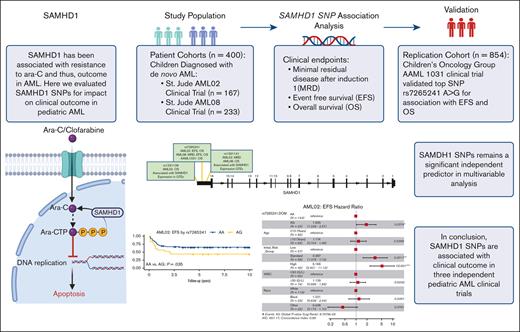
Key Points
SNPs within SAMHD1 are predictive of clinical outcome in 3 independent cohorts of children diagnosed with AML.
Future in-depth functional evaluation and validation in larger cohorts are needed to establish its prognostic relevance in AML treatment.
Abstract
Cytarabine arabinoside (Ara-C) has been the cornerstone of acute myeloid leukemia (AML) chemotherapy for decades. After cellular uptake, it is phosphorylated into its active triphosphate form (Ara-CTP), which primarily exerts its cytotoxic effects by inhibiting DNA synthesis in proliferating cells. Interpatient variation in the enzymes involved in the Ara-C metabolic pathway has been shown to affect intracellular abundance of Ara-CTP and, thus, its therapeutic benefit. Recently, SAMHD1 (SAM and HD domain–containing deoxynucleoside triphosphate triphosphohydrolase 1) has emerged to play a role in Ara-CTP inactivation, development of drug resistance, and, consequently, clinical response in AML. Despite this, the impact of genetic variations in SAMHD1 on outcome in AML has not been investigated in depth. In this study, we evaluated 25 single nucleotide polymorphisms (SNPs) within the SAMHD1 gene for association with clinical outcome in 400 pediatric patients with newly diagnosed AML from 2 clinical trials, AML02 and AML08. Three SNPs, rs1291128, rs1291141, and rs7265241 located in the 3′ region of SAMHD1 were significantly associated with at least 1 clinical outcome: minimal residual disease after induction I, event-free survival (EFS), or overall survival (OS) in the 2 cohorts. In an independent cohort of patients from the COG-AAML1031 trial (n = 854), rs7265241 A>G remained significantly associated with EFS and OS. In multivariable analysis, all the SNPs remained independent predictors of clinical outcome. These results highlight the relevance of the SAMHD1 pharmacogenomics in context of response to Ara-C in AML and warrants the need for further validation in expanded patient cohorts.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease characterized by the rapid progression of immature leukemic blasts interfering with normal blood cell production. The overall outcome for AML is dismal, with resistance, disease relapse, and toxicity being the most challenging obstacles in treating patients with AML. Although initial induction chemotherapy regimens induce remission in the majority of pediatric patients with AML, ∼30% of these patients will succumb to the disease as a result of relapse and refractory AML.1,2 One of the most important drugs in the treatment of AML is cytarabine arabinoside (Ara-C), which has been the mainstay of chemotherapy treatment for >50 years.3 Despite the widespread use of Ara-C in AML, considerable interpatient variation in clinical response, ideal dose, and tolerability underscores the need to optimize treatment strategies for childhood AML.
Ara-C is a prodrug that requires activation to ara-cytosine triphosphate (Ara-CTP) by a series of enzymes to exert its antileukemic effect4 (Figure 1A). Drug influx transporters SLC29A1 and SLC28A3 facilitate uptake of Ara-C by cells. Within the cells, the abundance of Ara-CTP is influenced by activating enzymes such as deoxycytidine kinase, nucleoside diphosphate kinase, ribonucleotide reductase, and inactivating enzymes such as cytidine deaminase, cytosolic 5′-nucleotidase II,5-7 and, more recently, SAMHD1.8 SAMHD1 is a deoxynucleoside triphosphate (dNTP) triphosphohydrolase that converts dNTPs into deoxyribonucleosides and inorganic triphosphate.9 Several studies establish SAMHD1 as an integral regulator of intracellular concentrations of dNTP pools in mammalian cells.9,10 Regulation of DNA precursors pools is important for DNA repair and replication; thus, nucleotide imbalance can lead to genomic instability and impaired cellular processes.11,12 SAMHD1 has been extensively studied in the setting of HIV-1 in which it has been shown to block viral replication in dendritic and myeloid cells.13 Mutations in SAMHD1 are associated with an inherited encephalopathy that affects newborn infants.14 In cancer, mutations in SAMHD1 have been reported in a myriad of cancer types and was shown to be recurrently mutated in AML and chronic lymphocytic leukemia.12
SAMHD1 summary. (A) Metabolic pathway of nucleoside analogs. (B) SAMHD1 gene schematic with top 3 SNPs. CDA, cytidine deaminase; CMPK1, cytidine monophosphate kinase 1; DCK, deoxycytidine kinase; DCTD, deoxycytidylate deaminase; hENT1, human equilibrative nucleoside transporter (hENT1/SLC29A1); NDPK, nucleoside diphosphate kinases; NT5C2, 5′nucleotidase; RRM1/RRM2, ribonucleotide reductase catalytic subunit M1/M2; SAMHD1, SAM domain and HD domain 1; GTEx, Genotype-Tissue Expression project; WT, wild type.
SAMHD1 summary. (A) Metabolic pathway of nucleoside analogs. (B) SAMHD1 gene schematic with top 3 SNPs. CDA, cytidine deaminase; CMPK1, cytidine monophosphate kinase 1; DCK, deoxycytidine kinase; DCTD, deoxycytidylate deaminase; hENT1, human equilibrative nucleoside transporter (hENT1/SLC29A1); NDPK, nucleoside diphosphate kinases; NT5C2, 5′nucleotidase; RRM1/RRM2, ribonucleotide reductase catalytic subunit M1/M2; SAMHD1, SAM domain and HD domain 1; GTEx, Genotype-Tissue Expression project; WT, wild type.
