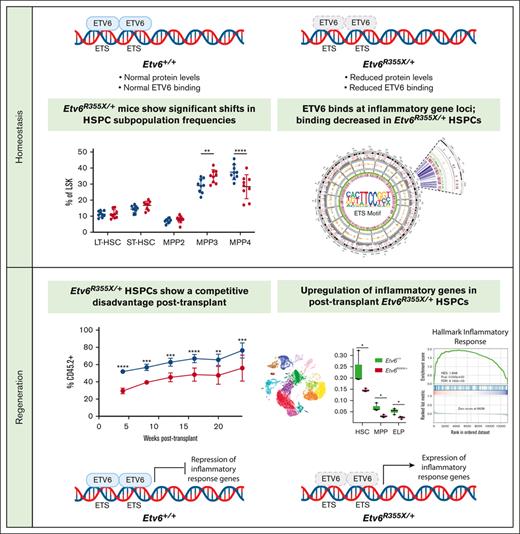
Key Points
Germline heterozygosity for a thrombocytopenia 5–associated ETV6 variant impairs the repopulating capacity of mouse HSPCs.
In HSPCs, ETV6 binds to Tnf and other inflammatory gene loci and represses their expression during stress hematopoiesis.
Abstract
ETS variant 6 (ETV6) encodes a transcriptional repressor expressed in hematopoietic stem and progenitor cells (HSPCs), where it is required for adult hematopoiesis. Heterozygous pathogenic germline ETV6 variants are associated with thrombocytopenia 5 (T5), a poorly understood genetic condition resulting in thrombocytopenia and predisposition to hematologic malignancies. To elucidate how germline ETV6 variants affect HSPCs and contribute to disease, we generated a mouse model harboring an Etv6R355X loss-of-function variant, equivalent to the T5-associated variant ETV6R359X. Under homeostatic conditions, all HSPC subpopulations are present in the bone marrow (BM) of Etv6R355X/+ mice; however, these animals display shifts in the proportions and/or numbers of progenitor subtypes. To examine whether the Etv6R355X/+ mutation affects HSPC function, we performed serial competitive transplantation and observed that Etv6R355X/+ lineage−sca1+cKit+ (LSK) cells exhibit impaired reconstitution, with near complete failure to repopulate irradiated recipients by the tertiary transplant. Mechanistic studies incorporating cleavage under target and release under nuclease assay, assay for transposase accessible chromatin sequencing, and high-throughput chromosome conformation capture identify ETV6 binding at inflammatory gene loci, including multiple genes within the tumor necrosis factor (TNF) signaling pathway in ETV6-sufficient mouse and human HSPCs. Furthermore, single-cell RNA sequencing of BM cells isolated after transplantation reveals upregulation of inflammatory genes in Etv6R355X/+ progenitors when compared to Etv6+/+ counterparts. Corroborating these findings, Etv6R355X/+ HSPCs produce significantly more TNF than Etv6+/+ cells post-transplantation. We conclude that ETV6 is required to repress inflammatory gene expression in HSPCs under conditions of hematopoietic stress, and this mechanism may be critical to sustain HSPC function.
Introduction
At the root of the blood system lies a heterogenous population of hematopoietic stem and progenitor cells (HSPCs), which reside in the bone marrow (BM) and give rise to diverse blood lineages. Maintaining this pool of self-renewing HSPCs is critical to uphold hematopoiesis during periods of stress such as bleeding or infection.1 ETS variant transcription factor 6 (ETV6) is a transcriptional repressor that is highly expressed in HSPCs,2 in which it is essential for development and maintenance of adult hematopoiesis.3-5 In 2015, our group6 and others7-9 identified germ line pathogenic ETV6 variants in families with predispositions to B–acute lymphoblastic leukemia (B-ALL) and thrombocytopenia, defining a new genetic syndrome known as thrombocytopenia 5 (T5). Subsequently, we performed targeted germ line ETV6 sequencing of remission blood samples from more than 4000 children with B-ALL and identified germ line ETV6 variants in ∼1% of cases.9 In vitro studies have revealed that pathogenic ETV6 variant proteins exhibit impaired repressor activity, reduced DNA binding, and abnormal subcellular localization.6-8,10 Overall, these studies indicate that T5-associated germ line ETV6 variants negatively affect the repressor activity of ETV6. Supporting this notion, transcriptional profiling of peripheral blood cells from patients with T5 has identified upregulation of interferon response genes.11 Nevertheless, little remains known about the mechanisms by which ETV6 regulates the HSPC compartment and how T5-associated ETV6 variants contribute to disease.
To address these questions, we used CRISPR–CRISPR-associated protein 9 (CRISPR-Cas9) gene editing to generate a novel mouse model harboring a pathogenic heterozygous germ line Etv6 variant, R355X. We chose to examine this variant because it is equivalent to the human T5-associated mutation R359X9 and is representative of the ∼50% of disease-causing mutations that truncate the ETV6 protein within or upstream of the ETS DNA–binding domain.10 Through the comprehensive study of this mouse model, we describe a role for ETV6 during regenerative hematopoiesis and show that the heterozygous Etv6R355X variant impairs HSPC function. Using integrated genomic and transcriptomic approaches, including cleavage under target and release under nuclease (CUT&RUN), assay for transposase accessible chromatin sequencing (ATAC-seq), high-throughput chromosome conformation capture (Hi-C), and single-cell RNA sequencing (RNA-seq) to interrogate mouse and human HSPCs and the mouse BM-derived stem cell-like HPC5 cell line, we identify new putative ETV6 targets, including the gene encoding tumor necrosis factor (TNF) as well as other genes involved in inflammatory signaling. Furthermore, we show increased TNF production in Etv6R355X/+ mouse HSPCs after BM transplantation. Together, these findings provide novel insights into the pathways regulated by ETV6 in HSPCs and demonstrate how a pathogenic T5-associated variant affects ETV6 function in the context of hematopoietic stress.
Methods
Mice
C57BL/6J, C57BL/6.SJL-PtprcaPep3b/BoyJ (CD45.1) mice were obtained from The Jackson Laboratory (Bar Harbor, Maine). Etv6R355X/+ mice were generated by zygotic injection at St Jude Children’s Research Hospital. To generate the ETV6:p.R>X variant, the conserved site in the mouse ortholog at position 355 within the ETS DNA–binding domain was targeted. A mixture of 100 ng/mL recombinant Cas9 protein, 50 ng/uL in vitro–transcribed genomic RNA, and 10 ng/μL single-stranded oligodeoxynucleotides (100 bp) was injected into proliferating C57BL/6J blastocysts, which were implanted into pseudopregnant C57BL/6J female mice. Founder mice were genotyped by Sanger sequencing and backcrossed for 3 generations before use. The levels of ETV6 protein in Etv6+/+and Etv6R355X/+ B220-enriched splenocytes were detected by western blotting, as described in the supplemental Methods. Experiments were carried out with approval from the St Jude Children’s Research Hospital institutional animal care and use committee.
Flow cytometry
Cells were stained for flow cytometric analysis in fluorescence-activated cell sorting staining buffer (phosphate-buffered saline supplemented with 10 g/L bovine serum albumin and 0.5 g/L sodium azide) for 30 minutes at 4°C using antibodies noted in the supplemental Methods. All flow cytometric assays were performed on an LSR Fortessa (BD Biosciences, San Jose, CA).
Blood cell analyses
Complete blood counts were obtained using a Forcyte Analyzer (Oxford Science Inc, Oxford, CT). Clot retraction assays were performed as previously described12 and further outlined in the supplemental Methods. For transmission electron microscopy, samples were prepared as in the supplemental Methods. Embedded samples were sectioned at 70 nm using a Leica (Wetzlar, Germany) ultramicrotome and examined in a TF20 transmission electron microscope (Thermo Fisher Scientific, Hillsboro, OR) at 80 kV. Digital micrographs were captured with an Advanced Microscopy Techniques (Woburn, MA) imaging system.
