TO THE EDITOR:
Introduction
CD19-directed immunotherapies have been extremely effective at inducing remission in patients with B-cell acute lymphoblastic leukemia (B-ALL), however, relapse remains a major challenge.1-3 The rarest but most problematic relapse event after CD19-directed immunotherapies is that of lineage switch (LS), in which neoplastic cells lose B-lymphoid–specific features and acquire a myeloid immunophenotype.4-8 Optimal salvage strategies for these patients are unknown and the absence of lymphoid targets reduces available immunotherapeutic options. Here, we present 2 cases of LS that are salvaged with myeloid-directed therapy but experience recurrent B-ALL. In both cases, patients were successfully rechallenged with CD19-directed immunotherapy without early recurrence of myeloid disease.
Methods
This is a retrospective case series reporting 2 patients with KMT2A-rearranged (KMT2A-r) infant B-ALL who experienced LS followed by subsequent B-ALL relapse after hematopoietic cell transplantation (HCT), successfully salvaged with CD19-directed immunotherapy. Each patient received treatment on a clinical trial protocol, with case 1 on PLAT-03 (#NCT03186118) and case 2 on PLAT-05 (#NCT0333691). PLAT-03 was a pilot study that incorporated periodic infusions of CD19-expressing antigen-presenting T cells after the initial infusion of a CD19–chimeric antigen receptor (CAR) T cell with a 4-1BB costimulatory domain (SCRI-CAR19).9 PLAT-05 is an ongoing phase 1 trial that tests the safety and efficacy of a CD19 × CD22 dual specific CAR T-cell product (SCRI-CAR19x22).10 Evaluation of leukemic blasts through multiparameter flow cytometry (MFC), fluorescence in situ hybridization, and G-banded karyotyping was performed in Clinical Laboratory Improvement Amendments–certified laboratories. For morphologic complete remissions (CRs), minimal residual disease (MRD) was evaluated by MFC and/or next-generation sequencing (NGS) of the IGH/IGL gene using Clonoseq (Adaptive Biotechnologies, Seattle, WA). MRD status is reported with methodology used (MFC-negative or -positive CR and/or NGS-negative or -positive CR). In addition, for 1 case, an RNA fusion assay using Archer FusionPlex (Seattle Children’s Hospital, Seattle, WA) and a targeted DNA-based NGS panel of cancer-related genes was performed via UW-OncoPlex (University of Washington, Seattle, WA).11 These studies were conducted with approval of the institutional review board of Seattle Children’s Hospital and in accordance with the Declaration of Helsinki.
Results/Discussion
Case 1
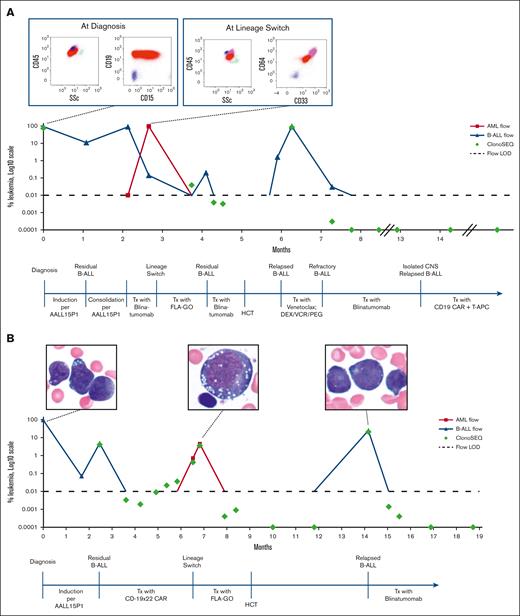
A 6-month-old female diagnosed with KMT2A-r, CNS2 infant B-ALL had persistent disease after induction chemotherapy on the Children’s Oncology Group study AALL15P1 (#NCT02828358). She experienced progressive medullary and central nervous system (CNS) disease during the subsequent consolidation phase of therapy. Salvage therapy was attempted with blinatumomab, but she experienced an LS during the first cycle (Table 1). She was able to achieve an MFC-positive CR after administration of FLA + GO (fludarabine, high-dose cytarabine, and gemtuzumab ozogamicin), with MRD notable for CD19+ B-ALL and no evidence of myeloid disease. She was rechallenged with blinatumomab and achieved MFC-negative but NGS-positive CR.8 She proceeded with cord blood HCT but relapsed with CD19+ B-ALL by day 28. Blinatumomab was initiated while tapering off immune suppression and she achieved an NGS-negative CR after 1 cycle. After a total of 5 cycles of blinatumomab, she was enrolled on the PLAT-03 trial owing to a CNS relapse of B-ALL. CNS disease was eradicated after infusion of SCRI-CAR19. Twenty-three months after infusion and 6 doses of antigen-presenting T cells, she showed evidence of ongoing persistence of her CAR T cells and remained in an NGS-negative CR (Figure 1A).