In 2017, 2 reports in Nature Medicine implicated SAMHD1 in Ara-CTP inactivation. The first report by Schneider et al,8 showed loss of SAMHD1 to enhance chemosensitivity to Ara-C in cell lines and mouse models. SAMHD1 expression by immunohistochemistry staining was reportedly highly variable and was associated with complete remission (CR) status after end of induction therapy, and event-free survival (EFS). Moreover, SAMHD1 messenger RNA (mRNA) expression was inversely associated with CR in publicly available The Cancer Genome Atlas (TCGA) database. The second report in the same year by Herold et al,15 also demonstrated the role of SAMHD1 in Ara-C sensitivity, as well as association with expression and response to Ara-C therapy using TCGA data from adult patients with AML and pediatric AML data from the Therapeutically Applicable Research To Generate Effective Treatments (TARGET) cohort, and concluded that SAMHD1 was a risk factor in patients that received Ara-C treatment diagnosed with de novo AML. Of note, in this report no significant difference was observed in the 10-year OS based on SAMHD1 expression in both cohorts; however, OS at 18 months in adults and 12 months in pediatric patients with AML differed significantly by SAMHD1 expression (based on the cut-off used to classify patients with high or low SAMHD1 expression). In another study by Rassidakis et al,16 it was reported that low SAMHD1 expression was significantly associated with OS and EFS in a subset of patients receiving high-dose Ara-C–based chemotherapeutic regimens during consolidation. In addition to Ara-C inactivation, computational modeling established other nucleoside analogs, such as clofarabine, fludarabine, and gemcitabine to be substrates of SAMHD1.17 As a result of these previous studies, SAMHD1 may present a barrier in treatment with various antimetabolite-based therapies.18
Previous studies that have focused on SAMHD1 expression levels highlight its potential as a promising prognostic biomarker, thus warranting further investigation with respect to the influence of genetic variation within SAMHD1 and its impact on clinical outcome in AML. To the best of our knowledge, there is only 1 report in literature in which Zhu et al19 evaluated 3 SAMHD1 expression quantitative trait loci single nucleotide polymorphisms (SNPs) selected from the GTEx database and showed a statistically nonsignificant trend between rs6102991 and reduced risk of non-CR in adult AML. In this study, we sought to investigate SNPs at the SAMHD1 locus for their association with clinical outcome end points in pediatric patients with newly diagnosed AML enrolled in 2 multisite clinical trial cohorts, with replication of the top SNP signals in a third clinical trial cohort.
Methods
Patient population
Cohort I
Multicenter AML02 clinical trial (clinicaltrials.gov, identifier #NCT00136084). The AML02 trial enrolled 232 pediatric and young adult patients with AML (aged 0.01-21.4 years), randomized to 2 treatment arms. Patients in arm A received high-dose cytarabine (3 g/m2, given every 12 hours on days 1, 3, and 5) along with daunorubicin and etoposide as initial chemotherapy with subsequent regimens optimized based on response and risk classification. Arm B followed the same regimen as arm A except patients received low-dose cytarabine (100 mg/m2 given every 12 hours on days 1-10). Details of the study design, eligibility, and clinical outcomes have previously been reported and showed no difference in clinical outcome end points by treatment arms.20 As per study protocol, initial risk assignment was as follows: patients with t(8;21), inv(16), or t(9;11) were classified as low-risk AML, whereas high-risk AML classification included presence of −7 or FLT3-internal tandem duplication (ITD) mutations, t[6;9], acute megakaryoblastic leukemia, treatment-related AML, or AML arising because of myelodysplastic syndrome. Standard risk was classified as the absence of low- or high-risk features. In this investigation, 167 patients with both clinical and genotype data were included.
Cohort II
Multicenter AML08 clinical trial (clinicaltrials.gov, identifier #NCT00703820). The AML08 trial enrolled 285 pediatric and young adult patients with AML (aged 0.03-19.9 years) randomized to 2 treatment arms. Patients in arm A were randomized to receive clofarabine and cytarabine (Clo/Ara-C; clofarabine 52 mg/m2 per day on days 1-5 and cytarabine 1 g/m2 per day on days 1-5), whereas patients in arm B received high-dose cytarabine, daunorubicin, and etoposide (HD-ADE; cytarabine 3 g/m2 given every 12 hours on days 1, 3, and 5; daunorubicin 50 mg/m2 on days 2, 4, and 6; and etoposide 100 mg/m2 per day on days 2-6) as induction I. Subsequent chemotherapeutic regimens were based on presenting features and minimal residual disease (MRD) evaluation and included low-dose ADE given alone or combined with sorafenib or vorinostat. Consolidation therapy was composed of 2 or 3 additional courses of chemotherapy or hematopoietic cell transplantation. Details of the study design, eligibility, and clinical outcomes were reported previously and showed no difference in EFS or OS across the 2 treatment arms, although day-22 MRD was slightly better in the HD-ADE arm.21 Patients with t(8;21) or inv(16) who were classified as being at low risk and high risk included t(6;9), t(8;16), t(16;21), −7, −5, or 5q-; French-American-British classification M0 or M6; French-American-British M7 without t(1;22)(p13;q13); treatment-related AML; and FLT3-ITD. Standard risk was classified as the absence of low- or high-risk features. This study included 233 patients with both clinical and genotype data.
