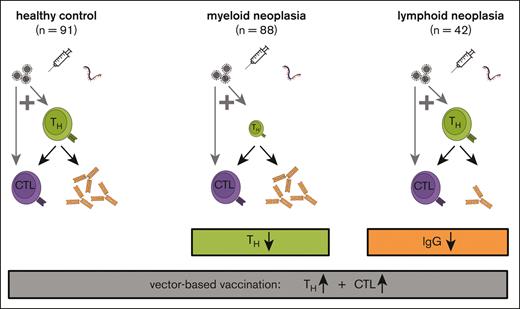
Key Points
Vector-based SARS-CoV-2 vaccines are advantageous for the generation of T-cell responses in patients with hematological neoplasia.
CD4+ and CD8+ T-cell responses cooccur in hematological neoplasms, whereas IgG titers are CD4+ T cells associated in myeloid neoplasia only.
Abstract
In order to elucidate mechanisms for severe acute respiratory syndrome coronavirus 2 vaccination success in hematological neoplasia, we, herein, provide a comprehensive characterization of the spike-specific T-cell and serological immunity induced in 130 patients in comparison with 91 healthy controls. We studied 121 distinct T-cell subpopulations and the vaccination schemes as putative response predictors. In patients with lymphoid malignancies an insufficient immunoglobulin G (IgG) response was accompanied by a healthy CD4+ T-cell function. Compared with controls, a spike-specific CD4+ response was detectable in fewer patients with myeloid neoplasia whereas the seroconversion rate was normal. Vaccination-induced CD4+ responses were associated to CD8+ and IgG responses. Vector-based AZD1222 vaccine induced more frequently detectable specific CD4+ responses in study participants across all cohorts (96%; 27 of 28), whereas fully messenger RNA-based vaccination schemes resulted in measurable CD4+ cells in only 102 of 168 participants (61%; P < .0001). A similar benefit of vector-based vaccination was observed for the induction of spike-specific CD8+ T cells. Multivariable models confirmed vaccination schemes that incorporated at least 1 vector-based vaccination as key feature to mount both a spike-specific CD4+ response (odds ratio, 10.67) and CD8+ response (odds ratio, 6.56). Multivariable analyses identified a specific CD4+ response but not the vector-based immunization as beneficial for a strong, specific IgG titer. Our study reveals factors associated with a T-cell response in patients with hematological neoplasia and might pave the way toward tailored vaccination schemes for vaccinees with these diseases. The study was registered at the German Clinical Trials Register as #DRKS00027372.
Introduction
On 11 March 2020 the World Health Organization declared the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreak a pandemic. Ever since, 2 major factors modified the individual risk of this disease and its general impact on health care systems: the advent of efficacious vaccines1-8 and the occurrence of novel virus variants.9,10 Both factors are interconnected by the ability of the host to mount a protective response against the circulating variants of the virus. Driven by the utmost importance of SARS-CoV-2, the characterization of infection11 and vaccine immunity12-14 recently attracted great interest. Animal models and correlative data suggest that antibodies might protect from reinfection with the same virus variant, whereas the quantity and activity of CD8+ T cells and CD4+ T cells determine the severity of the disease.9,11,15-24 Compared with protection by neutralizing antibodies, T-cell–mediated protection is considered less susceptible to immune evasion by novel virus variants.9,16,19,25-27 This has already been known for SARS-CoV-128,29 and is of particular importance for patients with hematological malignancies who are often unable to mount an antibody response.30-33 Although, more information on serological responses and their kinetics has been published, detailed analyses of CD8+ T-cell and CD4+ T-cell responses are scarce.34
Patients with myeloid and lymphoid neoplasms are more likely to experience a severe course of COVID-19 disease.34-40 Moreover, there are reports that vaccination might be less efficacious in these patient populations.31-33,36,41-55 Similar to the general population, research among individuals with hematological malignancies has focused on kinetics and predictors of antibody responses, whereas less information on T-cell responses is available. We56 and others57,58 have recently described that patients with hematological malignancies can mount T-cell responses after vaccination while lacking a simultaneous antibody response. This finding casts doubts on the assumption that seroconversion alone sufficiently mirrors the success of a vaccination. It also corroborates the hypothesis that mechanisms of protection might differ between patients with hematological malignancies and the general population.
The messenger RNA (mRNA) vaccines tozinameran (BNT162b2, Pfizer-BioNTech) and elasomeran (mRNA-1273, Moderna) as well as the adenoviral vector vaccine AZD1222 (ChAdOx1 nCoV-19, AstraZeneca)2,6,7 are most widely used in Western Europe and North America. Combinations of mRNA- and vector-based vaccines (heterologous schemes) reportedly more efficiently induce T-cell responses59-64 in healthy vaccinees than either principle.
In the future, vaccination strategies may be tailored based on the needs of particular patient groups. A better understanding of the adaptive immune responses to vaccination in patients is therefore urgently needed. Using extensive immune monitoring data from the prospective, multicentric ImV-HOng (OSHO#98) study, we, herein, comprehensively compare the immune system of patients with lymphoid and myeloid neoplasms who are SARS-CoV-2–vaccinated with that of healthy vaccinated controls. In-depth immunophenotyping data, demographics, clinical data, and the type of vaccine were fitted in multivariable regression models to study their association with vaccination-induced humoral and cellular immune responses. T-cell responses were assessed using a SARS-CoV-2 spike peptide mix to elicit polyfunctional CD8+ T cells and CD4+ T cells, which are considered key mediators of a successful T-cell response.65-67
Methods
Study cohort
ImV-HOng (OSHO#98, registered at the Paul-Ehrlich Institute [NIS-584] and the German Clinical Trials Register [DRKS00027372]) is a longitudinal, prospective, multicenter, noninterventional study to compare spike protein–specific humoral and T-cell responses between controls and patients with hematological neoplasms and solid tumors.56 The study was approved by the ethical review boards of all participating centers. All participants provided written informed consent.
We present an analysis of 228 participants from whom we received peripheral blood at Special Hematology Laboratory (Rostock University Medical Center) to evaluate the T-cell responses 120 days after the first SARS-CoV-2 vaccination. Patients who received their last disease-specific treatment within 6 months before first vaccination were considered to be under current cancer treatment. Additional details on sample eligibility and timing are provided in the supplemental Methods.
Detection of SARS-CoV-2–specific T cells
SARS-CoV-2–specific T cells were detected using intracellular cytokine staining after stimulation with peptides covering the full-length, wild-type spike protein of SARS-CoV-2 (SARS-CoV-2 Prot_S Complete, order number 130-127-953; Miltenyi Biotec, Bergisch Gladbach, Germany) as previously published (supplemental Table 1).56 In short, after adding brefeldin A, heparinized whole blood was either left unstimulated (negative control), stimulated with Staphylococcus enterotoxin B (SEB; positive control), or with the spike peptide mix for 4 hours at 37°C. After incubation, bulk lysis and surface and intracellular staining were performed per EuroFlow guidelines.68 The panel composition is given in supplemental Table 1. Samples were measured on BD FACS Lyric or Miltenyi MACS Quant 10 flow cytometers, which were aligned in accordance with EuroFlow standards and subjected to biannual EuroFlow quality control ring trials.69,70 Primary flow cytometry data were analyzed using Infinicyt (version 2.0.4b; Cytognos SL, Salamanca, Spain). Normalized proportions of SEB-activated and spike-specific T cells (expressed as percentages of the samples’ total CD4+ T cells and CD8+ T cells, respectively) were obtained by subtracting the respective frequencies of the negative control in the same sample from the raw frequencies. Fourteen unvaccinated and self-reportedly noninfected control participants were used to calculate the limit of detection (LOD) as the mean + 2 standard deviations. Samples >LOD (0.00459% for CD4+TNFα+IFNγ+IL-2+ T cells and 0.00287% for CD8+TNFα+IFNγ+IL-2+ T cells) were considered positive. We report on triple cytokine-positive, polyfunctional CD4+ cells and CD8+ cells (for gating refer to supplemental Figure 1; examples shown in supplemental Figure 2). The absolute numbers of spike-specific and SEB-responsive T cells per μL blood were calculated back from the numbers of T cells measured using immune monitoring tube 1.