Serial competitive transplantation experiments
For the primary transplant, 10 000 CD45.2+Etv6+/+ or Etv6R355X/+ lineage−sca1+cKit+ (LSK) test and 10 000 CD45.1+Etv6+/+ LSK competitor cells were transferred IV into a lethally-irradiated CD45.1/.2+Etv6+/+ recipient mouse. Peripheral blood chimerism was assessed by flow cytometry every 4 weeks. Sixteen weeks later, half of the recipient mice were euthanized, and the BM collected. CD45.1+ cells were depleted using anti-CD45.1 biotin with streptavidin beads on an autoMACs magnetic cell separator (Miltenyi Biotec, Carlsbad, CA). For the secondary transplant, 200 000 CD45.1-depleted Etv6+/+ or Etv6R355X/+ BM test cells were resuspended with 200 000 CD45.1+Etv6+/+ BM competitor cells collected from a new donor and transferred IV into a lethally-irradiated CD45.1/.2+Etv6+/+ recipient. Again, peripheral blood was collected and chimerism assessed by flow cytometry every 4 weeks. A tertiary transplant was then performed using the same methods as described for the secondary transplant. At 24 weeks after the primary, secondary, or tertiary transplant, the remaining recipients were euthanized and hematopoietic reconstitution within tissues assessed.
scRNA-seq and intracellular TNF production after transplantation
Ten thousand CD45.2+Etv6R355X/+ and 10 000 CD45.1+Etv6+/+ LSK cells were transferred IV into lethally-irradiated recipients. Six weeks later, recipient animals were euthanized, and the BM isolated for single-cell RNA sequencing (scRNA-seq) analysis and evaluation of intracellular TNF production. For scRNA-seq, CD45.1+ and CD45.2+ LSK cells were sort-purified from each recipient mouse. Subsequently, LSK cells and residual whole BM cells were washed 3 times with 1× phosphate-buffered saline (calcium- and magnesium-free) containing 0.04% weight/volume bovine serum albumin (Thermo Fisher Scientific) and manually counted. LSK and BM cells were mixed at a ratio of 1:1 to have a total desired recovery target ranging from 8000 to 10 000 total cells and were loaded onto a Chromium Next GEM Chip K (10X Genomics), with libraries prepared following the manufacturer’s protocol (see supplemental Methods for full details). Libraries were sequenced on the NovaSeq 6000 (Illumina) per the manufacturer's recommendations.
To measure intracellular TNF production, BM cells collected at 6 weeks after competitive transplantation were resuspended in RPMI 1640 medium containing 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% L-glutamine, at a concentration of 5 million cells per mL. Then, 1 mL of cell suspension per well was plated in a 24-well tissue culture plate for 24 hours. Five hours before collection, cells were treated with brefeldin A (final concentration, 3 μg/mL; Invitrogen, Waltham, MA) and protein transport inhibitor including monensin (final concentration, 112 μg/mL; monesin, BD Biosciences). Cells were surface-stained to detect specific HSPC populations (as described in the supplemental Methods), then fixed and permeabilized using the BD Cytofix/Cytoperm kit (BD Biosciences), followed by intracellular staining using TNF-allophycocyanin (MP6-XT22, Invitrogen) or Rat immunoglobulin G allophycocyanin (HRPN, TONBO).
CUT&RUN assay
One million HPC5 cells, 50 000 to 100 000 Etv6+/+ or Etv6R355X/+ LSK cells, or 500 000 CD34+ cells were subjected to CUT&RUN, as previously described13,14 using an anti-ETV6 antibody generated in our laboratory and a normal rat immunoglobulin G isotype control antibody (#2729, Cell Signaling Technologies; see supplemental Methods for full details).
Hi-C assay
Hi-C was performed using 5 million HPC5 mouse BM-derived progenitor cells per manufacturer’s instructions (ARIMA Genomics; as described in supplemental Methods).
ATAC-seq assay
ATAC-seq was performed following the Fast-ATAC protocol, using 100 000 HPC5 cells per sample, as previously described,15-17 with details included in the supplemental Methods.
Bioinformatics analyses
scRNA-seq analysis was performed following best practices, as described previously.18 Briefly, 10X Genomics sequencing output was processed by Cellranger version 7.1, followed by normalization, clustering, annotation, and integration using Scanpy19 in Python (supplemental Methods). CUT&RUN, ATAC-seq, and Hi-C analysis was performed using the in-house HemTools pipeline20 (supplemental Methods; code availability21).
Statistical analyses
Graphs were generated and statistical analyses performed using GraphPad Prism version 9 for Mac and R (version 4.1.2) software. Statistical analyses were as indicated in the figure legends.
Results
Generation and initial characterization of Etv6 R355X knockin mice
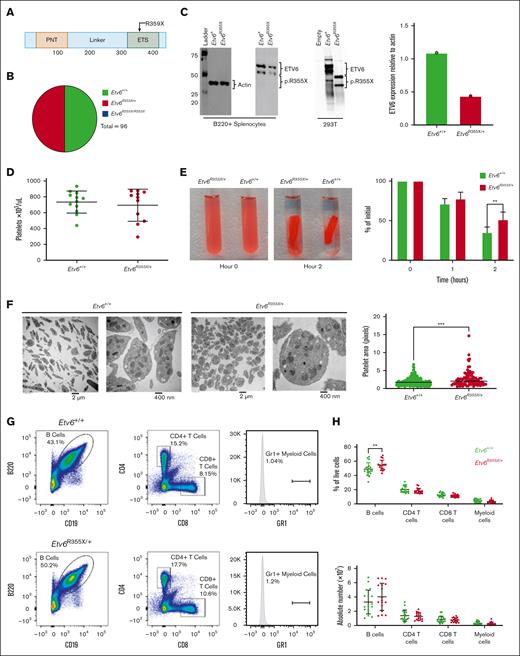
To better understand how germ line ETV6 variants affect hematopoiesis, we generated a knockin mouse model harboring an Etv6 R355X variant, which truncates the protein within the ETS domain and is comparable to a previously identified T5-associated ETV6 R359X variant (Figure 1A). Because ∼50% of T5-associated ETV6 variants are predicted to truncate ETV6 within, or upstream of, the DNA binding domain,10 we proposed that this model would recapitulate the human disease. We generated this mouse model via homology-directed repair mediated by the delivery of recombinant Cas9 protein complexed with targeting single-guide RNA and a 100–base pair single-strand DNA–repair template into mouse zygotes. After tracking the first 5 Etv6R355X/+ x Etv6R355X/+ breeding pairs, we observed no homozygous Etv6R355X/R355X pups, leading us to conclude that the Etv6R355X/R355X genotype is embryonic lethal. Among 96 viable pups, 48 were of genotype Etv6R355X/+ and 48 of genotype Etv6+/+, a ratio significantly divergent from the expected Mendelian ratio of 2:1 (Etv6R355X/+:Etv6+/+; P < .00001, χ2 analysis; Figure 1B). Etv6R355X/+ animals appeared healthy through 24 months of age, with no development of spontaneous malignancies. Western blot analysis of B220-enriched Etv6R355X/+ splenocytes revealed no truncated ETV6 protein. Furthermore, the levels of full-length ETV6 were half of those observed in Etv6+/+ cells (Figure 1C; western blotting demonstrates 2 bands because of an internal initiation site at codon 43). These findings are in agreement with our prior analysis of BM samples from individuals with ETV6R359X/+, which also showed reduced full-length ETV6 protein but no truncated product.10 From these data, we propose that the Etv6R355X variant results in ETV6 haploinsufficiency, although there may exist scant levels of protein below our level of detection.