Cohort III
Multicenter AAML1031 clinical trial (clinicaltrials.gov, identifier #NCT01371981). The AAML1031 trial led by the Children’s Oncology group enrolled patients with newly diagnosed AML aged from 0 to 29.5 years with previously untreated AML. Details of the study design, treatment arms, eligibility, and clinical outcome has been reported previously.22 The AAML1031 clinical trial consisted of 2 treatment arms for patients with non-FLT3-ITD AML, with each arm receiving the same cytarabine-based induction-I chemotherapy backbone with 4 courses of chemotherapy or 3 courses followed by allogeneic hematopoietic stem cell transplant in patients at high risk. Patients in arm A were randomized to receive standard therapy, which consisted of ADE 10 + 3 + 5 (cytarabine 100 mg/m2 per dose every 12 hours on days 1-10 plus daunorubicin 50 mg/m2 per dose on days 1, 3, and 5 plus etoposide 100 mg/m2 per dose on days 1-5) for induction-I chemotherapy cycle. Patients in the experimental arm received standard chemotherapy treatment plus bortezomib 1.3 mg/m2 per dose given on days 1, 4, and 8 of induction-I chemotherapy cycle. Subsequent chemotherapeutic cycles were determined by the patient’s risk classification based on cytogenetics, molecular markers, and MRD after induction-I. MRD detection was conducted using a “different from normal” algorithm as previously reported.22 It should be noted that for this trial, all patients with FLT3-ITD were treated with addition of sorafenib on a different arm and were not included in this study. As with the other 2 trials, no difference in clinical outcome end points by the 2 treatment arms for AAML1031. In total, 854 patients with both clinical and genotype data were included as a validation cohort in this study.
Clinical outcome end points
Flow cytometric studies were used to assess MRD after induction I in clinical trial cohorts. For AML02 and AML08, MRD positivity was defined as ≥1 leukemic cells per 1000 mononuclear bone marrow cells (ie, ≥0.1%). EFS was defined as the time from study enrollment to induction failure, relapse, secondary malignancy, death, or study withdrawal for any reason, with event-free patients censored on the date of last follow-up. OS was defined as the time from study enrollment to death, with living patients censored on the date of last follow-up. Appropriate Institutional Review Board approvals were obtained, informed consent was obtained from parents/guardians, and consents/assents from the individuals in accordance with the approved clinical trial protocols.
Genotyping and quality control (QC)
Genomic DNA from AML02 and AML08 were genotyped using the Illumina Omni 2.5M Exome Beadchip (Illumina, San Diego, CA) at the Hussman Institute for Human Genetics, University of Miami, Miami, FL, USA. Genotype calling was performed using Illumina’s Genome Studio software version 2011.1 (Illumina). QC procedural steps were performed using PLINK 1.9 software to obtain a high-quality data set for use in statistical analysis (PLINK, RRID:SCR_001757). Initially, 58 SNPs within ±10 kb of the SAMHD1 locus (chr20:35,510,227-35,590,246) were obtained from Illumina Omni 2.5M genotype data files consistent with Genome Reference Consortium Human Build 37 (Genome Reference Consortium, RRID:SCR_006553). SNPs with a call rate of <95% and minor allele frequency of <5% were excluded, resulting in 25 variants for subsequent analysis (supplemental Table 1). Sample QC consisted of exclusion of samples with <95% of SNP calls failing, diagnosis other than de novo AML, and samples with mismatch between genetic and reported sex. Hardy Weinberg Equilibrium was assessed for each SNP. Linkage disequilibrium (LD) was assessed using D′ and r2 values. After SNP and sample QC steps, 167 and 233 samples from patients with AML02 and AML08, respectively, were included for final analysis. For the AAML1031 cohort, genotype for SAMHD1 SNPs were obtained from next-generation sequencing data.
Gene expression profiling
The mRNA expression levels obtained from leukemic blasts at diagnosis from patients in the AML02 cohort was performed using GeneChip Human Genome U133A (Affymetrix, Santa Clara, CA) and processed as described previously.23 All gene expression levels were natural-log transformed before analysis. For the AML08 and AAML1031 cohort, SAMHD1 expression was obtained from RNA sequencing (RNA-seq) data. AAML1031 RNA-seq data are also available at the TARGET database (https://target-data.nci.nih.gov/Public/AML/mRNA-seq/L3/expression/). RPKM values were used for association analysis after log transformation.
Statistical analysis
Genotype deviations from Hardy Weinberg Equilibrium were assessed using an exact test. Wilcoxon rank-sum or Kruskal-Wallis tests were used for continuous variable comparisons between/among patient subgroups. χ2 or Fisher exact tests were used for testing association between categorical variables. The MRD positivity after induction I was modeled in logistic regression models with adjustment and without adjustment for initial risk assignment of the patient determined at diagnosis (ie, low, standard, and high risk). Of note, for the AAML1031 trial, protocol risk group assignment included MRD at end of induction I. Thus, for purposes of this analysis, diagnostic cytogenetic and molecular features were used to classify patients into risk groups using standard established criteria. Survival analyses were performed using survival and survminer packages in R4.1.0 (survival, RRID:SCR_021137; survminer, RRID:SCR_021094). Survival probabilities were estimated via the Kaplan Meier method. Cox proportional hazard models were used to examine the association of the SNPs with EFS and OS and calculated for each of the 3 modes of inheritance. Cox regression models were performed with or without adjusting for provisional risk group assignment at time of diagnosis for all 3 clinical trial cohorts. Significance levels for association of SNP with clinical outcome were set at P < .05. Multivariable Cox proportional hazard models were used for association with EFS and OS that included SNP genotype groups, risk group assignment, race, white blood cell count at diagnosis, and age as other study covariates. The 95% confidence interval (CI) of hazard ratios (HR) was calculated to quantitatively measure the effect on clinical outcome. All statistical analyses were performed using the statistical computing environment R (R Project for Statistical Computing, RRID:SCR_001905).