Immune monitoring
The immune monitoring panel comprised 3 eight-color tubes (supplemental Table 1) stained in accordance with the EuroFlow guidelines68 and acquired on a MACSQuant 10. In total, 121 distinct cell populations were identified using standardized gates and Infinicyt software (detailed in supplemental Figures 3-5). Numbers of cells per μL blood were derived from relative measurements of cell populations (of total leukocytes) via flow cytometry, and the total white blood count was determined with a Sysmex XP-300 cell counter (Sysmex Europe GmbH, Norderstedt, Germany).
SARS-CoV-2–specific IgG
SARS-CoV-2–specific immunoglobulin G (IgG) was measured using an enzyme-linked immunosorbent assay, as previously described56 (Roche Elecsys Anti–SARS-CoV-2 S assay, Roche Diagnostics International Ltd, Rotkreuz, Switzerland). The test detects antibodies to the receptor binding domain of the spike protein. The assay is calibrated so that 1 U/mL directly equals 1 binding antibody unit per mL (standardized according to World Health Organization).56 Titers ≥0.8 U/mL were considered positive.
Statistics
Statistical analyses were performed using R (version 4.2.1).71 Response levels below the LOD were set to the respective LOD if not stated otherwise. Kruskal-Wallis followed by Dunn tests were used to detect differences in central tendency. Welch one-way analysis of variance with subsequent Games-Howell tests were used to detect differences in means. If not stated otherwise, medians were reported for metric data. Count data were compared using Fisher exact test. Robust tree-based feature selection with Monte-Carlo methods was performed using the R package rmcfs (version 1.3.5) with standard cut-offs.72 Subsequent multiparametric logistic regression models were generated using the R package logistf (version 1.24.1) including all parameters selected by rmcfs in the full cohort or the 3 subcohorts.
Results
Clinical data
We studied 91 healthy control participants, 42 patients with lymphoid, and 88 patients with myeloid malignancies (Table 1; detailed in supplemental Table 2). The healthy control group was younger and comprised more women than the patient groups. Moreover, patients with myeloid neoplasms were significantly more often actively treated than patients with lymphoid neoplasms. We found no significant differences in terms of prior infections, vaccination type, or white blood count.
Demographic and clinical data for controls as well as patients with lymphoid and myeloid neoplasms
| Variable . | Healthy controls, n = 91∗ . | Lymphoid neoplasms, n = 42∗ . | Myeloid neoplasms, n = 88∗ . | P value† . |
|---|---|---|---|---|
| Age (y) | 52 (38; 62) | 66 (48; 74) | 62 (52; 67) | < .001 |
| Sex | .033 | |||
| Female | 54 (59%) | 17 (40%) | 37 (42%) | |
| Male | 37 (41%) | 25 (60%) | 51 (58%) | |
| WBC | 6.10 (5.30; 7.75) | 5.40 (4.10; 6.77) | 6.00 (4.27; 8.38) | .2 |
| SARS-CoV-2 vaccination scheme | .07 | |||
| Vector based | 13 (14%) | 4 (9.5%) | 11 (12%) | |
| 2× mRNA-based | 62 (68%) | 37 (88%) | 69 (78%) | |
| Only one‡ | 16 (18%) | 1 (2.4%) | 8 (9.1%) | |
| Prior SARS-CoV-2 infection | .7 | |||
| Yes | 3 (3.3%) | 1 (2.4%) | 5 (5.7%) | |
| No | 88 (97%) | 41 (98%) | 83 (94%) | |
| Current cancer treatment | .04 (lymphoid vs myeloid) | |||
| Yes | 0 (0%) | 21 (50%) | 60 (68%) | |
| No | 91 (100%) | 21 (50%) | 28 (32%) |
| Variable . | Healthy controls, n = 91∗ . | Lymphoid neoplasms, n = 42∗ . | Myeloid neoplasms, n = 88∗ . | P value† . |
|---|---|---|---|---|
| Age (y) | 52 (38; 62) | 66 (48; 74) | 62 (52; 67) | < .001 |
| Sex | .033 | |||
| Female | 54 (59%) | 17 (40%) | 37 (42%) | |
| Male | 37 (41%) | 25 (60%) | 51 (58%) | |
| WBC | 6.10 (5.30; 7.75) | 5.40 (4.10; 6.77) | 6.00 (4.27; 8.38) | .2 |
| SARS-CoV-2 vaccination scheme | .07 | |||
| Vector based | 13 (14%) | 4 (9.5%) | 11 (12%) | |
| 2× mRNA-based | 62 (68%) | 37 (88%) | 69 (78%) | |
| Only one‡ | 16 (18%) | 1 (2.4%) | 8 (9.1%) | |
| Prior SARS-CoV-2 infection | .7 | |||
| Yes | 3 (3.3%) | 1 (2.4%) | 5 (5.7%) | |
| No | 88 (97%) | 41 (98%) | 83 (94%) | |
| Current cancer treatment | .04 (lymphoid vs myeloid) | |||
| Yes | 0 (0%) | 21 (50%) | 60 (68%) | |
| No | 91 (100%) | 21 (50%) | 28 (32%) |
WBC, white blood cell count.
Median (Q1 and Q3); n (%).
Kruskal-Wallis rank sum test; Fisher exact test.
Summarizes in-label Ad26.COV2.S (Janssen) and off-label single-shot immunization using either mRNA vaccine.
Details on disease status and detection of spike-specific IgG before vaccination are provided in the supplemental Results.
SARS-CoV-2 specific CD8+ T-cell, CD4+ T-cell, and IgG responses in patient cohorts and healthy controls
We compared SARS-CoV-2 spike-specific CD4+ T-cell, CD8+ T-cell, and IgG responses between controls and both patient cohorts (Table 2) and additionally subdivided patients who were actively treated and those who were (at least currently) untreated (supplemental Table 3).