Disease characteristics
| . | Initial B-ALL diagnosis . | LS to AML . | Relapsed B-ALL . | |||
|---|---|---|---|---|---|---|
| Case 1 | ||||||
| Blast % | 93 B-lymphoblasts | 96 abnormal immature monocytes 0.14 B-lymphoblasts (immunophenotype similar to diagnosis) | 70 B-lymphoblasts | |||
| Flow cytometry immunophenotype of major blast population | CD19 | Positive | CD19 | Negative | CD19 | Positive |
| CD22 | Positive | CD22 | Negative | CD22 | Positive | |
| CD10 | Negative | CD10 | Negative | CD10 | Negative | |
| CD15 | Partial positive | CD15 | Positive | CD15 | Negative | |
| CD34 | Negative | CD34 | Negative | CD34 | Partial positive | |
| CD38 | Positive | CD38 | Positive | CD38 | Positive | |
| CD58 | Positive (increased) | CD58 | Positive | CD58 | Positive (increased) | |
| HLA-DR | Positive | HLA-DR | Positive | HLA-DR | Positive | |
| CD13 | Negative | CD13 | Negative | CD13/33 | Negative | |
| CD33 | Negative | CD33 | Positive | CD33 | Negative | |
| CD11b | Not evaluated | CD11b | Positive | CD11b | Not evaluated | |
| CD14 | Negative | CD14 | Partial positive | CD14 | Not evaluated | |
| CD64 | Not evaluated | CD64 | Positive | CD64 | Negative | |
| CD4 | Not evaluated | CD4 | Positive (dim) | CD4 | Not evaluated | |
| CD7 | Partial positive | CD7 | Negative | CD7 | Not evaluated | |
| CD45 | Positive (slightly dim) | CD45 | Positive | CD45 | Positive | |
| CD123 | Not evaluated | CD123 | Not evaluated | CD123 | Positive | |
| Cytogenetics | Karyotype: 46, XX, t(1;11)(p32;q23)[16]/46,XX[2] | Karyotype: 46, XX, t(1;11)(p32;q23)[5]/47,XX,t(1;11),+8[14]/46,XX[1] | Not performed | |||
| FISH: positive for a KMT2A rearrangement in 81% of nuclei (with loss of 3′ KMT2A signal) | FISH: positive for a KMT2A rearrangement in 95% of nuclei (loss of 3′ KMT2A signal) | |||||
| ClonoSEQ, % | IGH-SeqA = 60.9 IGH-SeqB = 70.0 IGL-SeqC = 76.2 IGL-SeqD = 60.9 IGH-SeqE = 0.6 IGH-SeqF = 1.8 IGH-SeqG = 0.0013 IGH-SeqH = ND | IGH-SeqA ≥ 99.9 IGH-SeqB ≥ 99.9 IGL-SeqC = 82.2 IGL-SeqD = ND IGH-SeqE = ND IGH-SeqF = ND IGH-SeqG = ND IGH-SeqH = ND | IGH-SeqA = 43.1 IGH-SeqB = 36.2 IGL-SeqC = 85.6 IGL-SeqD = ND IGH-SeqE = 68.4 IGH-SeqF = 67.4 IGH-SeqG = 68.5 IGH-SeqH = 68.5 | |||
| NGS | RNA fusion panel: positive for KMT2A-EPS15 fusion | DNA NGS panel: 1. Positive for the 3′ KMT2A deletion seen previously | Targeted testing (by DNA NGS): negative for NRAS p.G12C | |||
| DNA NGS panel: positive for 3′ KMT2A deletion | 2. New findings of trisomy 8, NRAS p.G12C, and low-level subclonal KRAS p.G13D. | |||||
| Case 2 | ||||||
| Blast % | 95.5 B-lymphoblasts | 4.5 abnormal immature monocytes | 25 B-lymphoblasts | |||
| Flow cytometry immunophenotype of major blast population | CD19 | Positive (increased) | CD19 | Negative | CD19 | Positive (increased) |
| CD22 | Positive | CD22 | Not evaluated | CD22 | Positive | |
| CD10 | Negative | CD10 | Not evaluated | CD10 | Negative | |
