TO THE EDITOR:
The prognosis of acute myeloid leukemia (AML) remains poor for both adult and pediatric patients, mainly due to high relapse rates that result from residual leukemic cells with stem cell features,1 including self-renewal capacity and increased resistance to treatment. Cellular drug resistance partly relies on active transport across cellular membranes undertaken by ATP-binding cassette (ABC) transport proteins. Of those, the ABCA3 transporter has been found particularly overexpressed in AML cells,2 especially in leukemic progenitors.3 ABCA3 expression has been previously shown to counteract the cytotoxicity of anthracyclines, etoposide, cytarabine, vincristine, and rituximab4-6 by strengthening drug efflux from leukemic cells or sequestrating cytostatic drugs to endosomal organelles.7 However, studies investigating the prognostic impact of ABCA3 expression in AML have yielded conflicting results, mainly due to sample size and methodological differences between studies.6,8-10 Indeed, some quantitative real-time polymerase chain reaction (RT-PCR)-based studies of ABCA3 transcripts used oligonucleotides that recognize constitutively expressed exons whereas others used TaqMan probes spanning exon junction 19-20,9,10 which was recently demonstrated as a target for aberrant alternative splicing in AML.11 Furthermore, the gene expression cutoff value used to discriminate high to low ABCA3-expressing patients has more often been empirically estimated, and the sample size of patient groups was frequently small, thereby precluding the use of multivariate analyses.
Here, we uniformly investigated the prognostic impact of ABCA3 expression on diagnosis in large cohorts of adult (ALFA0701 trial, 213 available samples12 ) and pediatric (ELAM02 trial, 270 available samples13 ) patients with AML by using a new quantitative RT-PCR approach that targets constitutive exons 6 and 7 shared by all alternative splice isoforms of ABCA3. The cutoff value between high and low expressers was determined using an algorithm calibrated for best event-free survival (EFS) determination (CutOff Finder14 ). All parents and patients included in the study gave signed informed consent for ancillary studies according to the declaration of Helsinki. ALFA0701 and ELAM02 were approved by the ethics committees of Saint-Germain en Laye and Saint-Antoine Paris University Hospital (Assistance Publique-Hôpitaux de Paris) in France, respectively, and by the institutional review board of the French Regulatory Agency. All authors had access to the primary clinical and biological data.
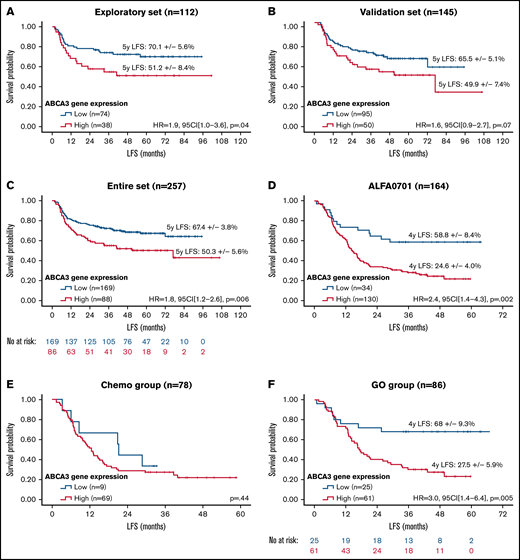
A total of 120 pediatric samples were randomly selected from the entire dataset (n = 270) to create an exploratory set. The remaining 150 samples were used for internal validation. The 2 cohorts were similar except for an excess of core-binding factor AML in the validation set (supplemental Table 1). In the exploratory set, a cutoff value of 1.20 arbitrary units (AU) defined a group of patients (n = 44/120) with high ABCA3 expression who displayed worse outcomes in terms of leukemia-free survival (LFS) (Figure 1A), overall survival (OS), and EFS (supplemental Figure 1A-B). Similar trends were obtained with the validation set for LFS (Figure 1B), OS, and EFS (supplemental Figure 1C-D) but with no statistical significance. This may stem from a lower number of standard-risk patients in the exploratory set (20 [17%]) than in the validation set (55 [36%]). Nevertheless, patients with high ABCA3 expression (n = 98/270; 36%; mean = 3.21 AU, median = 2.19 AU, standard deviation = 2.62 AU) displayed significantly reduced LFS (Figure 1C), OS, and EFS (supplemental Figure 1E-F). Multivariate analyses of pediatric samples (Table 1) revealed an independent prognostic impact of ABCA3 expression along with risk stratification, age, and number of white blood cells at diagnosis. No differences were noticed between high and low ABCA3 expressers in main clinical and biological characteristics (supplemental Table 2). In contrast, significantly worsened outcomes (lower complete response rates, higher relapse rates, and lower survival) were evidenced in the high expresser