Comprehensive comparative analysis of vaccination responses in patients and healthy controls
| Variable . | N . | Healthy control, n = 91 . | Lymphoid neoplasms, n = 42 . | P value healthy vs lymphoid neoplasms∗ . | Myeloid neoplasms, n = 88 . | P value healthy vs myeloid neoplasms∗ . |
|---|---|---|---|---|---|---|
| Spike-specific IgG detectable (n [%] of participants†) | 219 | 90 (99%) | 32 (78%) | < .001 | 86 (99%) | 1 |
| Spike-specific IgG levels (U/mL)‡ | 208 | 1554 (433; 3079) | 222 (92; 2115) | .02 | 942 (144; 2384) | .11 |
| Spike-specific CD4+ T cells detectable (n [%] of participants†) | 221 | 67 (74%) | 30 (71%) | .8 | 47 (53%) | .005 |
| Spike-specific CD4+ T cells (cells per μL blood)‡ | 144 | 0.166 (0.093; 0.251) | 0.165 (0.061; 0.248) | .7 | 0.111 (0.075; 0.238) | .7 |
| Spike-specific CD4+ T-cells (% of CD4+ T-cells)‡ | 144 | 0.012 (0.008; 0.02) | 0.024 (0.007; 0.035) | .12 | 0.018 (0.010; 0.027) | .26 |
| Activated CD4+ T cells by SEB detectable (n [%] of participants†) | 221 | 81 (89%) | 41 (97.6%) | .2 | 80 (91%) | .8 |
| Activated CD4+ T cells by SEB (cells per μL blood)‡ | 202 | 9.9 (3.9; 17.3) | 6.9 (4.3; 14.7) | .6 | 5.7 (2; 9.8) | .009 |
| Activated CD4+ T cells by SEB (% of CD4+)‡ | 202 | 0.82 (0.47; 1.25) | 1.71 (0.77; 2.81) | < .001 | 0.55 (0.34; 1.20) | .24 |
| Spike-specific CD8+ T-cells detectable (n [%] of particpants†) | 221 | 38 (42%) | 13 (31%) | .3 | 25 (28%) | .08 |
| Spike-specific CD8+ T cells (cells per μL blood)‡ | 76 | 0.038 (0.022; 0.085) | 0.035 (0.015; 0.057) | .5 | 0.050 (0.019; 0.168) | .5 |
| Spike-specific CD8+ T cells (% of CD8+)‡ | 76 | 0.008 (0.005; 0.017) | 0.009 (0.004; 0.012) | .9 | 0.014 (0.005; 0.027) | .4 |
| Activated CD8+ T cells by SEB detectable (n [%] of participants†) | 221 | 82 (90.1%) | 41 (97.6%) | .2 | 82 (93.2%) | .6 |
| Activated CD8+ T cells by SEB (cells per μL blood)‡ | 205 | 4 (1.9; 7.4) | 4.4 (2.7; 8.9) | .5 | 3 (0.9; 7.3) | .2 |
| Activated CD8+ T cells by SEB (% of CD8+)‡ | 205 | 0.86 (0.36; 1.50) | 1.15 (0.59; 2.01) | .14 | 0.76 (0.26; 1.65) | .6 |
| Variable . | N . | Healthy control, n = 91 . | Lymphoid neoplasms, n = 42 . | P value healthy vs lymphoid neoplasms∗ . | Myeloid neoplasms, n = 88 . | P value healthy vs myeloid neoplasms∗ . |
|---|---|---|---|---|---|---|
| Spike-specific IgG detectable (n [%] of participants†) | 219 | 90 (99%) | 32 (78%) | < .001 | 86 (99%) | 1 |
| Spike-specific IgG levels (U/mL)‡ | 208 | 1554 (433; 3079) | 222 (92; 2115) | .02 | 942 (144; 2384) | .11 |
| Spike-specific CD4+ T cells detectable (n [%] of participants†) | 221 | 67 (74%) | 30 (71%) | .8 | 47 (53%) | .005 |
| Spike-specific CD4+ T cells (cells per μL blood)‡ | 144 | 0.166 (0.093; 0.251) | 0.165 (0.061; 0.248) | .7 | 0.111 (0.075; 0.238) | .7 |
| Spike-specific CD4+ T-cells (% of CD4+ T-cells)‡ | 144 | 0.012 (0.008; 0.02) | 0.024 (0.007; 0.035) | .12 | 0.018 (0.010; 0.027) | .26 |
| Activated CD4+ T cells by SEB detectable (n [%] of participants†) | 221 | 81 (89%) | 41 (97.6%) | .2 | 80 (91%) | .8 |
| Activated CD4+ T cells by SEB (cells per μL blood)‡ | 202 | 9.9 (3.9; 17.3) | 6.9 (4.3; 14.7) | .6 | 5.7 (2; 9.8) | .009 |
| Activated CD4+ T cells by SEB (% of CD4+)‡ | 202 | 0.82 (0.47; 1.25) | 1.71 (0.77; 2.81) | < .001 | 0.55 (0.34; 1.20) | .24 |
| Spike-specific CD8+ T-cells detectable (n [%] of particpants†) | 221 | 38 (42%) | 13 (31%) | .3 | 25 (28%) | .08 |
| Spike-specific CD8+ T cells (cells per μL blood)‡ | 76 | 0.038 (0.022; 0.085) | 0.035 (0.015; 0.057) | .5 | 0.050 (0.019; 0.168) | .5 |
| Spike-specific CD8+ T cells (% of CD8+)‡ | 76 | 0.008 (0.005; 0.017) | 0.009 (0.004; 0.012) | .9 | 0.014 (0.005; 0.027) | .4 |
| Activated CD8+ T cells by SEB detectable (n [%] of participants†) | 221 | 82 (90.1%) | 41 (97.6%) | .2 | 82 (93.2%) | .6 |
| Activated CD8+ T cells by SEB (cells per μL blood)‡ | 205 | 4 (1.9; 7.4) | 4.4 (2.7; 8.9) | .5 | 3 (0.9; 7.3) | .2 |
| Activated CD8+ T cells by SEB (% of CD8+)‡ | 205 | 0.86 (0.36; 1.50) | 1.15 (0.59; 2.01) | .14 | 0.76 (0.26; 1.65) | .6 |
Significant (<0.05) P values are highlighted in bold.
P values calculated using Dunn post hoc test (continuous data) or Fisher exact test (categorical data).
Numbers and percentages of participants who show a response in spike-specific IgG, CD4+ T cells, and CD8+ T cells as well as in SEB-activated CD4+ T cells and CD8+ T cells are tabulated by patient and control groups.
Spike-specific IgG levels are provided in U/mL, spike-specific and SEB-activated CD4+ T cells and CD8+ T cells, respectively, are provided as percentage of the respective T-cell subpopulations and as counts of polyfunctional T cells per μL blood (medians, Q1 and Q3). These data were calculated based upon all participants with detectable IgG, polyfunctional CD4+ T cells, and CD8+ T cells, respectively.
Compared with healthy controls, fewer patients with lymphoid malignancies demonstrated a seroconversion after vaccination (99% vs 78%; P < .001). The reduced ability to generate SARS-CoV-2–specific IgG was similarly reflected by the antibody titers of responders: we measured median antibody titers of 222 U/mL in patients with lymphoid malignancies but sevenfold higher titers in healthy controls (1554 U/mL; P = .02). The reduced seroconversion rate as well as reduced antibody titers affected predominantly those patients with lymphoid neoplasms under active treatment. Patients with myeloid malignancies showed the same high seroconversion rate as the control cohort and a nonsignificant trend toward lower IgG antibody titers.
SARS-CoV-2–specific polyfunctional CD4+ T cells were detectable in a lower proportion of patients with myeloid neoplasms (53%) than in the control cohort (74%, P = .005), whereas no significant difference could be observed between controls and patients with lymphoid malignancies. Responding controls and patients did not differ in the numbers of spike-specific CD4+ T cells: we detected at median 0.166, 0.165, and 0.111 polyfunctional CD4+ T cells per μL. This pattern suggests that a particular subgroup of patients with myeloid neoplasms was unable to mount a polyfunctional spike-specific CD4+ T-cell response, whereas the remaining patients with myeloid neoplasms and the patients with lymphoid neoplasms showed CD4+ T-cell activation at the control level. Considering similar CD4+ T-cell response rates in patients receiving treatment and those who were not (supplemental Table 3), active treatment apparently does not explain subgroups with different vaccination results in myeloid malignancies.