Etv6R355X is homozygous lethal and heterozygotes display abnormal platelets and increased B cells. (A) Schematic diagram depicting the functional domains of ETV6 and location of the ETV6:p.R359X variant. (B) Genotypes generated by Etv6R355X/+ × Etv6R355X/+ mating, showing no Etv6R355X/R355X pups. (C) Western blot and quantification of ETV6 protein levels in B220-enriched splenocytes from Etv6+/+ and Etv6R355X/+ mice as well as 293 T cells transfected with empty vector, wild-type Etv6, or Etv6R355X. Etv6 contains an alternative initiation site at M43 resulting in 2 bands visible by western blot. (D) Peripheral blood platelet count in Etv6+/+ (n = 12) and Etv6R355X/+ (n = 11) mice. Data are from 2 combined experiments and show mean ± standard deviation (SD). (E) Representative images and quantification of ex vivo clot retraction using platelets from Etv6+/+ (n = 4) and Etv6R355X/+ (n = 4) mice, showing percent of initial clot area at 0, 1, and 2 hours. Data are representative of 2 experiments. (F) Transmission electron microscopy images of platelets from Etv6+/+ and Etv6R355X/+ mice and quantification of platelet area from Etv6+/+ (n = 192) platelets and Etv6R355X/+ (n = 92) platelets. (G) Representative gating strategy to identify leukocyte populations, showing representative plots from Etv6+/+ (top) and Etv6R355X/+ (bottom) mouse spleens. All populations are gated directly off live singlets. Gr1 labels both granulocytes and monocytes, gates are set using a tissue with a visible Gr1+ population (ie, BM; supplemental Figure 1B) and applied to all samples. (H) Frequency (top) and absolute number (bottom) of splenic leukocytes in Etv6+/+ (green, n = 18) and Etv6R355X/+ (red, n = 17) mice at 3 months of age. Data are from 5 combined experiments and show mean ± SD. ∗P < .05; ∗∗P < .001; and ∗∗∗P < .0001, as determined by unpaired t tests or two-way analysis of variance (ANOVA) with a Holm-Šidák multiple comparisons test.
Etv6R355X is homozygous lethal and heterozygotes display abnormal platelets and increased B cells. (A) Schematic diagram depicting the functional domains of ETV6 and location of the ETV6:p.R359X variant. (B) Genotypes generated by Etv6R355X/+ × Etv6R355X/+ mating, showing no Etv6R355X/R355X pups. (C) Western blot and quantification of ETV6 protein levels in B220-enriched splenocytes from Etv6+/+ and Etv6R355X/+ mice as well as 293 T cells transfected with empty vector, wild-type Etv6, or Etv6R355X. Etv6 contains an alternative initiation site at M43 resulting in 2 bands visible by western blot. (D) Peripheral blood platelet count in Etv6+/+ (n = 12) and Etv6R355X/+ (n = 11) mice. Data are from 2 combined experiments and show mean ± standard deviation (SD). (E) Representative images and quantification of ex vivo clot retraction using platelets from Etv6+/+ (n = 4) and Etv6R355X/+ (n = 4) mice, showing percent of initial clot area at 0, 1, and 2 hours. Data are representative of 2 experiments. (F) Transmission electron microscopy images of platelets from Etv6+/+ and Etv6R355X/+ mice and quantification of platelet area from Etv6+/+ (n = 192) platelets and Etv6R355X/+ (n = 92) platelets. (G) Representative gating strategy to identify leukocyte populations, showing representative plots from Etv6+/+ (top) and Etv6R355X/+ (bottom) mouse spleens. All populations are gated directly off live singlets. Gr1 labels both granulocytes and monocytes, gates are set using a tissue with a visible Gr1+ population (ie, BM; supplemental Figure 1B) and applied to all samples. (H) Frequency (top) and absolute number (bottom) of splenic leukocytes in Etv6+/+ (green, n = 18) and Etv6R355X/+ (red, n = 17) mice at 3 months of age. Data are from 5 combined experiments and show mean ± SD. ∗P < .05; ∗∗P < .001; and ∗∗∗P < .0001, as determined by unpaired t tests or two-way analysis of variance (ANOVA) with a Holm-Šidák multiple comparisons test.
Etv6R355X/+ mice exhibit platelet defects and increased B-cell proportions
Individuals with T5 display thrombocytopenia of varying degrees and abnormal platelet morphology and function.8,9,22 Although platelet numbers were similar in 3-month-old Etv6+/+ and Etv6R355X/+ mice (Figure 1D), Etv6R355X/+ platelets exhibited significantly delayed clot retraction (P = .0090 at 2 hours; Figure 1E) and an abnormally rounded shape with subtle but statistically increased area compared with that of Etv6+/+ cells (P < .0001, unpaired t test; Figure 1F). Three-month-old Etv6R355X/+ mice displayed no differences in proportions or numbers of mature cell lineages such as CD4+ or CD8+ T cells, or Gr1+ (marking both monocytes and granulocytes) myeloid cells; however, they exhibited a significant increase in the proportions of B220+CD19+ B cells in the spleen (P = .0032, unpaired t test; Figure 1G-H), BM, and peripheral blood (supplemental Figure 1A-C).
Etv6R355X/+ mice generate HSPC and committed progenitor populations
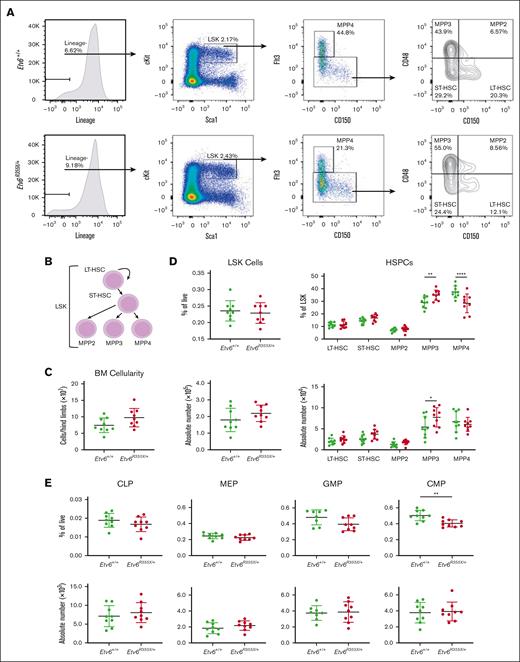
Mx1-Cre–mediated homozygous deletion of Etv6 leads to a rapid yet transient depletion of LSK cells.3 To examine whether the heterozygous Etv6R355X mutation affects the HSPC compartment, we quantified LSK cells and various HSPC subpopulations, including long-term hematopoietic stem cells (HSCs), short-term HSCs, and multipotent progenitor populations (MPPs; Figure 2A-B). Three-month-old Etv6+/+ and Etv6R355X/+ mice retained similar total BM cellularity (Figure 2C) as well as similar proportions and numbers of LSK cells (Figure 2D). However, Etv6R355X/+ animals exhibited a significant increase in the proportions and numbers of MPP3 cells (P = .008 and P = .0128, respectively, using two-way analysis of variance) and significant decrease in the proportions but not numbers of MPP4 cells (P < .0001, two-way analysis of variance; Figure 2D). Despite these shifts in heterogeneity, we observed no significant differences in cell cycling or cell death by flow cytometry (supplemental Figure 2).
Etv6R355X/+ mice generate all HSPC populations. (A) Representative gating strategy to identify HSPC subpopulations within LSK cells, including long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), MPP2, MPP3, and MPP4, showing representative plots from Etv6+/+ mouse BM (top row) and Etv6R355X/+ BM (bottom row). (B) Schematic diagram describing the hierarchical relationship between the earliest HSPC subpopulations. (C) Overall BM cellularity from both femurs of Etv6+/+ (green, n = 9) and Etv6R355X/+ (red, n = 9) mice. (D) Frequency (top) and absolute number (bottom) of LSK cells (left) and HSPC subpopulations (right) from Etv6+/+ (green, n = 9) and Etv6R355X/+ (red, n = 9) mice at 3 months of age. Data are representative of 4 independent experiments and show mean ± SD. (E) Frequency (top) and absolute number (bottom) of committed progenitor populations in Etv6+/+ (green, n = 9) and Etv6R355X/+ (red, n = 9) mice at 3 months of age. Data are representative of 2 independent experiments and show mean ± SD. ∗P < .05; ∗∗P < .001; and ∗∗∗P < .0001 were determined by unpaired t tests or two-way ANOVA with a Holm-Šidák multiple comparisons test. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocyte-erythroid progenitor.