group that could not be attributed to differences in hematopoietic stem-cell transplantation (HSCT) rates, a potentially confounding factor. Subgroup analysis based on age at diagnosis suggested that ABCA3 expression had a negative impact in patients older than 2 years (supplemental Figure 2A-B). According to the risk stratification used in the ELAM02 trial, ABCA3 expression prognostic impact seemed to be restricted to standard-risk patients (supplemental Figure 2C-E). Similar results were noticed when censoring patients receiving HSCT at the date of transplantation (not shown). LSC17 score, which relies on 17-gene expression signature for leukemic stem cells, has been shown to reliably separate patients into high- and low-risk groups with adult and pediatric AML.15,16 Using LSC17 scores previously described for 147 (54%) pediatric patients,15 we evidenced a positive correlation between LSC17 and ABCA3 levels (R2 = 0.08; P = .0006, Spearman rank). In line with published data,15 LSC17 appeared as a strong prognostic factor in the pediatric cohort (supplemental Figure 2F) whereas ABCA3 expression allowed stratification of patients who displayed intermediate LSC17 values (supplemental Figure 2G). Of note, as mentioned above, ABCA3 expression showed strong prognostic value in standard-risk (SR) patients, all of whom had core-binding factor AML, for which the LSC17 signature has not been validated.15
Survival impact of ABCA3 expression in AML. Kaplan-Meier curves of LFS in the pediatric exploratory set (A), pediatric validation set (B), pediatric entire dataset (C), and adult entire set (D) as well as restricted to patients treated with chemotherapy only (E) or GO) (F). No at risk, number of patients at risk. Patients with high (red curves) and low (blue curves) ABCA3 expression were separated with the best determined cutoff value of 1.20 AU. Hazard ratios (HR) were computed with 95% confidence intervals (95CI), log-rank P values set on the graphs. Quantitative RT-PCR analysis was carried out with iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions using oligonucleotides against exon 6 and 7 of ABCA3 (annotated according to FasterDB database (ABCA3_F: CCTTCAACCACAGCAAGGAG, ABCA3_R: TTGGGAAAAGCGGGAAAAGG). Relative quantification of the E6-E7 fragment of ABCA3 transcripts was performed according to the method described by Pfaffl using GUSb and ABL1 as references (GUSb_F: GCCTGGGTTTTGTGGTCATCTATTC, GUSb_R: CAGTAGCCACTTTCATGCCAACTC, ABL1_F: TGGTAGGGGAGAACCACTTG, and ABL1_R: GGTAGCAATTTCCCAAAGCA). Cycling reactions were performed with a C1000 Touch Thermal Cycler (Bio-Rad) and fluorescence signals measured with a CFX96 Real-Time System (Bio-Rad). Data were processed with CFX Manager software version 3.1 (Bio-Rad). RT-qPCR data were expressed in arbitrary units (AU).
Survival impact of ABCA3 expression in AML. Kaplan-Meier curves of LFS in the pediatric exploratory set (A), pediatric validation set (B), pediatric entire dataset (C), and adult entire set (D) as well as restricted to patients treated with chemotherapy only (E) or GO) (F). No at risk, number of patients at risk. Patients with high (red curves) and low (blue curves) ABCA3 expression were separated with the best determined cutoff value of 1.20 AU. Hazard ratios (HR) were computed with 95% confidence intervals (95CI), log-rank P values set on the graphs. Quantitative RT-PCR analysis was carried out with iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions using oligonucleotides against exon 6 and 7 of ABCA3 (annotated according to FasterDB database (ABCA3_F: CCTTCAACCACAGCAAGGAG, ABCA3_R: TTGGGAAAAGCGGGAAAAGG). Relative quantification of the E6-E7 fragment of ABCA3 transcripts was performed according to the method described by Pfaffl using GUSb and ABL1 as references (GUSb_F: GCCTGGGTTTTGTGGTCATCTATTC, GUSb_R: CAGTAGCCACTTTCATGCCAACTC, ABL1_F: TGGTAGGGGAGAACCACTTG, and ABL1_R: GGTAGCAATTTCCCAAAGCA). Cycling reactions were performed with a C1000 Touch Thermal Cycler (Bio-Rad) and fluorescence signals measured with a CFX96 Real-Time System (Bio-Rad). Data were processed with CFX Manager software version 3.1 (Bio-Rad). RT-qPCR data were expressed in arbitrary units (AU).