Fewer CD4+ T cells were activated upon T-cell receptor (TCR)-mediated, antigen-unspecific stimulation in patients with myeloid neoplasms compared with healthy participants (5.7 vs 9.9 polyfunctional CD4+ T cells per μL), whereas the percentages of responding CD4+ T cells were comparable (0.55% vs 0.82%). In contrast, we observed an increased percentage of CD4+ T cells activated by SEB in patients with lymphoid neoplasms as compared with healthy controls (1.71% vs 0.82%). This suggests that the CD4+ T cells in patients with lymphoid neoplasms are easier to activate, and this effect might compensate for the lower numbers of CD4+ T cells. The effect might be particularly relevant when patients are being actively treated (supplemental Table 3).
The proportion of participants with detectable SARS-CoV-2–specific polyfunctional CD8+ T cells and the numbers of these cells did not differ between controls and both patient cohorts. Moreover, CD8+ T cells from both patient groups could be as readily activated by SEB as those from control participants. Patients with lymphoid malignancies actively undergoing treatment might even possess an increased response to SEB (supplemental Table 3).
In summary, patients with lymphoid malignancies mounted an insufficient IgG response accompanied by a normal or even improved CD4+ T-cell function against SARS-CoV-2. Patients with myeloid neoplasia had normal spike-specific IgG levels, but a major subpopulation of these individuals presented with a defective CD4+ T-cell response. Spike-specific CD8+ T-cell immune responses were preserved in both patient groups.
In-depth immune phenotyping in patient cohorts and healthy controls
Having demonstrated differences in CD4+ T-cell and IgG response rates toward vaccination between patients with myeloid malignancies and those with lymphoid malignancies, we searched for mechanistic explanations and, therefore, characterized the composition of the immune system in the cohorts.
In contrast to the largely unaffected myeloid lineage, we observed a significant lymphopenia in both patients with myeloid malignancies and those with lymphoid malignancies (Table 3; supplemental Figure 6). T cells, B cells, and natural killer (NK)-cells were all significantly reduced in both patient cohorts compared with in healthy controls, with greater numerical reductions observed for patients with lymphoid neoplasia.
Differences in major leukocyte subsets per μL blood between patients with hematological neoplasia and healthy controls
| Population . | Healthy control∗ (n = 87) . | Lymphoid neoplasms∗ (n = 41) . | P value healthy vs lymphoid neoplasms† . | Myeloid neoplasms∗ (n = 86) . | P value healthy vs myeloid neoplasms† . |
|---|---|---|---|---|---|
| Myeloid cells | 3424 (2737; 4775) | 3461 (2680; 4286) | .7 | 4056 (2429; 6338) | .5 |
| Neutrophils | 2789 (2215; 3947) | 2771 (2167; 3707) | .8 | 3190 (1868; 5324) | .6 |
| Eosinophils | 83 (46; 151) | 72 (35; 102) | .2 | 68 (35; 165) | .4 |
| Monocyten | 412 (323; 612) | 417 (343; 538) | 1 | 404 (295; 633) | .9 |
| Lymphocytes | 2529 (2073; 3189) | 1556 (1150; 2536) | < .001 | 2031 (1462; 2864) | .002 |
| B cells | 304 (206; 462) | 160 (47; 542) | .03 | 251 (122; 384) | .03 |
| T cells | 1937 (1513; 2390) | 1220 (752; 1675) | < .001 | 1520 (993; 2150) | .002 |
| CD4+ T cells | 939 (643; 1293) | 421 (258; 694) | < .001 | 577 (355; 928) | < .001 |
| Naive CD4+ T cells | 398 (232; 641) | 100 (36; 194) | < .001 | 194 (88; 389) | < .001 |
| CM CD4+ T cells | 330 (218; 491) | 184 (115; 306) | .001 | 224 (141; 346) | .001 |
| EM CD4+ T cells | 89 (58; 144) | 77 (40; 116) | .06 | 55 (34; 88) | .002 |
| TE CD4+ T cells | 7 (3; 20) | 2 (2; 10) | .02 | 3 (2; 11) | .009 |
| CD8+ T cells | 444 (277; 581) | 380 (258; 532) | .4 | 284 (178; 443) | .001 |
| Naive CD8+ T cells | 106 (49; 164) | 35 (18; 111) | .002 | 53 (23; 103) | .002 |
| CM CD8+ T cells | 43 (23; 68) | 31 (17; 59) | .2 | 28 (15; 55) | .01 |
| EM CD8+ T cells | 89 (47; 173) | 95 (61; 150) | .7 | 57 (33; 118) | .02 |
| TE CD8+ T cells | 132 (50; 203) | 116 (72; 212) | .6 | 100 (33; 168) | .1 |
| NK cells | 293 (182; 425) | 209 (103; 292) | .003 | 231 (122; 355) | .04 |
| Population . | Healthy control∗ (n = 87) . | Lymphoid neoplasms∗ (n = 41) . | P value healthy vs lymphoid neoplasms† . | Myeloid neoplasms∗ (n = 86) . | P value healthy vs myeloid neoplasms† . |
|---|---|---|---|---|---|
| Myeloid cells | 3424 (2737; 4775) | 3461 (2680; 4286) | .7 | 4056 (2429; 6338) | .5 |
| Neutrophils | 2789 (2215; 3947) | 2771 (2167; 3707) | .8 | 3190 (1868; 5324) | .6 |
| Eosinophils | 83 (46; 151) | 72 (35; 102) | .2 | 68 (35; 165) | .4 |
| Monocyten | 412 (323; 612) | 417 (343; 538) | 1 | 404 (295; 633) | .9 |
| Lymphocytes | 2529 (2073; 3189) | 1556 (1150; 2536) | < .001 | 2031 (1462; 2864) | .002 |
| B cells | 304 (206; 462) | 160 (47; 542) | .03 | 251 (122; 384) | .03 |
| T cells | 1937 (1513; 2390) | 1220 (752; 1675) | < .001 | 1520 (993; 2150) | .002 |
| CD4+ T cells | 939 (643; 1293) | 421 (258; 694) | < .001 | 577 (355; 928) | < .001 |
| Naive CD4+ T cells | 398 (232; 641) | 100 (36; 194) | < .001 | 194 (88; 389) | < .001 |
| CM CD4+ T cells | 330 (218; 491) | 184 (115; 306) | .001 | 224 (141; 346) | .001 |
| EM CD4+ T cells | 89 (58; 144) | 77 (40; 116) | .06 | 55 (34; 88) | .002 |
| TE CD4+ T cells | 7 (3; 20) | 2 (2; 10) | .02 | 3 (2; 11) | .009 |
| CD8+ T cells | 444 (277; 581) | 380 (258; 532) | .4 | 284 (178; 443) | .001 |
| Naive CD8+ T cells | 106 (49; 164) | 35 (18; 111) | .002 | 53 (23; 103) | .002 |
| CM CD8+ T cells | 43 (23; 68) | 31 (17; 59) | .2 | 28 (15; 55) | .01 |
| EM CD8+ T cells | 89 (47; 173) | 95 (61; 150) | .7 | 57 (33; 118) | .02 |
| TE CD8+ T cells | 132 (50; 203) | 116 (72; 212) | .6 | 100 (33; 168) | .1 |
| NK cells | 293 (182; 425) | 209 (103; 292) | .003 | 231 (122; 355) | .04 |
CM, central memory cells (CD45RA−CCR7+); EM, effector memory cells (CD45RA−CCR7−); TE, terminal effector cells (CD45RA+CCR7−).
