHmut/NPM1mut with DNTM3Amut or FLT3-ITD. These patients may derive benefit from risk-adapted and age-adjusted therapy modifications including consideration of IDH inhibitors in combination with intensive chemotherapy, hypomethylating agents, Bcl-2 inhibition,22,50,52,57,58 and potentially HSCT for remission consolidation in younger or fit patients. Although rare in pediatric AML, our study demonstrates that subsets of IDHmut AML portend an inferior prognosis in pediatric patients and may also warrant consideration of risk-adapted therapy modification. In our study, patients aged >60 years with IDHmut all experienced poor outcomes, regardless of NPM1 mutational status, underscoring the need to study targeted therapy combinations with tolerable profiles for these patients. Our report on outcomes in subsets of IDHmut AML provides important historical comparator data as IDH inhibitors are studied for frontline use in adults and in the relapsed/refractory setting in pediatric patients.
Acknowledgments
This work was supported by funding from the National Cancer Institute’s National Clinical Trials Network (NCTN) Operations Center (grant U10CA180886), NCTN Statistics & Data Center grant U10CA180899, and the St. Baldrick’s Foundation.
Authorship
Contribution: S.Z.-L., K.T., R.B.G., R.E.R., T.A.A., and S.M. designed the study; R.B.G., T.A.A., M.O., Z.S., and J.W. provided biostatistical support for the study; S.Z.-L., K.T., R.E.R., S.M., R.B.G., T.A.A., M.O., and Z.S. analyzed and interpreted the data; S.Z.-L., K.T., R.B.G., and S.M. wrote the manuscript; S.Z.-L., K.T., R.B.G., A.L., and R.E.R. created manuscript figures; and all authors collected clinical and/or genetic data, reviewed the manuscript, and approved the final version of the manuscript.
Conflict-of-interest disclosure: M.O. reports consulting or an advisory role for GlycoMimetics, Cascadia Labs, Merck, Daiichi Sankyo, and BioSight. J.P.R. reports personal fees from Novartis, Bristol Myers Squibb, Takeda, Amgen, Cepheid, and Genentech outside the submitted work. M.S.T. reports grant/research support from AbbVie, Orsenix, Biosight, GlycoMimetics, Rafael Pharmaceuticals, and Amgen; a scientific advisory role for AbbVie, Orsenix, Biosight, Daiichi Sankyo Co., KAHR, Novartis Pharmaceuticals, and Innate Pharmaceuticals; and royalty from UpToDate. M.L. reports consulting or an advisory role for Omeros and Jazz Pharmaceuticals; reports research funding form Amgen, Astellas Pharma, Actinium Pharmaceuticals, Pluristem Therapeutics, AbbVie/Genentech, Tolero Pharmaceuticals, and AbbVie; and served on the data monitoring committee for BioSight. E.A. reports consultancy for AbbVie, Takeda, Pfizer, Novartis, and Amgen; speakers’ bureau role for AbbVie and Bristol Myers Squibb; received honoraria from Bristol Myers Squibb and Novartis; and received research funding from Takeda, Pfizer, and Novartis. O.A.-W. has served as a consultant for H3B Biomedicine, Foundation Medicine Inc., Merck, Prelude Therapeutics, and Janssen; is on the scientific advisory board of Envisagenics Inc., AIChemy, Harmonic Discovery Inc., and Pfizer Boulder; and has received prior research funding from H3B Biomedicine, Nurix Therapeutics, Minovia Therapeutics, and Loxo Oncology unrelated to the current manuscript. S.L. received honoraria from Syros, Agios, Daiichi Sankyo, Jazz Pharmaceuticals, Bristol Myers Squibb, Acceleron, Astellas, and Pfizer, and research funding from Onconova, Celgene, Biosight, Hoffman LaRoche, and Kura. H.E. reports consultancy or an advisory role for Agios, Astellas Pharma, Amgen, Celgene, Daiichi Sankyo, GlycoMimetics, Immunogen, Incyte, Jazz Pharmaceuticals, MacroGenics, Novartis, AbbVie/Genentech, Janssen Oncology, Pfizer, Trillium Therapeutics, Takeda, and Kura Oncology; a speakers’ bureau role for Agios, Celgene, Incyte, Jazz Pharmaceuticals, Novartis, and AbbVie/Genentech; received research funding from AbbVie, Agios (Inst), Amgen (Inst), Daiichi Sankyo (Inst), Forma Therapeutics (Inst), Gilead/Forty Seven (Inst), Immunogen (Inst), Jazz Pharmaceuticals (Inst), MacroGenics (Inst), Novartis (Inst), PTC Therapeutics (Inst), AbbVie (Inst), GlycoMimetics (Inst), and ALX Oncology (Inst); declares other relationship with GlycoMimetics and Celgene; and reports an uncompensated relationship with Daiichi Sankyo. R.L. serves on the supervisory board of Qiagen; is a scientific adviser to Imago, Mission Bio, Syndax, Zentalis, Ajax, Bakx, Auron, Prelude, C4 Therapeutics, and Isoplexis for which he receives equity support; received research support from Ajax and AbbVie; reports consultancy for Incyte, Janssen, Morphosys, and Novartis; and received honoraria from AstraZeneca and Kura for invited lectures, and from Gilead for grant reviews. The remaining authors declare no competing financial interests.
Correspondence: Sara Zarnegar-Lumley, Division of Hematology/Oncology, Department of Pediatrics, Vanderbilt University Medical Center, 2220 Pierce Ave, PRB 397, Nashville, TN 37232; e-mail: sara.zarnegar@vumc.org.
References
Author notes
∗S.Z.-L., T.A.A., S.M., and K.T. contributed equally to this work.
The data used for analysis in this manuscript are available in a variety of publicly available databases. The Database for Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap/) houses data for the AML TARGET (phs000465.v22.p8), Beat AML (phs001657.v2.p1), and The Cancer Genome Atlas (TCGA)-LAML (phs000178.v11.p8) projects. These data sets are also available through the Genomic Data Commons (GDC; https://portal.gdc.cancer.gov/). The Beat AML data set can also be viewed in an interactive browser available at: http://www.vizome.org/. In addition, the TCGA has an interactive browser available at: https://software.broadinstitute.org/software/igv/tcga. The ECOG E1900 mutation data are published here https://doi.org/10.1182/blood.V116.21.851.851. Transfer of the SWOG S0106 data into dbGaP is underway.
Data are available on request from the corresponding author, Sara Zarnegar-Lumley (sara.zarnegar@vumc.org).
The full-text version of this article contains a data supplement.