Results
The 167 pediatric and young adult patients with AML treated on the AML02 trial and the 233 patients treated on the AML08 clinical trial were included in this study. Of note, for 9 AML02 and 19 AML08 patients, data on MRD after induction I were not available. The 854 pediatric patients treated with AAML1031 were used as a validation cohort, of which 40 did not have postinduction MRD data and were excluded from MRD association analysis. For all the clinical cohorts tested, no difference in the outcome was observed across treatment arms as has been reported in the previous publications reporting results of the trial outcomes. The patient characteristics and demographic features for all cohorts are summarized in Table 1.
Patient characteristics across three clinical cohorts
| . | AML02 (N = 167) . | AML08 (N = 233) . | AAML1031 (N = 854) . | P value . |
|---|---|---|---|---|
| Feature | ||||
| Age at diagnosis, y | 10.2 (0.01-21.4) | 9.5 (0.01-19.9) | 8.8 (0.0-29.5) | .778 |
| Sex | .661 | |||
| Male | 94/167 (56.3%) | 123/233 (52.8%) | 448/854 (52.5%) | |
| Female | 73/167 (43.7%) | 110/233 (47.2%) | 406/854 (47.5%) | |
| Race | .031 | |||
| White | 116/167 (69.5%) | 166/233 (70.8%) | 608/854 (71.2%) | |
| Black | 29/167 (17.4%) | 41/233 (17.6%) | 102/854 (11.9%) | |
| Other | 22/167 (11.9%) | 26/233 (11.2%) | 144/854 (16.9%) | |
| Treatment arm | .427 | |||
| Arm A | 88/167 (52.7%) | 108/233 (52.4%) | 409/854 (47.9%) | |
| Arm B | 79/167 (47.3%) | 125/233 (47.6%) | 445/854 (52.1%) | |
| WBC (G/L) | 20 (0-514) | 26 (1-890) | 20 (1-713) | .824 |
| Cytogenetic risk group | <.00001 | |||
| Low | 57/167 (34.1%) | 57/233 (24.0%) | 316/854 (37%) | |
| Standard | 68/167 (40.7%) | 134/233 (57.9%) | 500/854 (58.5%) | |
| High | 42/167 (25.2%) | 42/233 (18.0%) | 25/854 (3%) | |
| Unknown | NA | NA | 13/854 (1.5%) |
| . | AML02 (N = 167) . | AML08 (N = 233) . | AAML1031 (N = 854) . | P value . |
|---|---|---|---|---|
| Feature | ||||
| Age at diagnosis, y | 10.2 (0.01-21.4) | 9.5 (0.01-19.9) | 8.8 (0.0-29.5) | .778 |
| Sex | .661 | |||
| Male | 94/167 (56.3%) | 123/233 (52.8%) | 448/854 (52.5%) | |
| Female | 73/167 (43.7%) | 110/233 (47.2%) | 406/854 (47.5%) | |
| Race | .031 | |||
| White | 116/167 (69.5%) | 166/233 (70.8%) | 608/854 (71.2%) | |
| Black | 29/167 (17.4%) | 41/233 (17.6%) | 102/854 (11.9%) | |
| Other | 22/167 (11.9%) | 26/233 (11.2%) | 144/854 (16.9%) | |
| Treatment arm | .427 | |||
| Arm A | 88/167 (52.7%) | 108/233 (52.4%) | 409/854 (47.9%) | |
| Arm B | 79/167 (47.3%) | 125/233 (47.6%) | 445/854 (52.1%) | |
| WBC (G/L) | 20 (0-514) | 26 (1-890) | 20 (1-713) | .824 |
| Cytogenetic risk group | <.00001 | |||
| Low | 57/167 (34.1%) | 57/233 (24.0%) | 316/854 (37%) | |
| Standard | 68/167 (40.7%) | 134/233 (57.9%) | 500/854 (58.5%) | |
| High | 42/167 (25.2%) | 42/233 (18.0%) | 25/854 (3%) | |
| Unknown | NA | NA | 13/854 (1.5%) |
Median (range); n/N (%). Arm A, AML02 received low dose cytarabine: LDAC/ AML08 received clofarabine + Ara-C/COG; AAML1031 received standard ADE; Arm B, AML02 and AML08 received high-dose cytarabine (HDAC)/COG; AAML1031 received standard ADE + bortezomib. For AAML1031 cohort only patients from treatment arms A and B were included in this study, which might contribute to lower number in high-risk group category due to exclusion of FLT3-ITD AML.
A, Ara-C or cytarabine; D, daunorubicin; E, etoposide.
Evaluation of SAMHD1 SNPs with clinical outcome in AML
Of the 25 SAMHD1 SNPs evaluated, 3 SNPs in the 3′ region of SAMHD1 (rs7265241, rs1291141, and rs1291128) were significantly associated with clinical outcome end points in the AML02 and AML08 cohorts, as summarized in Table 2. Table 2 includes survival analysis results for the top 3 SNPs in our study by cohort and mode of inheritance, with and without adjustment for initial risk group at diagnosis. Among these SNPs, rs1291128 is located in the downstream TLDC2 gene but occurred in LD with multiple SNPs on the SAMHD1 locus. Of these 3 SNPs, genotype data for only rs7265241 were available in 849 patients from the AAML1031 trial. Details on the association analysis in each cohort are described hereafter and Figure 1B shows the SNP map for these SNPs and highlights a few features.