Etv6R355X/+ mice generate all HSPC populations. (A) Representative gating strategy to identify HSPC subpopulations within LSK cells, including long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), MPP2, MPP3, and MPP4, showing representative plots from Etv6+/+ mouse BM (top row) and Etv6R355X/+ BM (bottom row). (B) Schematic diagram describing the hierarchical relationship between the earliest HSPC subpopulations. (C) Overall BM cellularity from both femurs of Etv6+/+ (green, n = 9) and Etv6R355X/+ (red, n = 9) mice. (D) Frequency (top) and absolute number (bottom) of LSK cells (left) and HSPC subpopulations (right) from Etv6+/+ (green, n = 9) and Etv6R355X/+ (red, n = 9) mice at 3 months of age. Data are representative of 4 independent experiments and show mean ± SD. (E) Frequency (top) and absolute number (bottom) of committed progenitor populations in Etv6+/+ (green, n = 9) and Etv6R355X/+ (red, n = 9) mice at 3 months of age. Data are representative of 2 independent experiments and show mean ± SD. ∗P < .05; ∗∗P < .001; and ∗∗∗P < .0001 were determined by unpaired t tests or two-way ANOVA with a Holm-Šidák multiple comparisons test. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; GMP, granulocyte-monocyte progenitor; MEP, megakaryocyte-erythroid progenitor.
We also assessed the proportions and numbers of more committed cell populations, including common lymphoid progenitors, megakaryocyte-erythroid progenitors, granulocyte-monocyte progenitors, and common myeloid progenitors (gating strategy shown in supplemental Figure 3). The only difference observed was a decrease in the proportions, but not numbers, of Etv6R355X/+ vs Etv6+/+ common myeloid progenitors (P = .0017, unpaired t test; Figure 2E). Together, these data reveal that Etv6R355X/+ mice generate each of the expected stem and progenitor cell subtypes within the BM, with only subtle shifts in the proportions and/or numbers of specific populations.
Etv6R355X/+ HSPCs are functionally impaired
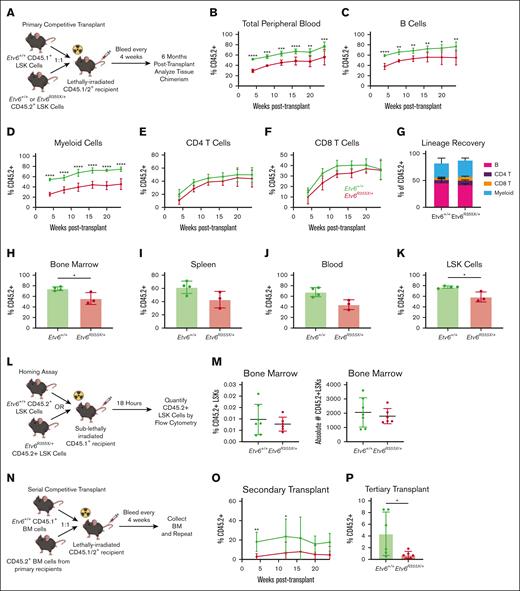
Having demonstrated that HSPCs are present in the BM of Etv6R355X/+ mice, we sought to investigate their functional capacity through serial competitive transplantation. For each primary transplant, we transferred 10 000 CD45.2+Etv6+/+ or Etv6R355X/+ test LSK cells along with 10 000 CD45.1+Etv6+/+ competitor LSK cells via IV injection into a lethally-irradiated CD45.1/.2+Etv6+/+ recipient (Figure 3A). Blood was collected every 4 weeks and analyzed to assess the percent chimerism and leukocyte recovery. At 24 weeks after transplantation, we collected the BM, peripheral blood, and splenocytes, which we analyzed by flow cytometry. Compared with Etv6+/+ LSK cells, Etv6R355X/+ cells displayed significantly reduced contribution to hematopoiesis, observed as early as 4 weeks after transfer (Figure 3B). This disadvantage affected B220+CD19+ B cells (Figure 3C) and Gr1+ myeloid cells (Figure 3D), with no significant difference between Etv6+/+ and Etv6R355X/+ T cells (Figure 3E-F). We did not observe any significant lineage skewing within the Etv6R355X/+ population (Figure 3G), and the reduced chimerism was observed across all hematopoietic tissues and within LSK cells (Figure 3H-K).
Etv6R355X/+ HSPCs show reduced long-term serial repopulation capacity. (A) Schematic diagram describing the competitive BM transplantation assay used for this study. Percent of CD45.2+ peripheral blood cells (B), B220+CD19+ B cells (C), Gr1+ myeloid cells (D), CD4+ T cells (E), and CD8+ T cells (F) after primary transplantation of 10 000 CD45.2+Etv6+/+ (green) or Etv6R355X/+ (red) LSK cells along with 10 000 CD45.1+Etv6+/+ LSK cells into lethally-irradiated recipient mice (n = 10-12 recipients per genotype per experiment). Data are representative of 3 experiments and show mean ± SD. (G) Percent of CD45.2+ peripheral blood Gr1+ myeloid cells, B220+ B cells, CD4+ T cells, and CD8+ T cells at 6 months after competitive transplantation. Percent of total BM cells (H), splenocytes (I), peripheral blood cells (J), and LSK cells (K) that are CD45.2+ at 6 months after transplantation within remaining recipients (note: half of the recipients were used for serial transplantation depicted in panel N). Data are representative of 3 experiments and show mean ± SD. (L) Schematic diagram describing the LSK cell homing assay used for this study. (M) Frequency (left) and absolute number (right) of CD45.2+ LSK cells in the BM of recipients at 18 hours after adoptive transfer. Data are representative of 2 independent experiments and show mean ± SD. (N) Schematic description of the serial BM transplantation assay used for this study. (O) Percent of CD45.2+ peripheral blood cells after the secondary transplantation of 200 000 CD45.2+Etv6+/+ (green) or Etv6R355X/+ (red) BM cells, harvested from primary recipient mice, along with 200 000 fresh CD45.1+Etv6+/+ BM cells, into lethally-irradiated recipient mice (n = 9 recipients per genotype). Data are representative of 2 experiments and show mean ± SD. (P) Percent of CD45.2+ peripheral blood cells 4 weeks after tertiary transplantation of 200 000 CD45.2+Etv6+/+ (green) or Etv6R355X/+ (red) BM cells harvested from secondary recipients, along with 200 000 fresh CD45.1+Etv6+/+ BM cells, into lethally-irradiated recipient mice. Data are representative of 2 experiments (total n = 10 recipients per group) and show mean ± SD. ∗P < .05; ∗∗P < .001; and ∗∗∗P < .0001, determined by multiple unpaired t tests.
Etv6R355X/+ HSPCs show reduced long-term serial repopulation capacity. (A) Schematic diagram describing the competitive BM transplantation assay used for this study. Percent of CD45.2+ peripheral blood cells (B), B220+CD19+ B cells (C), Gr1+ myeloid cells (D), CD4+ T cells (E), and CD8+ T cells (F) after primary transplantation of 10 000 CD45.2+Etv6+/+ (green) or Etv6R355X/+ (red) LSK cells along with 10 000 CD45.1+Etv6+/+ LSK cells into lethally-irradiated recipient mice (n = 10-12 recipients per genotype per experiment). Data are representative of 3 experiments and show mean ± SD. (G) Percent of CD45.2+ peripheral blood Gr1+ myeloid cells, B220+ B cells, CD4+ T cells, and CD8+ T cells at 6 months after competitive transplantation. Percent of total BM cells (H), splenocytes (I), peripheral blood cells (J), and LSK cells (K) that are CD45.2+ at 6 months after transplantation within remaining recipients (note: half of the recipients were used for serial transplantation depicted in panel N). Data are representative of 3 experiments and show mean ± SD. (L) Schematic diagram describing the LSK cell homing assay used for this study. (M) Frequency (left) and absolute number (right) of CD45.2+ LSK cells in the BM of recipients at 18 hours after adoptive transfer. Data are representative of 2 independent experiments and show mean ± SD. (N) Schematic description of the serial BM transplantation assay used for this study. (O) Percent of CD45.2+ peripheral blood cells after the secondary transplantation of 200 000 CD45.2+Etv6+/+ (green) or Etv6R355X/+ (red) BM cells, harvested from primary recipient mice, along with 200 000 fresh CD45.1+Etv6+/+ BM cells, into lethally-irradiated recipient mice (n = 9 recipients per genotype). Data are representative of 2 experiments and show mean ± SD. (P) Percent of CD45.2+ peripheral blood cells 4 weeks after tertiary transplantation of 200 000 CD45.2+Etv6+/+ (green) or Etv6R355X/+ (red) BM cells harvested from secondary recipients, along with 200 000 fresh CD45.1+Etv6+/+ BM cells, into lethally-irradiated recipient mice. Data are representative of 2 experiments (total n = 10 recipients per group) and show mean ± SD. ∗P < .05; ∗∗P < .001; and ∗∗∗P < .0001, determined by multiple unpaired t tests.