Significant (<0.05) P values are highlighted in bold.
Median (Q1 and Q3); n (%); T-cell maturation parameters (CCR7, CD45RA) could not be assessed in 1 healthy control, 3 patients with lymphoid, and 1 patient with myeloid neoplasia because of technical reasons.
P values calculated using Dunn post hoc test.
We found a reduction in the number of CD4+ T cells in both patient cohorts. Within the CD4+ T-cell compartment, all maturation steps, except the effector memory CD4+ T cells in lymphoid neoplasia were significantly reduced. Naive CD4+ T cells were diminished the most: patients with lymphoid neoplasms exhibited only a quarter of the number of naive CD4+ T cells compared with that in the healthy controls (100 vs 398 cells per μL). Similarly, the controls had twice as many naive CD4+ T cells than patients with myeloid neoplasms (398 vs 194 cells per μL). A significant overall reduction of CD8+ T cells was demonstrated in patients with myeloid neoplasms, whereas patients with lymphoid neoplasms presented nearly normal CD8+ T-cell numbers. Nevertheless, naive CD8+ T cells were reduced in both patient cohorts when compared with the control population.
A systematic comparison of both patient cohorts with the normal control group and between each other (supplemental Figure 6) revealed that both patient cohorts presented similar reductions of many different T-cell subpopulations. In lymphoid, compared with myeloid malignancies, fewer T cells, naive regulatory T cells, and naive CD4+ T cells were detected. Patients with lymphoid neoplasms presented not only fewer CD4+ T cells expressing inhibitory molecules TIM373 and LAG3,74 but also fewer CD4+ T cells expressing the activating NKG2D molecule.75
A complete list of all 121 investigated parameters, including descriptive statistics, is provided in supplemental File 2.
Taken together, the immune system of patients with lymphoid or myeloid malignancies resembled each other with an overall lymphopenia and a pronounced reduction in naive CD4+ T cells, which might reflect a contraction of the reservoir of CD4+ T cells that are able to respond to vaccination. Both patient cohorts differed between each other in the numbers of naive CD4+ T cells and regulatory T cells as well as in the expression of inhibitory and activating receptors on CD4+ T cells.
Parameters associated with spike-specific CD4+ T-cells, CD8+ T-cell, and IgG responses
We investigated whether immunophenotypic or demographic parameters or the type of vaccination were significantly associated with detectable polyfunctional spike-specific CD4+ T cells and CD8+ T cells or with above-median spike-specific IgG titers (Figure 1; detailed in supplemental File 2).
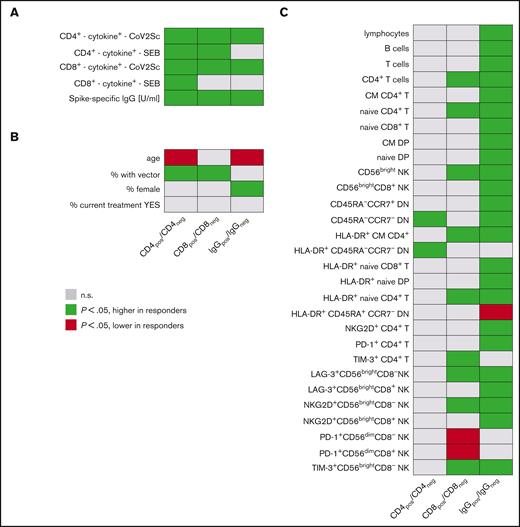
Parameters associated with vaccination responses. Univariate analyses of (A) vaccination response parameters, (B) clinicodemographic features, and (C) immune signatures in study participants based on CD4+ T-cell, CD8+ T-cell, and IgG responses. (left) Groups of study participants with detectable spike-specific CD4+ response (n = 144) compared with those without detectable CD4+ response (n = 77). Analogously, (middle) comparison of participants with spike-specific CD8+ response (n = 76) with CD8+ nonresponders (n = 145); (right) participants with specific IgG levels above median (n = 109) vs participants with lower spike-specific IgG (n = 110). Each line represents a vaccination response parameter, clinicodemographic feature, or specific population from in-depth immunophenotyping (expressed as cells per μL). Green cells denote higher age, higher IgG titers, higher percentages of women, and of recipients of heterologous vaccination schemes, as well as higher cell numbers per μL blood for all other features in CD4+, CD8+, and IgG responders. Red cells indicate lower respective values in responders. Shown are all parameters with at least 1 of the 3 P values < .05 and for comparison, also the percentage of patients under active treatment. A total of 121 immunophenotypic parameters were analyzed. CD4pos/neg, with/without detectable spike-specific CD4+ T-cell response; CD8pos/neg, with/without detectable spike-specific CD8+ T-cell response; CM, central memory; CoV2Sc, after stimulation with spike peptides; cytokine+, polyfunctional activated T cells; DN, CD4−CD8− double-negative T cells; DP, CD4+CD8+ double-positive T cells; EM, effector memory; IgGpos/neg, with/without spike-specific IgG levels above median; TE, terminal effector.
Parameters associated with vaccination responses. Univariate analyses of (A) vaccination response parameters, (B) clinicodemographic features, and (C) immune signatures in study participants based on CD4+ T-cell, CD8+ T-cell, and IgG responses. (left) Groups of study participants with detectable spike-specific CD4+ response (n = 144) compared with those without detectable CD4+ response (n = 77). Analogously, (middle) comparison of participants with spike-specific CD8+ response (n = 76) with CD8+ nonresponders (n = 145); (right) participants with specific IgG levels above median (n = 109) vs participants with lower spike-specific IgG (n = 110). Each line represents a vaccination response parameter, clinicodemographic feature, or specific population from in-depth immunophenotyping (expressed as cells per μL). Green cells denote higher age, higher IgG titers, higher percentages of women, and of recipients of heterologous vaccination schemes, as well as higher cell numbers per μL blood for all other features in CD4+, CD8+, and IgG responders. Red cells indicate lower respective values in responders. Shown are all parameters with at least 1 of the 3 P values < .05 and for comparison, also the percentage of patients under active treatment. A total of 121 immunophenotypic parameters were analyzed. CD4pos/neg, with/without detectable spike-specific CD4+ T-cell response; CD8pos/neg, with/without detectable spike-specific CD8+ T-cell response; CM, central memory; CoV2Sc, after stimulation with spike peptides; cytokine+, polyfunctional activated T cells; DN, CD4−CD8− double-negative T cells; DP, CD4+CD8+ double-positive T cells; EM, effector memory; IgGpos/neg, with/without spike-specific IgG levels above median; TE, terminal effector.
Participants with detectable spike-specific CD4+ T cells (CD4pos) were more frequently vaccinated with at least 1 dose of vector-based AZD1222 vaccine when compared with nonresponders (27 of 144 vs 1 of 77; P < .001). CD4pos individuals at median had higher spike-specific IgG titers (1500 vs 176 U/mL; P < .001) and more spike-specific CD8+ T cells (0.019 vs 0.016 cells per μL; P = .007). These CD4+ T-cell responders also showed a stronger activation upon antigen-unspecific, TCR-dependent stimulation using SEB (CD4+ T cells: 8.0 vs 4.0 cells per μL and CD8+ T cells: 4.3 vs 1.9 cells per μL). CD4pos participants were younger than nonresponders (55 vs 62 years; all P < .001) and had significantly more frequent CD4−CD8− double-negative T cells76 lacking both CD45RA and CCR7 and an increased frequency of HLA-DR+ among those cells.