| CD15 | Not evaluated | CD15 | Positive | CD15 | Partial positive | |
| CD34 | Positive | CD34 | Negative | CD34 | Positive | |
| CD38 | Positive | CD38 | Positive | CD38 | Positive | |
| CD58 | Positive (increased) | CD58 | Not evaluated | CD58 | Positive (increased) | |
| HLA-DR | Positive | HLA-DR | Positive | HLA-DR | Positive | |
| CD13 | Negative | CD13 | Negative | CD13 | Negative | |
| CD33 | Negative | CD33 | Positive | CD33 | Negative | |
| CD11b | Not evaluated | CD11b | Not evaluated | CD11b | Not evaluated | |
| CD14 | Not evaluated | CD14 | Negative | CD14 | Not evaluated | |
| CD64 | Negative | CD64 | Positive | CD64 | Not evaluated | |
| CD7 | Negative | CD7 | Negative | CD7 | Not evaluated | |
| CD56 | Not evaluated | CD56 | Positive | CD56 | Negative | |
| CD45 | Positive (dim) | CD45 | Positive | CD45 | Positive | |
| CD123 | Negative | CD123 | Negative | CD123 | Not evaluated | |
| Cytogenetics | 46, XX, t(4;11)(q21;q23)[20]; | Karyotype not performed | 46, X, t(X;10)(q13;p13),t(4;11)(q21;q23),t(4;15)(p16;q22),t(8;14)(q22;q32)[15]//46,XY[5] | |||
| FISH: positive for KMT2A rearrangement in 100% of nuclei | FISH: positive for KMT2A rearrangement in 10.5% of nuclei | FISH: positive for KMT2A rearrangement in 30% of nuclei | ||||
| ClonoSEQ, % | (performed 2.5 months after diagnosis) IGH-SeqA = 4.0 IGH-SeqB = 3.8 IGH-SeqC = 0.036 | IGH-SeqA = 3.6 IGH-SeqB = 0.019 IGH-SeqC = 3.1 | IGH-SeqA = 20.9 IGH-SeqB = ND IGH-SeqC = ND | |||
| NGS | Not performed | Not performed | Not performed | |||
| . | Initial B-ALL diagnosis . | LS to AML . | Relapsed B-ALL . | |||
|---|---|---|---|---|---|---|
| Case 1 | ||||||
| Blast % | 93 B-lymphoblasts | 96 abnormal immature monocytes 0.14 B-lymphoblasts (immunophenotype similar to diagnosis) | 70 B-lymphoblasts | |||
| Flow cytometry immunophenotype of major blast population | CD19 | Positive | CD19 | Negative | CD19 | Positive |
| CD22 | Positive | CD22 | Negative | CD22 | Positive | |
| CD10 | Negative | CD10 | Negative | CD10 | Negative | |
| CD15 | Partial positive | CD15 | Positive | CD15 | Negative | |
| CD34 | Negative | CD34 | Negative | CD34 | Partial positive | |
| CD38 | Positive | CD38 | Positive | CD38 | Positive | |
| CD58 | Positive (increased) | CD58 | Positive | CD58 | Positive (increased) | |
| HLA-DR | Positive | HLA-DR | Positive | HLA-DR | Positive | |
| CD13 | Negative | CD13 | Negative | CD13/33 | Negative | |
| CD33 | Negative | CD33 | Positive | CD33 | Negative | |
| CD11b | Not evaluated | CD11b | Positive | CD11b | Not evaluated | |
| CD14 | Negative | CD14 | Partial positive | CD14 | Not evaluated | |
| CD64 | Not evaluated | CD64 | Positive | CD64 | Negative | |
| CD4 | Not evaluated | CD4 | Positive (dim) | CD4 | Not evaluated | |
| CD7 | Partial positive | CD7 | Negative | CD7 | Not evaluated | |
| CD45 | Positive (slightly dim) | CD45 | Positive | CD45 | Positive | |
| CD123 | Not evaluated | CD123 | Not evaluated | CD123 | Positive | |