To determine whether the observed repopulation defect was due to an inability of Etv6R355X/+ cells to traffic to the BM, we performed a homing assay in which 25 000 CD45.2+Etv6+/+ or Etv6R355X/+ LSK cells were transferred IV into a sublethally irradiated CD45.1+Etv6+/+ mouse; 18 hours later, we quantified the CD45.2+ LSK cells in the BM (Figure 3L). We found no difference in the frequency or number of Etv6+/+ or Etv6R355X/+ cells that homed to the BM (Figure 3M). Finally, we completed serial transplantation by transferring 200 000 CD45.2+Etv6+/+ or Etv6R355X/+ BM cells from primary recipients to a second cohort of lethally-irradiated CD45.1/.2+Etv6+/+ recipient mice, along with 200 000 fresh CD45.1+Etv6+/+ BM cells for support (Figure 3N). Once again, Etv6R355X/+ cells exhibited significantly reduced engraftment after secondary transplantation (Figure 3O). After the tertiary competitive transplant, only 1 of 10 recipients (pooled from 2 independent experiments) displayed >1% peripheral blood reconstitution with CD45.2+Etv6R355X/+ cells compared with 9 of 10 that received CD45.2+Etv6+/+ cells (Figure 3P). Thus, Etv6R355X/+ HSPCs effectively home to the BM but exhibit a competitive disadvantage after transplantation and a significantly reduced long-term repopulating capacity.
ETV6 target genes are differentially bound in Etv6R355X/+ HSPCs
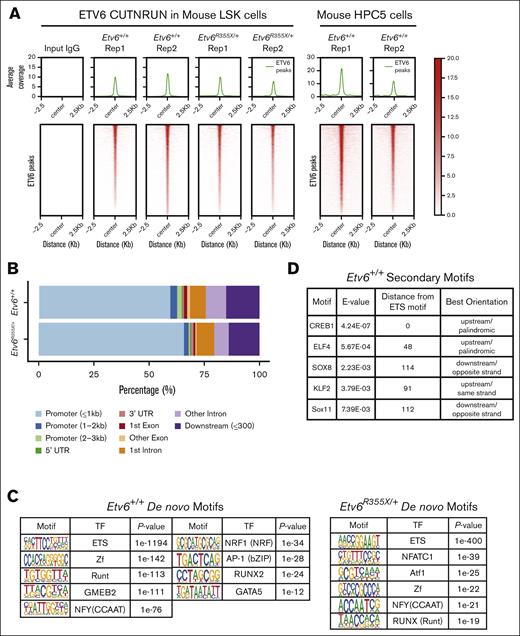
To explore how ETV6 regulates HSPC function, we sought to identify ETV6 target genes within hematopoietic progenitors. Using an anti-ETV6 polyclonal antibody generated against the N-terminal pointed domain of the protein, we performed CUT&RUN, a technique that identifies protein-DNA interactions by combining the antibody-targeted cleavage of the DNA around protein-DNA interaction sites, followed by sequencing of the captured DNA fragments.14 First, we completed CUT&RUN using LSK cells from Etv6+/+ and Etv6R355X/+ mice and from the mouse BM progenitor–derived cell line HPC523 (Figure 4A). From these data, we observed that >50% of ETV6 occupancy peaks were at promoter regions across both Etv6 wild-type and heterozygous mutant samples (Figure 4B), which is consistent with the role of ETV6 as a transcription factor. Overall, we identified ETV6 binding near 2165 genes in Etv6+/+ LSK cells, which included 937 shared between 2 replicates or 1128 that passed filtering in a single replicate (supplemental Figure 4A; supplemental Tables 1 and 2). In addition, we identified ETV6 occupancy peaks near 1681 genes in HPC5 cells (supplemental Table 3). Among the occupancy peaks in HPC5 cells, 932 genes overlapped with those identified in LSK cells (supplemental Figure 4B). De novo and known motif enrichment analysis revealed a significant enrichment in the ETS motif (P < 1e−1194; Figure 4C; supplemental Figure 4C), a primary DNA binding sequence for ETV6 and one of its known binding partners FLI1.24 We observed ETS motifs cooccurring with previously described RUNX motifs,25 further validating these data (supplemental Figure 4D). Furthermore, we identified motifs representative of known ETV6 binding partners, such as IRF8,2 as well as novel putative cofactors, such as nuclear transcription factor Y, E2 transcription factor, or thanatos-associated domain-containing apoptosis-associated protein (Figure 4C; supplemental Figure 4C-D). To detect other proteins potentially cooperating with ETV6 in LSK cells, we performed secondary motif analysis, which revealed binding motifs near ETV6 occupancy sites for other essential hematopoietic transcription factors including CREB1,26 ELF4,27 and KLF228 (Figure 4D).
ETV6 binds to inflammatory gene loci in mouse and human hematopoietic progenitors. (A) Peak occupancy heatmap of CUT&RUN peaks in Etv6+/+ and Etv6R355X/+ LSK cells and Etv6+/+ HPC5 cells using an immunoglobulin G (IgG) control or anti-ETV6 antibody. (B) Genomic distribution of ETV6 binding sites in Etv6+/+ and Etv6R355X/+ LSK cells identified by CUT&RUN. (C) Hypergeometric optimization of motif enrichment (HOMER) de novo motif enrichment analysis of CUT&RUN peaks in Etv6+/+ and Etv6R355X/+ LSK cells, showing the most enriched motifs, ordered according to the P value. (D) Secondary motif analysis using the SpaMo tool (MEME suite) shows neighboring motifs, their distance, and location with respect to ETS motif. (E) (Left) Volcano plot showing differential peak enrichment analysis using HOMER. Significantly downregulated (blue; P value < 1e−05; fold-change < 2) or upregulated (red; P value < 1e−05; fold-change > 2) ETV6-binding peaks are shown. Dotted lines indicate P value < 1e−05 and |fold-change > 2 threshold. (Right) Overrepresentation analysis of 813 genes near 853 significantly downregulated ETV6 binding peaks in Etv6R355X/+ LSK cells compared with Etv6+/+ LSK cells using MSigDB version 7.5.1 Hallmark gene sets. (F) Significant differentially expressed genes from bulk RNA-seq of Etv6+/+ and Etv6R355X/+ LSK cells obtained from 3-month-old mice (false discovery rate < 0.05). ETV6 target genes identified from CUT&RUN are annotated in orange. (G) Boxplots for RNA-seq expression of the 9 ETV6 targets, showing significant differential expression in Etv6+/+ vs Etv6R355X/+ LSK cells. (H) Overrepresentation analysis of ETV6 binding targets from LSK and HPC5 cells across MSigDB version 7.5.1 Hallmark gene sets. (I) Peak occupancy heatmap of CUT&RUN peaks in human ETV6+/+ granulocyte colony–stimulating factor (G-CSF)-mobilized CD34+ cells using an IgG control or anti-ETV6 antibody (left). Genomic distribution of ETV6 binding sites in ETV6+/+ CD34+ cells identified by CUT&RUN (top right). Overrepresentation analysis of ETV6 binding targets in ETV6+/+ CD34+ cells across MSigDB version 7.5.1 Hallmark gene sets (bottom right).