Significant association with SNP and survival outcome across cohorts
| SAMHD1 SNPs . | EFS . | OS . | ||
|---|---|---|---|---|
| HR (95% CI) . | P value . | HR (95% CI) . | P value . | |
| rs7265241 A>G | AML02 (n = 167) | |||
| Unadjusted analysis† | 1.926 (1.046-3.547) | .035 | 2.410 (1.223-4.771) | .012 |
| Risk-group adjusted analysis† | 1.860 (1.010-3.428) | .046 | 2.235 (1.216-4.779) | .021 |
| AML08 (n = 233) | ||||
| Unadjusted analysis† | 1.264 (0.952-1.678) | .104 | 1.439 (1.043-1.986) | .027 |
| Risk-group adjusted analysis† | 1.823 (1.061-3.131) | .030 | 1.532 (1.042-1.987) | .010 |
| AAML1031 (n = 849) | ||||
| Unadjusted analysis‡ | 2.722 (1.538-4.816) | .001 | 2.214 (1.253-3.912) | .006 |
| Risk-group adjusted analysis‡ | 2.581 (1.458-4.567) | .001 | 1.202 (1.253-3.753) | .009 |
| rs1291141 T>G | AML02 (n = 167) | |||
| Unadjusted analysis∗ | 1.204 (0.817-1.775) | .348 | 1.221 (0.772-1.933) | .393 |
| Risk-group adjusted analysis∗ | 1.309 (0.881-1.947) | .183 | 1.396 (0.761-1.959) | .166 |
| AML08 (n = 233) | ||||
| Unadjusted analysis‡ | 1.247 (0.911-1.707) | .167 | 1.548 (1.122-2.138) | .008 |
| Risk-group adjusted analysis‡ | 1.160 (0.847-1.589) | .355 | 1.427 (1.122-2.139) | .031 |
| rs1291128 T>C | AML02 (n = 163) | |||
| Unadjusted analysis† | 0.932 (0.699-1.240) | .628 | 0.778 (0.564-1.073) | .126 |
| Risk-group adjusted analysis† | 0.859 (0.644-1.146) | .300 | 0.716 (0.563-1.075) | .043 |
| SAMHD1 SNPs . | EFS . | OS . | ||
|---|---|---|---|---|
| HR (95% CI) . | P value . | HR (95% CI) . | P value . | |
| rs7265241 A>G | AML02 (n = 167) | |||
| Unadjusted analysis† | 1.926 (1.046-3.547) | .035 | 2.410 (1.223-4.771) | .012 |
| Risk-group adjusted analysis† | 1.860 (1.010-3.428) | .046 | 2.235 (1.216-4.779) | .021 |
| AML08 (n = 233) | ||||
| Unadjusted analysis† | 1.264 (0.952-1.678) | .104 | 1.439 (1.043-1.986) | .027 |
| Risk-group adjusted analysis† | 1.823 (1.061-3.131) | .030 | 1.532 (1.042-1.987) | .010 |
| AAML1031 (n = 849) | ||||
| Unadjusted analysis‡ | 2.722 (1.538-4.816) | .001 | 2.214 (1.253-3.912) | .006 |
| Risk-group adjusted analysis‡ | 2.581 (1.458-4.567) | .001 | 1.202 (1.253-3.753) | .009 |
| rs1291141 T>G | AML02 (n = 167) | |||
| Unadjusted analysis∗ | 1.204 (0.817-1.775) | .348 | 1.221 (0.772-1.933) | .393 |
| Risk-group adjusted analysis∗ | 1.309 (0.881-1.947) | .183 | 1.396 (0.761-1.959) | .166 |
| AML08 (n = 233) | ||||
| Unadjusted analysis‡ | 1.247 (0.911-1.707) | .167 | 1.548 (1.122-2.138) | .008 |
| Risk-group adjusted analysis‡ | 1.160 (0.847-1.589) | .355 | 1.427 (1.122-2.139) | .031 |
| rs1291128 T>C | AML02 (n = 163) | |||
| Unadjusted analysis† | 0.932 (0.699-1.240) | .628 | 0.778 (0.564-1.073) | .126 |
| Risk-group adjusted analysis† | 0.859 (0.644-1.146) | .300 | 0.716 (0.563-1.075) | .043 |
MOI, mode of inheritance; MAF, minor allele frequency.
additive
dominant
recessive
rs7265241
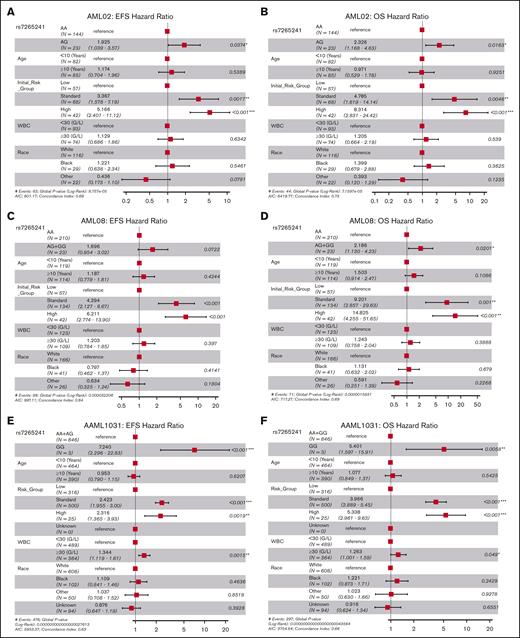
For this SNP, presence of the variant G allele of rs7265241 was associated with lower EFS (unadjusted HR, 1.93; 95% CI, 1.05-3.55; P = .035 and risk-group adjusted HR, 1.86; 95% CI, 1.01-3.42; P = .046, Figure 2A) and lower OS (unadjusted HR, 2.41; 95% CI, 1.22-4.77; P = .012 and risk-group adjusted HR, 2.24; 95% CI, 1.22-4.78; P = .021, Figure 2B) in the AML02 cohort. Within the AML08 cohort, we observed consistent association with the presence of variant G allele being associated with lower EFS (risk-adjusted HR, 1.82; 95% CI, 1.06-3.13; P = .030, Figure 2C) and with inferior OS (unadjusted HR, 1.44; 95% CI, 1.04-1.99; P = .027, and risk-group adjusted HR, 1.53; 95% CI, 1.04-1.99; P = .010, Figure 2D). Within the AAML1031 cohort, presence of the variant G allele was associated with lower EFS (unadjusted HR, 2.72; 95% CI, 1.54-4.82); P = .001 and risk-group adjusted HR, 2.58; 95% CI, 1.46-4.57; P = .001, Figure 2E) and lower OS (unadjusted HR, 2.21; 95% CI, 1.25-3.91; P = .006 and risk-group adjusted HR, 2.12; 95% CI, 1.20-3.75; P = .009, Figure 2F). With respect to association with MRD1, presence of the variant G allele was associated with MRD positivity after induction-I chemotherapy in the AML08 trial (unadjusted OR, 2.06; 95% CI, 1.25-3.39; P = .005; risk-adjusted OR, 2.74; 95% CI, 1.54-4.68; P = .0004, Figure 2G), but a similar association in AML02 or AAML1031 cohorts was not observed for MRD1. In a multivariable analysis of OS, adjusting for age, risk group, race, white blood cell count, SNP rs7265241 remained as an independent predictor across all 3 cohorts (AML02, P = .0163; AML08, P = .020; and AAML1031, P = .006), for EFS it retained its significance in AML02 (P = .037) and AAML1031 (P < .001) but not in AML08 (P = .0722) (Figure 3).