| Cytogenetics | Karyotype: 46, XX, t(1;11)(p32;q23)[16]/46,XX[2] | Karyotype: 46, XX, t(1;11)(p32;q23)[5]/47,XX,t(1;11),+8[14]/46,XX[1] | Not performed | |||
| FISH: positive for a KMT2A rearrangement in 81% of nuclei (with loss of 3′ KMT2A signal) | FISH: positive for a KMT2A rearrangement in 95% of nuclei (loss of 3′ KMT2A signal) | |||||
| ClonoSEQ, % | IGH-SeqA = 60.9 IGH-SeqB = 70.0 IGL-SeqC = 76.2 IGL-SeqD = 60.9 IGH-SeqE = 0.6 IGH-SeqF = 1.8 IGH-SeqG = 0.0013 IGH-SeqH = ND | IGH-SeqA ≥ 99.9 IGH-SeqB ≥ 99.9 IGL-SeqC = 82.2 IGL-SeqD = ND IGH-SeqE = ND IGH-SeqF = ND IGH-SeqG = ND IGH-SeqH = ND | IGH-SeqA = 43.1 IGH-SeqB = 36.2 IGL-SeqC = 85.6 IGL-SeqD = ND IGH-SeqE = 68.4 IGH-SeqF = 67.4 IGH-SeqG = 68.5 IGH-SeqH = 68.5 | |||
| NGS | RNA fusion panel: positive for KMT2A-EPS15 fusion | DNA NGS panel: 1. Positive for the 3′ KMT2A deletion seen previously | Targeted testing (by DNA NGS): negative for NRAS p.G12C | |||
| DNA NGS panel: positive for 3′ KMT2A deletion | 2. New findings of trisomy 8, NRAS p.G12C, and low-level subclonal KRAS p.G13D. | |||||
| Case 2 | ||||||
| Blast % | 95.5 B-lymphoblasts | 4.5 abnormal immature monocytes | 25 B-lymphoblasts | |||
| Flow cytometry immunophenotype of major blast population | CD19 | Positive (increased) | CD19 | Negative | CD19 | Positive (increased) |
| CD22 | Positive | CD22 | Not evaluated | CD22 | Positive | |
| CD10 | Negative | CD10 | Not evaluated | CD10 | Negative | |
| CD15 | Not evaluated | CD15 | Positive | CD15 | Partial positive | |
| CD34 | Positive | CD34 | Negative | CD34 | Positive | |
| CD38 | Positive | CD38 | Positive | CD38 | Positive | |
| CD58 | Positive (increased) | CD58 | Not evaluated | CD58 | Positive (increased) | |
| HLA-DR | Positive | HLA-DR | Positive | HLA-DR | Positive | |
| CD13 | Negative | CD13 | Negative | CD13 | Negative | |
| CD33 | Negative | CD33 | Positive | CD33 | Negative | |
| CD11b | Not evaluated | CD11b | Not evaluated | CD11b | Not evaluated | |
| CD14 | Not evaluated | CD14 | Negative | CD14 | Not evaluated | |
| CD64 | Negative | CD64 | Positive | CD64 | Not evaluated | |
| CD7 | Negative | CD7 | Negative | CD7 | Not evaluated | |
| CD56 | Not evaluated | CD56 | Positive | CD56 | Negative | |
| CD45 | Positive (dim) | CD45 | Positive | CD45 | Positive | |
| CD123 | Negative | CD123 | Negative | CD123 | Not evaluated | |
| Cytogenetics | 46, XX, t(4;11)(q21;q23)[20]; | Karyotype not performed | 46, X, t(X;10)(q13;p13),t(4;11)(q21;q23),t(4;15)(p16;q22),t(8;14)(q22;q32)[15]//46,XY[5] | |||
| FISH: positive for KMT2A rearrangement in 100% of nuclei | FISH: positive for KMT2A rearrangement in 10.5% of nuclei | FISH: positive for KMT2A rearrangement in 30% of nuclei | ||||
| ClonoSEQ, % | (performed 2.5 months after diagnosis) IGH-SeqA = 4.0 IGH-SeqB = 3.8 IGH-SeqC = 0.036 | IGH-SeqA = 3.6 IGH-SeqB = 0.019 IGH-SeqC = 3.1 | IGH-SeqA = 20.9 IGH-SeqB = ND IGH-SeqC = ND | |||