ETV6 binds to inflammatory gene loci in mouse and human hematopoietic progenitors. (A) Peak occupancy heatmap of CUT&RUN peaks in Etv6+/+ and Etv6R355X/+ LSK cells and Etv6+/+ HPC5 cells using an immunoglobulin G (IgG) control or anti-ETV6 antibody. (B) Genomic distribution of ETV6 binding sites in Etv6+/+ and Etv6R355X/+ LSK cells identified by CUT&RUN. (C) Hypergeometric optimization of motif enrichment (HOMER) de novo motif enrichment analysis of CUT&RUN peaks in Etv6+/+ and Etv6R355X/+ LSK cells, showing the most enriched motifs, ordered according to the P value. (D) Secondary motif analysis using the SpaMo tool (MEME suite) shows neighboring motifs, their distance, and location with respect to ETS motif. (E) (Left) Volcano plot showing differential peak enrichment analysis using HOMER. Significantly downregulated (blue; P value < 1e−05; fold-change < 2) or upregulated (red; P value < 1e−05; fold-change > 2) ETV6-binding peaks are shown. Dotted lines indicate P value < 1e−05 and |fold-change > 2 threshold. (Right) Overrepresentation analysis of 813 genes near 853 significantly downregulated ETV6 binding peaks in Etv6R355X/+ LSK cells compared with Etv6+/+ LSK cells using MSigDB version 7.5.1 Hallmark gene sets. (F) Significant differentially expressed genes from bulk RNA-seq of Etv6+/+ and Etv6R355X/+ LSK cells obtained from 3-month-old mice (false discovery rate < 0.05). ETV6 target genes identified from CUT&RUN are annotated in orange. (G) Boxplots for RNA-seq expression of the 9 ETV6 targets, showing significant differential expression in Etv6+/+ vs Etv6R355X/+ LSK cells. (H) Overrepresentation analysis of ETV6 binding targets from LSK and HPC5 cells across MSigDB version 7.5.1 Hallmark gene sets. (I) Peak occupancy heatmap of CUT&RUN peaks in human ETV6+/+ granulocyte colony–stimulating factor (G-CSF)-mobilized CD34+ cells using an IgG control or anti-ETV6 antibody (left). Genomic distribution of ETV6 binding sites in ETV6+/+ CD34+ cells identified by CUT&RUN (top right). Overrepresentation analysis of ETV6 binding targets in ETV6+/+ CD34+ cells across MSigDB version 7.5.1 Hallmark gene sets (bottom right).
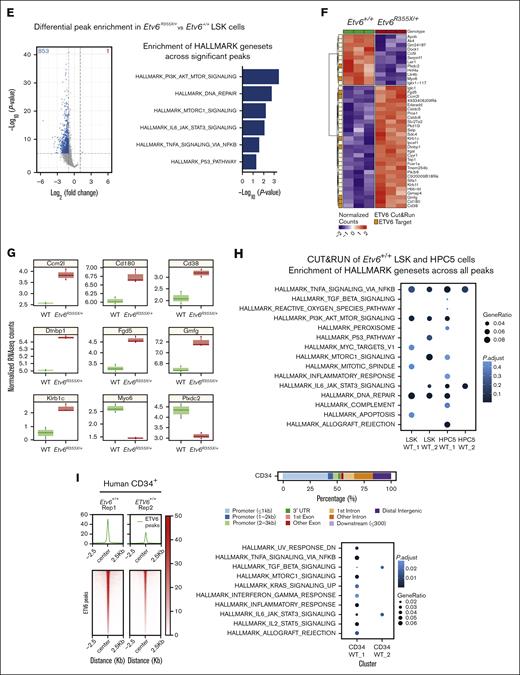
Next, we completed differential peak analysis comparing ETV6 occupancy within Etv6+/+ and Etv6R355X/+ LSK cells, which revealed a partial or complete loss of 853 ETV6-binding peaks near 813 genes and a potential gain of ETV6 binding at 1 peak in Etv6R355X/+ cells (Figure 4E, left; supplemental Table 4). We observed almost no differences in the motif enrichment in ETV6-binding sites within Etv6R355X/+ vs Etv6+/+ cells, except for nuclear factor of activated T cells 1 (NFATC) motif enrichment (Figure 4C). NFATc1 is an inflammation-induced transcription factor,29 and this may suggest a potential gain-of-function effect of the ETV6R355X variant protein through NFATc1 binding. Despite the loss of ETV6 binding at several loci, bulk RNA-seq of sort-purified LSK cells from 3-month-old mice identified few differentially expressed genes, including only 29 significantly upregulated and 12 downregulated genes (Figure 4F; false discovery rate < 0.05). Among the 29 upregulated genes, 7 appeared to be direct ETV6 targets based on CUT&RUN data, including key B-cell proliferation and immune response genes, such as Cd180 and Cd38. Among the 12 downregulated genes, 2 represented putative ETV6 targets (Figure 4G; supplemental Tables 5-7). Taken together, these CUT&RUN data identify novel putative ETV6 target genes in LSK cells; however, changes in gene expression are minimal in Etv6R355X/+ cells when examined under homeostatic conditions, likely because of residual levels of wild-type ETV6 protein.
ETV6 targets inflammatory response pathway genes in mouse and human HSPCs
Analysis of CUT&RUN data demonstrates that ETV6 directly binds near genes from the TNF signaling via NF-κB, PI3K/AK strain transforming/mammalian target of rapamycin, interleukin-6 (IL-6)/JAK/STAT3 signaling, and DNA-repair pathways (Figure 4H; supplemental Table 8), with genes from several of these pathways showing significant loss of or depletion in ETV6 binding in Etv6R355X/+ LSK cells (Figure 4E, right; supplemental Table 9). To determine whether ETV6 might target similar loci in human cells, we completed the CUT&RUN analysis of granulocyte colony–stimulating factor–mobilized CD34+ cells, which revealed that genes adjacent to ETV6 binding sites were also involved in the inflammatory response, including the TNF signaling via NF-κB and IL-6/JAK/STAT3 pathways (Figure 4I; supplemental Tables 10 and 11). Genes within these pathways included Relt, a hematopoietic-specific member of the TNF superfamily,30 and Nlrp1b, a member of the inflammasome machinery31 as well as genes involved in HSC activation, such as Cd4732 (supplemental Table 10). Notably, we observed ETV6 binding near genes reported to be upregulated in peripheral blood mononuclear cells from patients with T5 who were heterozygous for ETV6 P214L (Ccl3),11 MPP4 cells and HSCs from mice homozygous for an Etv6 P216 variant (which is orthologous to human ETV6 P214L; Cnn3, Plxdc2, Pik3ip1, Gmfg, Egr1, and Msrb3),33 and human peripheral blood mononuclear cells in which ETV6 was silenced (Tnf, Tlr4, Il1b, and Myd88).11
To define ETV6 target genes within the larger structure of the 3-dimensional genome, we integrated CUT&RUN data with ATAC-seq and Hi-C data, with the latter generated using HPC5 cells. Hi-C analysis identified 3357 high-resolution chromatin loops involving >5800 genes, which overlapped with 339 and 251 genes with ETV6 binding sites from LSK and HPC5 cells, respectively. We, then, performed pathway enrichment analysis using Hallmark gene sets focusing on ETV6 occupancy sites contained within Hi-C loops and observed enrichment of several gene sets associated with inflammation, including the TNF signaling via NF-κB, IL-6/JAK/STAT3, and IL-2/STAT5 pathways (Figure 5A; supplemental Tables 12 and 13). One set of chromatin interactions on chromosome 17 contained 5 genes in the TNF pathway (Lst1, Ltb, Tnf, Lta, and Nfkbil1), with 3 of these genes (Tnf, Nfkbil1, and Lst1) residing within open chromatin regions and showing ETV6 binding in Etv6+/+ but potentially reduced binding in Etv6R355X/+ LSK cells. Notably, Nfkbil1 represents 1 of the genes with the greatest ETV6 occupancy identified through the CUT&RUN analysis of Etv6+/+ LSK cells (Figure 5B-C).
ETV6 binds to a chromosomal locus encompassing Tnf with upregulation of TNF production in Etv6R355X/+ HSPCs after transplantation. (A) Overrepresentation analysis of ETV6 targets within chromatin loops and Hallmark gene sets. (B) Regulatory activity at the Tnf locus showing topologically associated domain on chromosome 17 identified by integrative analysis incorporating Hi-C loops at this locus in HPC5 cells; CUT&RUN of Etv6+/+ LSK cells, Etv6R355X/+ LSK cells, and Etv6+/+ HPC5 cells; and ATAC-seq data generated from the HPC5 cells. (C) ETV6 binding sites from CUT&RUN in Etv6+/+ and Etv6R355X/+ LSK cells and HPC5 cells, and ATAC-seq from HPC5 cells at Tnf, Nfkbil1, and Lst1 loci encompassed by the chromatin loop at the Tnf locus. (D) Schematic description of the experimental design to examine TNF expression in HSPCs after competitive transplantation. (E) Representative histogram of mean fluorescence intensity (MFI) of TNF staining in Etv6+/+ and Etv6R355X/+ LSK cells harvested from the same recipient animal 6 weeks after transplantation compared with phorbol myristate acetate/ionomycin-stimulated or unstimulated BM as well as BM stained with an IgG control antibody. (F) MFI of intracellular TNF within each HSPC subpopulation (n = 6). Data are representative of 3 experiments and show mean ± SD. ∗P < .05; ∗∗P < .001; and ∗∗∗P < .0001, determined by multiple paired t tests.