A vector-based vaccine was more frequently used in participants with detectable spike-specific CD8+ T cells (CD8pos, 20 of 76) compared with nonresponders (8 of 145; P < .001). CD8pos participants had higher IgG titers (1621 vs 623 U/mL; P < .001). As expected from the data mentioned above, we detected more spike-specific CD4+ T cells in CD8pos individuals (median: 0.16 vs 0.07 cells per μL; P < .001). SEB stimulation induced a stronger CD4+ T-cell response in CD8pos individuals (median 8.3 vs 5.5 cells per μL, P = .006), whereas the CD8+ T-cell response to SEB was comparable (medians 3.2 vs 4.3 cells per μL; P = .1). Furthermore, we found a significantly higher number of CD4+ T cells and naive CD4+ T cells (median, 357 vs 208 cells per μL; P = .006) in CD8pos individuals. Moreover, the numbers of PD-1+CD56dimCD8+ T cells and CD56dimCD8+ NK cells were significantly lower in the CD8pos group, whereas the number of CD56bright NK cells was elevated (supplemental File 2).
Given the remarkable benefit for the vector-based AZD1222 to induce spike-specific T-cell vaccination responses, we specifically compared study participants by vaccination scheme (Table 4). Vaccinees who received schemes that included vector-based AZD1222 were marginally younger than the remaining participants but exhibited no other clinical or demographic features suggestive of a higher likelihood for a successful immunization. When analyzed based on the participant cohort, AZD1222 induced a detectable, specific CD4+ T-cell response in 100% (13 of 13) of healthy controls, 100% (4 of 4) of patients with lymphoid neoplasms, and 90% (10 of 11) of patients with myeloid neoplasms, whereas fully mRNA-based vaccination schemes resulted in measurable CD4+ T-cell responses in 73% (45 of 62; P = .03), 68% (25 of 37; P = .3), and 46% (32 of 69; P = .008), of the participant cohorts, respectively. Similarly, vector-based prime-boost vaccination also more frequently induced spike-specific CD8+ T-cell responses in controls (vector, 11 of 13 [85%] vs mRNA, 21 of 62 [34%]; P = .001), lymphoid malignancies (vector 2 of 4 [50%] vs mRNA, 11 of 37 [30%]; P = .6), and myeloid malignancies (vector, 7 of 11 [64%] vs mRNA, 16 of 69 [23%]; P = .01). However, quantitatively, spike-specific T cells per μL blood in responders did not differ significantly between mRNA-based vaccination schemes and AZD1222 immunization.
Demographic, clinical, and vaccination response data for participants grouped based on the vaccination scheme
| Variable . | Vector based, n = 28∗ . | Only 1†, n = 25∗ . | 2× mRNA-based, n = 168∗ . | P value‡ . |
|---|---|---|---|---|
| Age (y) | 53 (45; 60) | 55 (48; 61) | 60 (47; 69) | .05 |
| Sex | .2 | |||
| Female | 13 (46%) | 8 (32%) | 87 (52%) | |
| Male | 15 (54%) | 17 (68%) | 81 (48%) | |
| WBC | 5.5 (4.5; 6.1) | 5.7 (4.7; 7.1) | 6.3 (4.8; 8.2) | .1 |
| Study cohort | .07 | |||
| Healthy control | 13 (46%) | 16 (64%) | 62 (37%) | |
| Lymphoid neoplasms | 4 (14%) | 1 (4%) | 37 (22%) | |
| Myeloid neoplasms | 11 (39%) | 8 (32%) | 69 (41%) | |
| Prior SARS-CoV-2 infection | < .001 | |||
| Yes | 0 (0%) | 7 (28%) | 2 (1.2%) | |
| No | 28 (100%) | 18 (72%) | 166 (98.8%) | |
| Current cancer treatment | .4 | |||
| Yes | 11 (39%) | 6 (24%) | 64 (38%) | |
| No | 17 (61%) | 19 (76%) | 104 (62%) | |
| Spike-specific IgG detectable (n [%] of participants) | 26 (93%) | 23 (96%) | 159 (95%) | .9 |
| Spike-specific CD4+ T cells detectable (n [%] of participants) | 27 (96%) | 15 (60%) | 102 (61%) | .001 |
| Spike-specific CD8+ T cells detectable (n (%) of participants) | 20 (71%) | 8 (32%) | 48 (29%) | < .001 |
| Variable . | Vector based, n = 28∗ . | Only 1†, n = 25∗ . | 2× mRNA-based, n = 168∗ . | P value‡ . |
|---|---|---|---|---|
| Age (y) | 53 (45; 60) | 55 (48; 61) | 60 (47; 69) | .05 |
| Sex | .2 | |||
| Female | 13 (46%) | 8 (32%) | 87 (52%) | |
| Male | 15 (54%) | 17 (68%) | 81 (48%) | |
| WBC | 5.5 (4.5; 6.1) | 5.7 (4.7; 7.1) | 6.3 (4.8; 8.2) | .1 |
| Study cohort | .07 | |||
| Healthy control | 13 (46%) | 16 (64%) | 62 (37%) | |
| Lymphoid neoplasms | 4 (14%) | 1 (4%) | 37 (22%) | |
| Myeloid neoplasms | 11 (39%) | 8 (32%) | 69 (41%) | |
| Prior SARS-CoV-2 infection | < .001 | |||
| Yes | 0 (0%) | 7 (28%) | 2 (1.2%) | |
| No | 28 (100%) | 18 (72%) | 166 (98.8%) | |
| Current cancer treatment | .4 | |||
| Yes | 11 (39%) | 6 (24%) | 64 (38%) | |
| No | 17 (61%) | 19 (76%) | 104 (62%) | |
| Spike-specific IgG detectable (n [%] of participants) | 26 (93%) | 23 (96%) | 159 (95%) | .9 |
| Spike-specific CD4+ T cells detectable (n [%] of participants) | 27 (96%) | 15 (60%) | 102 (61%) | .001 |
| Spike-specific CD8+ T cells detectable (n (%) of participants) | 20 (71%) | 8 (32%) | 48 (29%) | < .001 |
Vaccination success is expressed as percentage of study participants with detectable spike-specific IgG, CD4+, and CD8+ responses, respectively. WBC, white blood cell count.
Significant (<0.05) P values are highlighted in bold.
Median (Q1 and Q3); n (%).
Summarizes in-label Ad26.COV2.S (Janssen) and off-label single-shot immunization using either mRNA vaccine.
Kruskal-Wallis rank sum test; Fisher exact test.
Finally, we compared participants with (IgG+) and without spike-specific IgG titers above the overall median titer 120 days after vaccination. Individuals who were IgG+ were younger (median: 54 vs 63.5 years; P < .001) and more often female (65 of 109 vs 42 of 110; P = .002). Spike-specific CD4+ T cells were roughly twice as frequent in IgG+ compared with IgG− patients and controls (medians: 0.13 vs 0.07 cells per μL blood; P < .001). A stronger IgG response was associated with higher B-cell counts (median: 318 vs 196 cells per μL). Total lymphocyte and T-cell numbers followed this pattern, as did the numbers of various T-cell and NK-cell subpopulations (supplemental File 2). Of note, the numbers of naive CD4+ T cells were also increased in individuals who were IgG+ compared with those who were IgG− (median 334 vs 186 cells per μL; P < .001).