| NGS | Not performed | Not performed | Not performed | |||
AML, acute myeloid leukemia; FISH, fluorescence in situ hybridization; LS, lineage switch; ND, not detected; NGS, next-generation sequencing.
Patient clinical courses. (A) Clinical course of case 1 bone marrow evaluation of leukemic blasts by MFC and by NGS of the IGH/IGL gene through clinical course, with MFC diagrams at diagnosis and at LS. (B) Clinical course of case 2 bone marrow evaluation of leukemic blasts by MFC and by NGS of the IGH/IGL gene through clinical course, with histology images at diagnosis, at LS, and at relapse aftertransplant. AML, acute myeloid leukemia; Blina, blinatumomab; DEX, dexamethasone; LOD, level of detection; PEG, pegaspargase; T-APCs, T-cell antigen-presenting cells; Tx, treatment; SSc, side scatter; VCR, vincristine.
Patient clinical courses. (A) Clinical course of case 1 bone marrow evaluation of leukemic blasts by MFC and by NGS of the IGH/IGL gene through clinical course, with MFC diagrams at diagnosis and at LS. (B) Clinical course of case 2 bone marrow evaluation of leukemic blasts by MFC and by NGS of the IGH/IGL gene through clinical course, with histology images at diagnosis, at LS, and at relapse aftertransplant. AML, acute myeloid leukemia; Blina, blinatumomab; DEX, dexamethasone; LOD, level of detection; PEG, pegaspargase; T-APCs, T-cell antigen-presenting cells; Tx, treatment; SSc, side scatter; VCR, vincristine.
Case 2
A 4-month-old female diagnosed with KMT2A-r infant B-ALL had persistent MFC-positive MRD and CNS2 status after AALL15P1 induction and modified consolidation therapy. She was enrolled on PLAT-05 and received SCRI-CAR19×22. She achieved an MFC-negative CR with persistent NGS-MRD by day 21 after infusion. In the setting of ongoing CAR T-cell persistence 5 months after infusion, she experienced an LS (Table 1). Salvage therapy with FLA + GO resulted in an MFC-negative CR with persistent NGS-MRD, which was consolidated with a cord blood HCT. Relapse of CD19+ B-ALL was noted 5 months after HCT. She was initiated on blinatumomab and achieved an MFC-negative CR after the first cycle and NGS-negative CR after the second cycle. The patient remained in an NGS-negative CR after 6 courses of blinatumomab and transitioned to maintenance chemotherapy (Figure 1B).
Discussion
We present 2 patients with KMT2A-r infant B-ALL with persistent disease after upfront treatment. Owing to dismal outcomes associated with persistent disease, experimental immunotherapies were pursued.12 Both patients experienced LS after CD19-directed immunotherapy, which was successfully salvaged with intensive myeloid-directed therapy and HCT. In each case, patients experienced recurrent CD19+ B-ALL after clearance of their myeloid disease. Remarkably, both were successfully rechallenged with CD19-directed immunotherapy and have not experienced recurrent disease despite prolonged immunotherapeutic pressure.