ETV6 binds to a chromosomal locus encompassing Tnf with upregulation of TNF production in Etv6R355X/+ HSPCs after transplantation. (A) Overrepresentation analysis of ETV6 targets within chromatin loops and Hallmark gene sets. (B) Regulatory activity at the Tnf locus showing topologically associated domain on chromosome 17 identified by integrative analysis incorporating Hi-C loops at this locus in HPC5 cells; CUT&RUN of Etv6+/+ LSK cells, Etv6R355X/+ LSK cells, and Etv6+/+ HPC5 cells; and ATAC-seq data generated from the HPC5 cells. (C) ETV6 binding sites from CUT&RUN in Etv6+/+ and Etv6R355X/+ LSK cells and HPC5 cells, and ATAC-seq from HPC5 cells at Tnf, Nfkbil1, and Lst1 loci encompassed by the chromatin loop at the Tnf locus. (D) Schematic description of the experimental design to examine TNF expression in HSPCs after competitive transplantation. (E) Representative histogram of mean fluorescence intensity (MFI) of TNF staining in Etv6+/+ and Etv6R355X/+ LSK cells harvested from the same recipient animal 6 weeks after transplantation compared with phorbol myristate acetate/ionomycin-stimulated or unstimulated BM as well as BM stained with an IgG control antibody. (F) MFI of intracellular TNF within each HSPC subpopulation (n = 6). Data are representative of 3 experiments and show mean ± SD. ∗P < .05; ∗∗P < .001; and ∗∗∗P < .0001, determined by multiple paired t tests.
Etv6R355X/+ mouse HSPCs exhibit increased TNF production after transplantation
The CUT&RUN, ATAC-seq, and Hi-C findings revealed that ETV6 binds to TNF and other inflammatory gene loci in mouse and human hematopoietic progenitor cells, suggesting that it might regulate their expression. However, we saw no major differences in the proinflammatory gene expression between Etv6+/+ and Etv6R355X/+ cells under homeostatic conditions (Figure 4E), nor did we see alterations in the levels of TNF in the BM or peripheral blood (not shown). Because TNF is an important regulator of HSPCs during stress hematopoiesis,34 we chose to examine how TNF production might be perturbed in Etv6R355X/+ cells under stress conditions. Therefore, we performed a competitive transplant, transferring 10 000 CD45.1+Etv6+/+ and 10 000 CD45.2+Etv6R355X/+ LSK cells into lethally-irradiated CD45.1/2+Etv6+/+ recipient mice. After 6 weeks, we collected BM cells and assessed intracellular TNF production within the HSPC subpopulations (Figure 5D; supplemental Figure 5). Through this analysis, we observed significantly increased TNF in Etv6R355X/+ vs Etv6+/+ LSK cells, long-term HSCs, short-term HSCs, and MPP3 and MPP4 cells, at levels that appeared higher than those in BM cells stimulated in vitro with phorbol myristate acetate/ionomycin (Figure 5E-F). Thus, during regenerative stress, Etv6R355X/+ HSPCs produce more TNF compared with Etv6+/+ cells. However, the ablation of TNF is not sufficient to rescue the observed competitive defect of Etv6R355X/+ LSK cells, suggesting that the elevated levels of TNF are not solely responsible for this phenotype (supplemental Figure 6).
Etv6R355X/+ HSPCs exhibit increased inflammatory gene expression after transplantation
Etv6R355X/+ HSPCs are present in the BM of 3-month-old mice, and these animals remain healthy for up to 24 months or longer. However, we observed a significant competitive disadvantage and elevated TNF production after transplantation. To determine the transcriptional consequences of ETV6 perturbation during regenerative hematopoiesis, we performed scRNA-seq at 6 weeks after competitive transplantation. Toward this end, data across samples were integrated and clustered to identify 22 distinct cell populations (Figure 6A; supplemental Figures 7 and 8). We annotated clusters representing HSCs, MPPs, early lymphoid progenitors (ELPs), and other myeloid-biased progenitors, neutrophils at various stages of differentiation, and several smaller mature cell populations. All clusters were represented in Etv6+/+ and Etv6R355X/+ replicates at varying proportions (supplemental Figure 7A).
Etv6R355X/+ HSCs are reduced and have increased expression of inflammatory response genes when examined 6 weeks after transplantation. (A) Uniform manifold approximation and projection visualization of 22 cell populations identified from the posttransplant Etv6+/+ and Etv6R355X/+ LSK-enriched BM, colored by cell population. (B) Proportions of cells in each of the 22 clusters (left). Box plots depicting the proportions of Etv6R355X/+ or Etv6+/+ cells within clusters that show significant reductions across 3 replicates at 6 weeks after transplantation (right). ∗P value < .05, t test. (C) Summary of significant differentially expressed genes from all Etv6R355X/+ clusters compared with Etv6+/+ from scRNA-seq data (false discovery rate < 0.05) (left). Upregulated (red) and downregulated (blue) genes marked by |log2 fold-change (FC)| >1. Volcano plots for HSC, MPP, and ELP clusters are shown on the right: each dot represents a gene, colored by P value < 10e−6 (blue), log2 FC > 1 (green), or both (red). (D) Gene-set enrichment analysis (GSEA) analysis using custom gene sets of 813 genes, exhibiting significant depletion of ETV6 binding from anti-ETV6 CUT&RUN in Etv6R355X/+ LSK compared with Etv6+/+ LSK cells in HSC (top) and MPP (bottom) clusters. (E) Top 10 significant gene sets from GSEA of Etv6R355X/+ vs Etv6+/+ HSC, MPP, and ELP cell clusters using Hallmark gene sets (MSigDb version 7.5.1).
Etv6R355X/+ HSCs are reduced and have increased expression of inflammatory response genes when examined 6 weeks after transplantation. (A) Uniform manifold approximation and projection visualization of 22 cell populations identified from the posttransplant Etv6+/+ and Etv6R355X/+ LSK-enriched BM, colored by cell population. (B) Proportions of cells in each of the 22 clusters (left). Box plots depicting the proportions of Etv6R355X/+ or Etv6+/+ cells within clusters that show significant reductions across 3 replicates at 6 weeks after transplantation (right). ∗P value < .05, t test. (C) Summary of significant differentially expressed genes from all Etv6R355X/+ clusters compared with Etv6+/+ from scRNA-seq data (false discovery rate < 0.05) (left). Upregulated (red) and downregulated (blue) genes marked by |log2 fold-change (FC)| >1. Volcano plots for HSC, MPP, and ELP clusters are shown on the right: each dot represents a gene, colored by P value < 10e−6 (blue), log2 FC > 1 (green), or both (red). (D) Gene-set enrichment analysis (GSEA) analysis using custom gene sets of 813 genes, exhibiting significant depletion of ETV6 binding from anti-ETV6 CUT&RUN in Etv6R355X/+ LSK compared with Etv6+/+ LSK cells in HSC (top) and MPP (bottom) clusters. (E) Top 10 significant gene sets from GSEA of Etv6R355X/+ vs Etv6+/+ HSC, MPP, and ELP cell clusters using Hallmark gene sets (MSigDb version 7.5.1).