Association of rs7265241 and clinical outcome end points in 3 independent cohorts. Kaplan-Meier curves showing association of rs7265241 genotypes with EFS (A: AML02 trial, C: AML08 trial, and E: AAML1031 trial), OS (B: AML02 trial, D: AML08 trial, and F: AAML1031 trial), and MRD (G: AML08 trial).
Association of rs7265241 and clinical outcome end points in 3 independent cohorts. Kaplan-Meier curves showing association of rs7265241 genotypes with EFS (A: AML02 trial, C: AML08 trial, and E: AAML1031 trial), OS (B: AML02 trial, D: AML08 trial, and F: AAML1031 trial), and MRD (G: AML08 trial).
Forest plots of multivariable Cox proportional hazard models for rs7265241. Model for rs7265241 with EFS and OS in AML02 (A-B), AML08 (C-D), and AAML1031 (E-F). All models included age, risk-group assignment, white blood cell count (WBC), and race. HR are shown within the plots with their respective 95% CIs and P values listed.
Forest plots of multivariable Cox proportional hazard models for rs7265241. Model for rs7265241 with EFS and OS in AML02 (A-B), AML08 (C-D), and AAML1031 (E-F). All models included age, risk-group assignment, white blood cell count (WBC), and race. HR are shown within the plots with their respective 95% CIs and P values listed.
rs1291141
For rs1291141 T>G SNP, the reference allele is not the major allele in our patient population, which is consistent with larger reference genome panels such as HapMap and TopMed.24,25 Genotype data for this SNP were not available in AAML1031. Patients homozygous for the variant allele (TT genotype) had significantly poor OS (TT vs GT + GG; unadjusted HR, 1.55; 95% CI, 1.12-2.14; P = .008; risk-group adjusted HR, 1.43; 95% CI, 1.12-2.14; P = .031, Figure 4A) in the AML08 cohort but not in the AML02 cohort (Table 2). Instead, we observed significant association with higher MRD positivity in homozygous TT genotype as compared with other genotype groups (TT vs GT + GG, risk-group adjusted OR, 2.12; 95% CI, 1.15-3.92; P = .017, Figure 4B) in the AML02 cohort but not in the AML08 cohort. In multivariable analysis adjusting for age, risk group, race, and white blood cell count, SNP rs1291141 remained as an independent predictor for OS in the AML02 and AML08 cohorts (Figure 4C-D). SNP rs1291141 occurs in LD with 27 additional SNPs on the SAMHD1 locus (supplemental Table 2).
Association of rs1291141 with clinical outcome. Kaplan-Meier overall survival curve in AML08 (A). Association with MRD1 in AML02 (B). Forest plots of multivariable Cox proportional hazard models that include age, risk-group assignment, WBC, and race for OS in patients treated on AML02 (C) and AML08 (D).
Association of rs1291141 with clinical outcome. Kaplan-Meier overall survival curve in AML08 (A). Association with MRD1 in AML02 (B). Forest plots of multivariable Cox proportional hazard models that include age, risk-group assignment, WBC, and race for OS in patients treated on AML02 (C) and AML08 (D).
rs1291128
The third SNP, rs1291128 T>C SNP was evaluated in the AML02 and AML08 cohorts and was associated with improved OS in AML02 (risk-group adjusted HR, 0.716; 95% CI, 0.563-1.075; P = .043, Figure 5A) but no significant association was observed in the AML08 cohort. SNP rs1291128 occurs in LD with 11 additional SNPs on SAMHD1 locus (supplemental Table 2). In multivariable analysis, SNP rs1291128 remained as an independent predictor for OS in AML02 (Figure 5B).
Association of rs1291128 and rs4812641 with clinical outcome. Kaplan-Meier survival plot for rs1291128 with OS in AML02 (A) and multivariable forest plot for OS in patients treated on AML02 (B). Association of rs4812641 with MRD1 (C), OS (D) and the corresponding multivariable forest plot (E) in AML02. Forest plots are of multivariable Cox proportional hazard models that includes age, risk-group assignment, WBC, and race.