LS has been previously reported in patients after cytotoxic chemotherapy and/or HCT,13,14 but the frequency has increased with the broader use of immunotherapy.4-8 The incidence of LS ranges anywhere from 1% to 3% after CD19-CAR but is higher in patients with a KMT2A-r, including those with infant ALL.15,16 LS has also been well documented after treatment with blinatumomab.6,17-19
The prognosis in patients with KMT2A-r that experience an event after CD19-directed immunotherapy, including LS, is dismal.15,16 The reason for these poor outcomes is multifactorial and includes the heavy pretreatment, including HCT, patients have often received before CD19-directed immunotherapies. These exposures limit further cytotoxic options because of disease resistance and cumulative toxicity. Furthermore, the phenotypic switch shifts the antigens available for effective immunotherapeutic targeting. Despite this, if patients can tolerate myeloid-directed therapy, there is potential therapeutic benefit with subsequent HCT. In our 2 cases, the LS event occurred after minimal disease–directed therapy, which may have contributed to the higher tolerance and favorable responses to myeloid-directed therapy and subsequent HCT, consistent with previous reports.4
We also report on the unique phenomenon of recurrent B-ALL after successful treatment of the LS. Although spontaneous reversion after cessation of blinatumomab has been previously described, we are unaware of any patient being successfully rechallenged with CD19-directed immunotherapy.6 Our patients’ B-ALL recurred after myeloid-directed therapy or HCT and demonstrated sustained responses to CD19-directed immunotherapy without early recurrence of LS. Both of our cases had very short intervals between the initial CD19-directed immunotherapy and LS and, although late LS events have been described, their extended follow-up without LS is encouraging.
The mechanism behind LS is not completely understood and may vary based on disease characteristics. One proposed mechanism involves disease plasticity in which a single genotype results in a fluid phenotype informed by therapeutic pressures. Alternatively, a nonlineage committed progenitor cell harboring the leukemogenic alteration may exist, which can propagate a heterogeneous disease similarly dependent on therapeutic context.20 Irrespective of the mechanism, it is compelling to speculate that a patient already predisposed to LS is destined to experience recurrent LS with repetitive immunotherapy. Our report challenges this concern, however, whether the responsible clone was eliminated because of intensive myeloid-directed chemotherapy or is more sensitive to the graft-versus-leukemia effect of HCT remains to be elucidated. In addition, it may be continued graft-versus-leukemia surveillance that extinguishes any subsequent LS events that may occur, whereas the ongoing CD19-directed immunotherapy controls the lymphoid component.
This report confirms that LS may be a salvageable entity but requires a combined modality approach. Patients can achieve elimination of their myeloid disease after intensive myeloid-directed therapy coupled with HCT but remain at risk for leukemia relapse. Those who experience relapse restricted to their lymphoid disease could be successfully rechallenged with CD19-directed immunotherapy with durable remissions. Prospective studies exploring manners to prevent and treat LS are needed.
Acknowledgments: B.M.L. is supported by a Fellow Grant from the Rally Foundation for Childhood Cancer Research. K.J.L. is supported by a Leukemia Lymphoma Society Career Development Award.
Contribution: B.M.L., C.A., and A.J.L. curated clinical data and wrote the manuscript; K.J.L., K.M.C., and S.D.B. curated clinical data, created the figure, and read and approved the final manuscript; and K.J.L., C.S., K.M.C., and R.G. read, edited, and approved the final manuscript.
Conflict-of-interest disclosure: R.G. receives royalty payments from Juno Therapeutics for licensed IP and has received honorarium for advisory roles from Novartis, Crispr Therapeutics and Sobi. The remaining authors declare no competing financial interests.
Correspondence: Brittany M. Lee, Seattle Children's Research Institute, M/S CURE-4, PO Box 5371, Seattle, WA 98145-5005; e-mail: brittany.lee@seattlechildrens.org.
References
Author notes
Data are available on request from the corresponding author, Brittany M. Lee (brittany.lee@seattlechildrens.org).
The full-text version of this article contains a data supplement.