In general, strong vaccination responses of CD8+ T cells, CD4+ T cells, and IgG cooccur when all participants are analyzed together and when the quantitative strength of the responses (ie, titers and cell counts) are considered. Furthermore, a responsive immune system can be characterized by sufficient numbers of naive T cells77 and the responsiveness of T cells after a TCR-mediated, antigen-independent stimulation. Importantly, spike-specific T-cell responses were detected in a higher fraction of study participants when vector-based vaccinations were used.
Multivariable response models
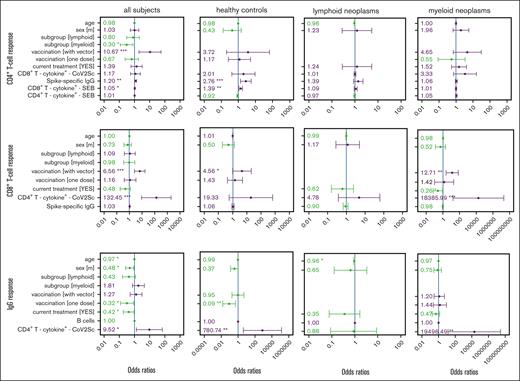
Lastly, we identified parameters that were independently associated with SARS-CoV-2–specific CD4+ and CD8+ T-cell and IgG vaccination responses using multivariable logistic regression models. We included parameters identified as important using a Monte Carlo–based feature selection approach (supplemental Figure 7) along with parameters significantly associated with the response in univariable analyses: age, sex, and the vaccination type. Owing to published evidence on the significance of the current treatment status,33,78 the initial models were also fitted to include this parameter.
The generic model identified vaccination with at least 1 vector-based vaccine dose (odds ratio [OR], 10.7), the CD8+ antigen-unspecific T-cell responsiveness (OR, 1.05), and spike-specific IgG titers (OR, 1.2) as significantly associated with a spike-specific CD4+ T-cell response (Figure 2). Patients with myeloid neoplasms were significantly less likely to mount a detectable CD4+ T-cell response (OR, 0.3). Exclusively for healthy controls, the model retained a positive association with IgG titers (OR, 2.76) and with unspecific CD8+ T-cell responsiveness (OR, 1.39). With lower participant numbers, the models for each subgroup separately did not identify any other parameter as being significantly associated with CD4+ T-cell response. However, the chance of developing a spike-specific CD4+ T-cell response was numerically higher when at least 1 vector-base vaccine dose was given to control participants (OR, 3.72) and patients with myeloid neoplasms (OR, 4.65).
Multivariable regression of factors associated with response to vaccination against SARS-CoV-2 in patients with hematological neoplasms and in controls. Parameters rated important by the feature selection algorithm (supplemental Figure 7) in 1 of the 4 groups were included in every model. ORs and the 95% confidence intervals are indicated. Reference groups for the categorical parameters were belonging to the healthy control group, female sex, 2 mRNA-based vaccine doses, and no current treatment. The CD4+ T-cell, CD8+ T-cell, and IgG vaccination response models were calculated for the total study population as well as for the 3 study cohorts, separately. Parameters in green show an OR <1 and parameters in violet an OR >1. The size of vaccination groups “only 1” (n = 1) and “with vector” (n = 4) precluded an analysis of vaccination scheme in lymphoid neoplasia models. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
Multivariable regression of factors associated with response to vaccination against SARS-CoV-2 in patients with hematological neoplasms and in controls. Parameters rated important by the feature selection algorithm (supplemental Figure 7) in 1 of the 4 groups were included in every model. ORs and the 95% confidence intervals are indicated. Reference groups for the categorical parameters were belonging to the healthy control group, female sex, 2 mRNA-based vaccine doses, and no current treatment. The CD4+ T-cell, CD8+ T-cell, and IgG vaccination response models were calculated for the total study population as well as for the 3 study cohorts, separately. Parameters in green show an OR <1 and parameters in violet an OR >1. The size of vaccination groups “only 1” (n = 1) and “with vector” (n = 4) precluded an analysis of vaccination scheme in lymphoid neoplasia models. ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
Similarly, the CD8+ T-cell response was also significantly associated with a vaccination scheme that included at least 1 vector-based dose as compared with 2 doses of a mRNA-based vaccine (OR, 6.56). Furthermore, the numbers of spike-specific CD4+ T cells were associated with a CD8+ T-cell response. The subgroup-specific models underlined the statistically significant beneficial effect of using at least 1 vector-based vaccine for the control group (OR, 4.56) and patients with myeloid neoplasms (OR, 12.71). In contrast to our observations for the CD4+ T-cell response, for patients with myeloid neoplasms, ongoing therapy, when being vaccinated, lowered the chance of developing a measurable CD8+ T-cell response.
The serological response was negatively affected by older age, being male, currently receiving therapy, and by having received only a single immunization. In contrast, having measurable spike-specific CD4+ T cells was positively associated with an IgG response. Of note, the numbers of B cells were not associated with an IgG response in the multivariable models.
In summary, the multivariable models confirmed a strong interdependence of spike-specific CD8+ T-cell, CD4+ T-cell, and IgG responses to vaccination across the study cohorts of patients and controls. Of particular interest was the observation of a higher likelihood of mounting spike-specific CD8+ and CD4+ T-cell responses upon vector-based vaccination. None of the immunophenotypic parameters showed an association to the cellular or humoral vaccination immunity independent of the respective other spike-specific immune response outcomes and the vaccination scheme.
Discussion
In order to elucidate immunological mechanisms for vaccination success and failure, we, herein, provide a comprehensive characterization of the specific CD8+ T-cell, CD4+ T-cell, and serological immunity induced by SARS-CoV-2 vaccination in 130 patients with hematological malignancies and 91 healthy controls. To the best of our knowledge, our study is the first to measure polyfunctional spike-specific T-cell responses using a dedicated assay in a large cohort of patients with lymphoid or myeloid neoplasms.
The flow cytometry assay used in this study allows the separate analysis of spike-specific CD8+ T-cell and CD4+ T-cell responses, unlike, for example, the enzyme-linked immunospot assay used by others.44,58,79 Taking advantage of this technical feature, we could demonstrate (Table 2) that the reduced T-cell response in myeloid malignancies affects CD4+ T cells only, whereas the CD8+ T-cell response can be detected with the same frequency and strength as those in controls and patients with lymphoid malignancies. The reduction in CD4+ T-cell responsiveness appears to be restricted to a distinct subgroup of patients with myeloid malignancies, because the counts of SARS-CoV-2–specific CD4+ T cells in responders equal the counts in healthy controls. T-cell responses without seroconversion were detectable in a sizable fraction of patients with lymphoid malignancies.56 That dissociation of humoral and cellular vaccination immunity in hematological malignancies has very recently also been described by other groups.44,58,79,80
To explain the differences in responses between myeloid and lymphoid neoplasia, we hypothesized that certain myeloid neoplasms and their treatment might specifically impair the CD4+ T-cell compartment, whereas the treatment of lymphoid malignancies might interfere with a B cell–mediated vaccination response. We sought to elucidate mechanistic links between vaccination success and disease categories using in-depth immunophenotyping of 121 distinct cell populations. However, these investigations mainly revealed similarities between both patient groups: lymphopenia affecting T, B, and NK cells as well as a profound reduction of the naive compartment in CD8+ T cells and CD4+ T cells. We identified only a few, relatively small CD4+ T-cell populations that were increased in myeloid neoplasm in comparison with lymphoid neoplasms. Thus, immunophenotyping did not offer a clear explanation for the impaired CD4+ T-cell immunity co-occurring with preserved CD8+ T-cell responses in a fraction of patients with myeloid malignancies.