Association of rs1291128 and rs4812641 with clinical outcome. Kaplan-Meier survival plot for rs1291128 with OS in AML02 (A) and multivariable forest plot for OS in patients treated on AML02 (B). Association of rs4812641 with MRD1 (C), OS (D) and the corresponding multivariable forest plot (E) in AML02. Forest plots are of multivariable Cox proportional hazard models that includes age, risk-group assignment, WBC, and race.
rs6102991
A previous study in adult AML reported rs6102991 A>G to be associated with reduced risk of non-CR in adult AML, although it did not reach statistical significance19. We sought to investigate this SNP in our pediatric AML cohorts for association with clinical outcome, however the rs6102991 SNP is not included on the Infinium Omni2.5Exome-8 Kit microarray. Of the 25 SNPs we investigated in this analysis, SNP rs1291133 is in LD (r2 = 0.7941) with rs6102991 and was investigated for association with clinical outcome in our present analysis. We did not observe a statistically significant association for rs1291133 SNP with MRD, EFS, or OS in our pediatric AML cohorts. In an effort to look deeper into this, we expanded our search for SNPs beyond the ±10 kb SAMHD1 locus and identified rs4812641 that is located >10 kb from the SAMHD1 locus but with better LD (r2 = 0.8943) with rs6102991. Consistent with previous findings, we observed significant association of this SNP with reduced MRD positivity (OR, 0.591; 95% CI, 0.364-0.958; P = .033; and risk group adjusted OR, 0.554; 95% CI, 0.324-0.948; P = .031, Figure 5C) and increased OS (risk group adjusted HR, 0.735; 95% CI, 0.603-1.109; P = .047) in the AML02 cohort but such an association was not observed in the AML08 trial (Figure 5D). In multivariable analysis, SNP rs4812641 remained as an independent predictor for OS in AML02 (Figure 5E).
Evaluation of SAMHD1 mRNA levels
Gene expression was analyzed separately by continuous variable, median, and quartile, with and without risk-group adjustment in all 3 cohorts. No difference in SAMHD1 expression levels in diagnostic specimens was observed by race or gender. Within the AML02 (n = 163) and AML08 (n = 147) clinical trial cohorts, patients within the high-risk group had lower SAMHD1 expression as compared with standard and low risk groups (supplemental Figure 1A). When analyzing SAMHD1 expression vs SNPs, no statistically significant associations were observed, however, within AML02, patients heterozygous for rs7265241 had slightly higher SAMHD1 levels (Wilcox P = .056, supplemental Figure 1B). We did not observe any significant association with SAMHD1 gene expression and clinical outcomes of MRD, EFS, or OS. Within the GTEx database, variant T-allele for rs1291141 and variant T-allele for rs1291128 was associated with higher SAMHD1 levels in whole blood, consistent with association of these alleles with poor outcome in our study (supplemental Figure 1C).
Discussion
Prognostic markers that improve risk stratification or optimize chemotherapeutic dosing regimens is of paramount importance in improving clinical outcome in childhood AML. Within the last decade, newer targeted drugs that affect certain mutated genes in AML have become an important treatment option for a small fraction of patients with AML; however, for the foreseeable future, the backbone of chemotherapy for the majority of patients with AML still relies on cytarabine-based chemotherapy. Recently, SAMHD1 has arisen as an emerging candidate of relevance to drug response in AML. The results of 2 studies using various in vitro and in vivo mouse models published in 2017 in the same issue of Nature Medicine highlighted the role of SAMHD1 in the development of resistance to cytarabine.8,15 These, and few other follow-up studies, investigated SAMHD1 protein using immunohistochemistry (IHC) or publicly available mRNA expression data from adult (TCGA database, n = 145) and pediatric AML (TARGET database, n = 147).8,15,26,27 To date, only 1 study has investigated SNPs within SAMHD1 and association with clinical outcome, based on 3 expression quantitative trait loci SNPs (rs6102991, rs2872906, and rs6029941) from the GTEx database and reported that rs6102991 showed a nonstatistically significant trend toward CR status.19
In this study, we investigated 25 SNPs within the SAMHD1 locus and identified 3 rs7265241, rs1291128, and rs1291141, that were associated with clinical outcome end points in children diagnosed with de novo AML across multiple clinical trials. In multivariable analysis, the SNPs remained independent predictors of outcome in the respective cohorts after adjusting for age, race, risk group, and white blood cell counts. Of particular interest is rs7265241, for which the presence of variant G allele was associated with poor OS in >1200 patients across 3 clinical trials (AML02, n = 167; AML08, n = 233; and AML1031, n = 849), making this one of the largest cohorts ever tested for SAMHD1 SNPs. Although at this time we do not have an in-depth understanding of the functional consequence of this SNP within SAMHD1, it does occur in high LD with 11 additional SNPs on the SAMHD1 locus (supplemental Table 2) and in the GTEx database another variant, rs117729827, which occurs in partial LD with rs7265241 was significantly associated with gene expression. To reproduce the results from the only study previously published, we identified rs4812641 (>10 kb from the SAMHD1 locus) occurring in LD with previously reported SNP rs610299 and observed consistent and significant association with outcome in the AML02 cohort.
Although we did not observe any association of these SNPs with mRNA expression from the AML cohorts tested, variants for both SNPs were associated with higher SAMHD1 expression in whole blood tissue in the GTEx database (supplemental Figure 1C). It is important to note, SAMHD1 expression is comparatively low in whole blood from normal healthy individuals as compared with other tissues. Interestingly, data from the TCGA database also show significantly higher SAMHD1 expression levels in AML samples as compared with corresponding normal tissues (supplemental Figure 1D). Lastly, differences in treatment regimens (eg, induction, maintenance, supportive care, etc) and initial risk group at time of diagnosis (such as exclusion of FLT3-ITD in AAML1031 cohort included in this study) may have contributed to variation in response and overall treatment outcomes within our discovery cohorts and replication cohort.