Compositional analysis of clusters showed significant depletion of cells in the HSC, MPP, and ELP clusters of Etv6R355X/+ compared with in those of Etv6+/+ samples (t test P value < .05; Figure 6B). Within the significantly depleted populations, we observed 28 (HSC), 48 (MPP), and 203 (ELP) differentially expressed genes (false discovery rate cutoff <0.05; |log2 fold-change| > 1), with the majority of differentially expressed genes being increased in number, consistent with the role of ETV6 as a transcriptional repressor (Figure 6C; supplemental Table 14). Furthermore, we found a significant enrichment of putative target genes that were differentially bound by ETV6 via CUT&RUN of Etv6R355X/+ vs Etv6+/+ LSK cells (supplemental Table 4) in the Etv6R355X/+ HSC and MPP clusters (Figure 6D). This observation suggests that within these populations, gene expression differences were due to perturbation of ETV6-mediated gene regulation. In contrast, the differentially expressed genes in the ELP cluster did not contain enrichment of any putative ETV6 targets. This latter finding implies that gene expression effects in ELPs might be due to perturbed ETV6 function occurring earlier in hematopoiesis.
To investigate the biological pathways affected by the Etv6R355X mutation during hematopoietic reconstitution, we performed gene-set enrichment analysis, using Hallmark gene sets across all scRNA-seq clusters. In the HSC cluster, inflammatory and IL-6/JAK/STAT3 signaling pathways were significantly upregulated. Similarly, within the MPP cluster, allograft rejection and IL-6/JAK/STAT3 signaling pathways were significantly upregulated (Figure 6E; supplemental Table 15). Further along the developmental lineage, we observed significant upregulation of interferon-gamma–response pathway and downregulation of cell proliferation–related MYC and E2 transcription factor target pathways in the ELP cluster (Figure 6E). Altogether, these data reveal that ETV6 plays an important role during stress hematopoiesis, with heterozygosity for a T5-associated Etv6 mutation leading to the depletion of immature progenitors, paired with increased expression of inflammation-related genes, many of which are putative ETV6 targets.
Discussion
Herein, we describe a novel mouse model of the human disorder T5, a poorly understood condition caused by pathogenic ETV6 variants. We used this model to define how heterozygosity for Etv6R355X affects the frequencies, functions, and transcriptional landscape of HSPCs. Through this analysis, we demonstrate that ETV6R355X/+ mice can generate all HSPC subpopulations under homeostatic conditions; however, ETV6R355X/+ cells exhibit impaired repopulation capacity after serial competitive transplantation. Furthermore, we establish a robust data set of endogenous putative ETV6 target genes in mouse and human hematopoietic progenitors and show how binding near these genes is altered in Etv6 mutant cells. Finally, we define the transcriptional and cellular consequences of ETV6 perturbation in HSPCs during regenerative hematopoiesis, highlighting an important role for this gene in repressing inflammatory gene pathways, with many of the affected genes representing ETV6 targets.
Previous scRNA-seq studies of peripheral blood mononuclear cells from individuals with T5 showed enrichment for interferon response gene signatures.11 Furthermore, humans harboring T5 ETV6 variants have increased numbers of circulating CD34+ cells,22 which are known to enter the circulation in response to inflammation.35 TNF signaling via NF-κB and other inflammation-related pathways were enriched in our ETV6 CUT&RUN studies of human CD34+ cells. Therefore, in patients with T5, enhanced inflammatory signaling through cytokines such as TNF may cause HSPC abnormalities similar to those observed in our mouse model. However, the levels of most proinflammatory cytokines tested, including TNF, are not elevated in the blood of patients with T511 or in the blood or whole BM of Etv6R355X/+ mice (supplemental Figure 9); therefore, it remains possible that these cytokines are being “consumed” locally and, thus, their levels are too low to detect. Alternatively, it is possiblethat proinflammatory signaling pathways are being induced in a cytokine-independent and cell-autonomous manner, as previously suggested.11 Of note, within mouse LSK cells, we did observe ETV6 binding at a putative distant enhancer of the Il12a gene (supplemental Figure 10A), a cytokine that induces TNF and has suppressive effects on hematopoiesis,36 and in the BM of Etv6R355X/+ mice we observed significantly increased levels of IL-12 (p70; supplemental Figure 9). In addition, we saw increased expression of Il12a within the Etv6R355X/+ HSC and MPP clusters after transplantation (supplemental Figure 10B; supplemental Table 14). Thus, ETV6 likely regulates other inflammatory cytokines, and it remains to be determined which ones are most important in different hematopoietic contexts.
Approximately 30% of individuals with T5 develop lymphoid or myeloid leukemia, with the period of risk extending across the entire lifespan. Chronic TNF or other inflammatory signaling is proposed to provide a selective pressure for oncogenic genetic events.33 For example, recent work has shown that inflammatory exposures create long-term impairment of HSC self-renewal and accelerate hematopoietic aging,37 and reduce the fitness of B-cell progenitors, creating an environment that selects for cells harboring oncogenic mutations.38 Thus, it is possible that in the setting of a germ line pathogenic ETV6 variant, improper repression of TNF or other proinflammatory pathway genes predisposes to malignant transformation through this mechanism.
The link between hematologic malignancy and inflammation is not a new concept. It is well recognized that there is a role of inflammatory drivers in myelodysplastic syndromes and myeloid malignancies.39-41 Recently, in a mouse model of a pediatric myeloid neoplasm predisposing Samd9L germ line mutation, reduced stem cell fitness was observed, and this was exacerbated with inflammatory stimuli and led to BM hypocellularity.42 Furthermore, germ line RUNX1 mutations result in familial platelet disorder with predisposition to myeloid malignancy, and it has been shown that hematopoietic-specific loss of RUNX1 in mice induces increased production of proinflammatory mediators.43 However, other myeloid predisposing syndromes, such as GATA2 deficiency, often result in immunodeficiency and a loss of proinflammatory signaling.44,45 Although patients harboring ETV6 germ line variants most often develop B-ALL, they occasionally develop myeloid malignancies, in line with this link between inflammation and myeloid neoplasia. Of note, germ line IKZF1 mutations, which also result in B-ALL predisposition, are associated with immunodeficiency46,47 as well as autoimmunity.48 Altogether, there is mounting evidence that dysregulation of inflammatory factors and immune responses contributes to leukemia predisposition; however, these phenotypes are complex and future studies are needed to better understand the underlying mechanisms.
Acknowledgments
The authors thank the members of the Nichols Laboratory for scientific discussions and input. The authors also gratefully acknowledge the members of the St. Jude Children’s Research Hospital Flow Cytometry and Cell Sorting Shared Resource, Animal Resource Center, Hartwell Sequencing Center, Electron Microscopy Core Facility, Transgenic Core Facility, Veterinary Pathology Core Facility, and the Center for Applied Bioinformatics, without whose assistance this project would not have been possible. Figures 1A, 2B, 3A,L,N, and 5D were created with BioRender.com.
This work was supported by American Lebanese Syrian Associated Charities, the National Heart, Lung, and Blood Institute at the National Institutes of Health (F31HL154645; M.B.), and the National Cancer Institute at the National Institutes of Health (R01CA241452; K.E.N. and J.J.Y.).
Authorship
Contribution: M.B., N.O., and K.E.N. wrote the manuscript; M.B., R.B.-D., R.F., I.I., P.B., X.Z., H.S.T., K.C.V., A.N.S., and S.A. gathered data; R.F., I.I., P.B., X.Z., C.L., K.C.V., A.N.S., D.C.S., M.T.K., and S.A. provided critical technical support; M.B., N.O., R.B.-D., P.O., and H.S.T. analyzed the data; M.B., N.O., R.B.-D., K.C.V., S.L.M.-F., M.J.W., J.J.Y., and K.E.N. interpreted data; and all authors critically reviewed the manuscript and agreed to submit the manuscript for publication.
Conflict-of-interest disclosure: K.E.N. reports research funding from Incyte Pharmaceuticals. J.J.Y. reports research funding from Takeda Pharmaceutical Company. M.J.W. serves on advisory boards for Cellarity Inc, Novartis, Graphite Bio, Dyne Therapeutics, and Forma Therapeutics, and owns equity in Cellarity Inc. The remaining authors declare no competing financial interests.
Correspondence: Kim E. Nichols, Department of Oncology, St. Jude Children’s Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105; e-mail: kim.nichols@stjude.org.
References
Author notes
Single-cell RNA sequencing, CUT&RUN, ATAC-seq, and Hi-C data are deposited in the Gene Expression Omnibus database (accession number GSE213597) and Sequence Read Archive (BioProject number PRJNA880871).
All postanalysis data files are in the supplemental Tables.
The full-text version of this article contains a data supplement.