In order to identify immunophenotypic predictors for the observed different immune responses by study cohort, we developed multivariable models for the prediction of detectable spike-specific CD8+ T cells and CD4+ T cells as well as above-median IgG levels. Our analyses revealed that the CD8+ T-cell response strongly depended on the CD4+ T-cell response reaching statistical significance in multivariable models of the total study population and patients with myeloid malignancies and was also evident for healthy controls and patients with lymphoid malignancies. This observation suggests that a strong CD8+ T-cell response is supported by an extensive spike-specific CD4+ T-cell activation (Figure 2). Moreover, the CD8+ T-cell but not the CD4+ T-cell response is negatively affected by the current treatment. In line with our observation, a negative impact of the current treatment on T-cell responses in general had been demonstrated before.32,79,80 In our study, we could refine this observation as being only significant for the CD8+ T-cell response.
Reduced titers of anti-SARS-CoV-2 spike-protein IgG as well as neutralizing antibodies in patients with hematological neoplasms has previously been described to be likely mediated by B-cell depletion or suppression.31,81 This is in line with our multivariable model showing a strong significant effect of current treatment on the serological response. In our cohort, the numbers of B cells showed no independent association with the serological response. We speculate that in our models the same effect might be captured by the parameter current treatment, which would lead to a severe B-cell depletion in many lymphoid malignancies. The multivariable approach also identified a strong spike-specific CD4+ T-cell response as a beneficial parameter for a strong serological immunization response for both individuals with myeloid neoplasia and healthy controls. That interaction was completely lost in lymphoid malignancies.
Our data endorsed a model with a central role of spike-specific CD4+ T-cell support for both CD8+ T-cell and IgG immunization responses. Patients without an operational B-cell compartment, such as those with treated lymphoid malignancies, cannot mount an IgG response even in the presence of a strong CD4+ T-cell activation. They therefore have to rely upon CD8+ T cells for protection from severe COVID-19.25
For both the CD4+ T-cell and CD8+ T-cell responses, we found a significant beneficial effect of vector-based vaccination schemes. The healthy controls, but more importantly the group of patients with myeloid malignancy, appeared to benefit from such a schedule in terms of response rates (with low patient numbers receiving vector-based vaccines, the relationship could not be established for lymphoid neoplasia). Very recently, it had been shown that in healthy vaccinees, a heterologous prime-boost immunization resulted in higher T-cell reactivities as compared with homologous vaccination using mRNA vaccines only.60,61,63,64 To our knowledge, no such studies had been performed with focus on individuals who are immune compromised. If confirmed in additional studies, our results suggest revisiting heterologous vaccination schemes for those vulnerable patient groups.
Published data on T-cell responses to SARS-CoV-2 vaccination in patients with hematological neoplasms are sparse and sometimes conflicting.32,44,45,57,58,66,79,80,82-86 The majority of studies used enzyme-linked immunospot assays to assess T-cell responses and thus were not able to distinguish between CD4+ T-cell and CD8+ T-cell responses or measure polyfunctional responses. In contrast, we had established an intracellular cytokine staining assay using minimal sample manipulation to calculate the exact numbers of spike-specific CD4+ T cells and CD8+ T cells. We were able to detect spike protein–specific polyfunctional CD4+ T-cell responses in 74% of all healthy participants and spike protein–specific polyfunctional CD8+ T-cell responses in 42% of all healthy donors. The frequencies of spike-specific polyfunctional CD4+ T cells ranged from 0.00459% (our LOD) to 0.5% among all CD4+ T cells and from 0.00287% to 0.6% among all CD8+ T cells, and thus match respective values published, for example by Cohen et al.65 Seminal work by Sahin et al87 reported CD4+ and CD8+ T-cell responses in 95% and 76%, respectively, of all participants, 7 days after secondary immunization with tozinameran, with frequencies ranging from 0.02% to 0.1% and from 0.01% to 1.44% for interferon-γ+ CD4+ and CD8+ T cells, respectively. These studies indicate that CD8+ T-cell responses are less frequently detectable than CD4+ T-cell responses. Our results are within these published ranges.7
Beside the technically advanced assay used to quantify T-cell responses, another strength of our study is the large control cohort that was treated and investigated exactly like the patient cohorts. As a major drawback, we acknowledge that our patient cohorts are heterogeneous in terms of diseases and treatments. This heterogeneity and the resulting low number of study participants with particular clinicodemographic and disease features most probably explain the lack of statistical significance of certain associations in our multivariable analyses. Thus, our cohort is a cross-sectional representation of the patient’s spectrum that allows the generation of hypotheses for patient groups but not disease or treatment-specific analyses.
Although there are convincing data to link spike-specific IgG responses in vaccinees with protection from disease, such correlations between spike-specific CD4+ T-cell and CD8+ T-cell counts as a quantitative correlate of protection against symptomatic or severe COVID-19 remain to be established.16,25 In an attempt to clarify this in the future, we are currently assessing long-term clinical outcomes in our study participants.
In conclusion, our study untangles the links and differences between the 3 arms of the adaptive immune system in a large, healthy control group compared with those in patients with malignancies of the lymphoid and myeloid lineage. These clear differences in responses reported herein call for tailored vaccination plans that might be additionally refined using the monitoring of polyfunctional T cells. Moreover, we provide preliminary evidence that the use of vector-based vaccines increases T-cell responses, which might be important to protect patients who are immune compromised from severe COVID-19.3,16,25
Acknowledgments
The authors thank Patrick Brennan for English proof reading.
This work received a grant from the East German Study Group for Hematology and Oncology (OSHO e.V.; project number: OSHO#98).
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Authorship
Contribution: S.B., H.K.A.-A., and R.E. designed the research; S.B. and R.E. analyzed and interpreted the data and drafted the manuscript; and all authors performed research and carefully revised and approved the final version of the manuscript.
Conflict-of-interest disclosure: H.K.A.-A. consults for Novartis, Bristol Myers Squibb, Takeda, Pfizer, and AbbVie; receives honoraria from Novartis, Bristol Myers Squibb, Takeda, Pfizer, and AbbVie; and receives research funding from Novartis, Bristol Myers Squibb, and Incyte. S.B. consults for Roche; reports honoraria from Roche, AbbVie, Novartis, AstraZeneca, Amgen, and Janssen; and receives research funding from Janssen, Miltenyi Biotec, Roche, Genentech, and AbbVie. C.J. receives honoraria from Novartis, Amgen, Janssen, and AbbVie and received research funding from Janssen, Miltenyi Biotec, Roche, and Centogene. The remaining authors declare no conflicting financial interests.
Correspondence: Sebastian Böttcher, Clinic III (Hematology, Oncology and Palliative Medicine), Special Hematology Laboratory, Rostock University Medical Center, Ernst-Heydemann-Strasse 6, 18057 Rostock, Germany; e-mail: sebastian.boettcher@med.uni-rostock.de.
References
Author notes
All R scripts and accompanying raw data are available upon reasonable request to the corresponding author, Sebastian Böttcher (sebastian.boettcher@med.uni-rostock.de).
The full-text version of this article contains a data supplement.