With respect to association of SAMHD1 mRNA expression with clinical outcome, we did not observe any significant association with the SNPs investigated in our study. A few points worth mentioning are that the sample size of our cohorts limited our ability to perform any meaningful subgroup analyses. Gene expression was measured via array-based and RNA-seq technologies, therefore a direct comparison between gene expression in each clinical trial cohort could not be made. In contrast to our results, previous studies implied association of higher SAMHD1 expression with poor outcome. Two studies analyzed SAMHD1 mRNA expression levels from public databases. The first study by Schneider et al,8 reported no correlation between SAMHD1 mRNA levels and CR rates when tested in the whole cohort (n = 145); however, in a subset of patients that received Ara-C (n = 82), high SAMHD1 expression was observed. It is important to note that of the 63 patients excluded in the subset, 55 were removed from the non-CR group and predominantly had lower expression, thereby introducing a bias. Moreover, association with EFS or OS was not reported in the study. The second study by Herold et al,15 also evaluated SAMHD1 mRNA expression levels in TCGA (n = 147) and TARGET (n = 145) databases. No significant association with 10-year EFS and OS, nor with remission status or relapse risk was reported. However, low SAMHD1 expression was associated with better OS at 12 months in adult (TCGA) and 18 months in pediatric (TARGET) patients with AML. Of note, the cut-off to classify patients into high and low SAMHD1 expressers was set at 105 reads per kilobase of transcript per million reads mapped (RPKM) for TCGA, resulting in 44.8% patients in the high SAMHD1 group. For the TARGET cohort, the threshold for high expression was set at 1 288 288 RPKM, resulting in only 25% of patients in the high SAMHD1 group.
There are 2 studies that have analyzed SAMHD1 protein levels in leukemic blasts from patients with AML. Schneider et al8 evaluated SAMDH1 protein levels fixed in paraffinized bone marrow using IHC in leukemic blasts isolated at the time of diagnosis and reported low SAMHD1 levels to be associated with better remission rates, EFS, and OS in their cohort. Another study by Rassidakis et al,16 evaluated SAMHD1 protein levels by IHC and reported a trend with EFS and OS in the whole cohort and intermediate risk group. When restricting to patients who received high-dose Ara-C (HDAC) in consolidation therapy, significant association between SAMHD1 levels and EFS and OS was observed, implying SAMHD1’s significance during postremission consolidation therapies. Recently, a study by Zhang et al28 showed that non-POU domain–containing octamer-binding protein (NONO) interacts with, and stabilizes, SAMHD1 in AML cell lines and in an AML xenograft model. Interestingly, this study confirmed that SAMHD1 protein level expression was positively correlated with NONO protein levels in THP-1 and HL60 cells; however, association with NONO and SAMHD1 mRNA levels were not observed, thus indicating posttranslational regulation of SAMHD1. The results from previous studies highlights the variability in SAMHD1 expression, heterogeneity of various treatment arms that was not accounted for, and different thresholds used to classify patients as low or high SAMHD1 expressers.
Moreover, another aspect that adds to the complexity of the relationship observed between SAMHD1 expression and outcome toward Ara-C–based regimens (specifically high dose), is its role in other cellular processes. For instance, alternative splicing reportedly results in splice variants of unknown biological relevance that are coexpressed with full length transcripts.29 Recent data indicate roles for SAMHD1 as a tumor suppressor (because of its antiproliferative properties), as a negative regulator of innate immune response, or in DNA replication repair pathways.12,30 Data on the role of SAMHD1 in disease resistance and antiproliferative qualities, and at what time point patient stratification occurs based on the biomarker, remain to be fully elucidated; however, recent time-dependent analysis of TCGA data suggests that patients with high SAMHD1 levels might have better response after induction and consolidation therapies, whereas those with low levels might have better response after HDAC consolidation.31SAMHD1 has been shown to be frequently mutated in other leukemias and lymphomas and that deleterious mutations may potentiate growth of cancer cells.12 So far there are accumulating data suggesting that SAMHD1 is a tumor suppressor and its implication with drug resistance, thus warranting the need for future studies focused on identifying the optimal balance/threshold to inform meaningful clinical decision making.
In conclusion, we report SAMHD1 SNPs that show consistent association with clinical outcome based on data from 3 independent pediatric AML clinical trials. To the best of our knowledge, this is the biggest investigation of SAMHD1 SNPs in pediatric AML and the promising results warrant future studies focused on in-depth functional evaluation of SAMHD1 pharmacogenomics with validation of the observed results by performing mechanistic studies in independent larger patient cohorts.
Acknowledgments
The authors thank Campana, Coustan-Smith, and Raimondi for MRD and cytogenetics data, and Downing for the AML02 gene expression data.
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health (R01-CA132946) and the American Cancer Society award/St. Baldrick’s Foundation (SAP-21-061-01). St. Jude clinical trials are supported by the American Lebanese Syrian Associated Charitie and the AAML1031 study is supported by NCTN Operations Center Grant (U10CA180886), NCTN Statistics & Data Center (U10CA180899), and St. Baldrick's Foundation. R.J.M. is supported by an institutional research training award from the National Human Genome Research Institute, National Institutes of Health (T32HG008958).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authorship
Contribution: R.J.M. and J.K.L. contributed to study design and generated data; R.J.M., J.K.L., X.C., H.W., A.H.E., T.A.A., and S.B.P. performed data analysis and interpretation; J.K.L., J.M.K., R.C.R., J.E.R., X.M., S.M., R.A., E.A.K., and R.E.R. contributed genotype, expression, or clinical data; and all authors provided critical review of the manuscript and have approved it for publication.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jatinder K. Lamba, Department of Pharmacotherapy and Translational Research, College of Pharmacy, University of Florida, Gainesville, FL 32610; e-mail: jlamba@cop.ufl.edu.
References
Author notes
AAML1031 RNA sequencing data are available at TARGET database (https://target-data.nci.nih.gov/Public/AML/mRNA-seq/L3/expression).
Data are available on request from the corresponding author, Jatinder K. Lamba (jlamba@cop.ufl.edu).
The full-text version of this article contains a data supplement.