Multivariate analyses for OS, EFS, and LFS in the pediatric (upper panel) and adult (lower panel) datasets
| . | OS . | EFS . | LFS . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ELAM02 entire set . | HR . | 95CI . | P . | HR . | 95CI . | P . | HR . | 95CI . | P . |
| ABCA3 gene expression (>1.20 AU) | 1.93 | 1.18-3.14 | .009 | 1.92 | 1.31-2.82 | <.001 | 1.81 | 1.21-2.71 | .004 |
| Risk group* | 1.62 | 1.12-2.36 | .011 | 1.32 | 0.98-1.76 | .06 | — | — | — |
| Age (>6 y) | 2.27 | 1.25-4.13 | .007 | 2.04 | 1.30-3.22 | .002 | 1.99 | 1.24-3.22 | .005 |
| Leukocytes (>30 g/L) | 2.02 | 1.24-3.29 | .005 | 1.78 | 1.22-2.60 | .003 | 1.53 | 1.02-2.29 | .041 |
| OS | EFS | LFS | |||||||
| ALFA0701 entire set | HR | 95CI | P | HR | 95CI | P | HR | 95CI | P |
| ABCA3 gene expression (>1.20 AU) | 1.7 | 0.7-4.0 | .20 | 2.6 | 1.3-6.5 | .01 | 2.6 | 1.1-5.9 | .03 |
| CD33 expression (>90% blasts +) | 0.5 | 0.3-1.0 | .04 | 0.6 | 0.3-0.9 | .03 | 0.5 | 0.3-1.0 | .05 |
| Normal karyotype | — | — | — | 0.6 | 0.4-1.0 | .06 | — | — | — |
| ELN risk stratification | 2.2 | 1.3-3.5 | .002 | — | — | — | — | — | — |
| . | OS . | EFS . | LFS . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ELAM02 entire set . | HR . | 95CI . | P . | HR . | 95CI . | P . | HR . | 95CI . | P . |
| ABCA3 gene expression (>1.20 AU) | 1.93 | 1.18-3.14 | .009 | 1.92 | 1.31-2.82 | <.001 | 1.81 | 1.21-2.71 | .004 |
| Risk group* | 1.62 | 1.12-2.36 | .011 | 1.32 | 0.98-1.76 | .06 | — | — | — |
| Age (>6 y) | 2.27 | 1.25-4.13 | .007 | 2.04 | 1.30-3.22 | .002 | 1.99 | 1.24-3.22 | .005 |
| Leukocytes (>30 g/L) | 2.02 | 1.24-3.29 | .005 | 1.78 | 1.22-2.60 | .003 | 1.53 | 1.02-2.29 | .041 |
| OS | EFS | LFS | |||||||
| ALFA0701 entire set | HR | 95CI | P | HR | 95CI | P | HR | 95CI | P |
| ABCA3 gene expression (>1.20 AU) | 1.7 | 0.7-4.0 | .20 | 2.6 | 1.3-6.5 | .01 | 2.6 | 1.1-5.9 | .03 |
| CD33 expression (>90% blasts +) | 0.5 | 0.3-1.0 | .04 | 0.6 | 0.3-0.9 | .03 | 0.5 | 0.3-1.0 | .05 |
| Normal karyotype | — | — | — | 0.6 | 0.4-1.0 | .06 | — | — | — |
| ELN risk stratification | 2.2 | 1.3-3.5 | .002 | — | — | — | — | — | — |
Cox proportional hazard models factoring variables with a P ≤ .10 in univariate analyses.
HR, hazard ratio; CI95, 95% confidence interval. --, no data.
Riskgroup values are defined according to the ELAM02 and ALFA0701 risk stratification.
In adult patients with AML, ABCA3 expression was assessed in total RNA derived from 213 samples harvested at diagnosis. The cutoff value of ABCA3 expression was similar to that of pediatric AML (1.20 AU), suggesting that this value might be considered a reference threshold. However, the median expression value of ABCA3 was higher (3.47 vs 0.89; Mann-Whitney P < .0001), and the high ABCA3 expressers were more frequent among adults than children (n = 173/213, 81% vs n = 98/270, 36%). High ABCA3 expression had an adverse prognostic impact on LFS (Figure 1D) in all tested adult patients. Such prognostic impact did not pertain to the chemotherapy group (Figure 1E), in accordance with previous studies published by our team11 and others,10 but it was exclusive to patients receiving gemtuzumab-ozogamycin (GO) (Figure 1F), who showed shorter LFS in association with an excess of leukemia-related deaths in high ABCA3 expressers (54% vs 22%; P = .007, Fisher’s exact test). In contrast, the death rates due to toxicity and HSCT were not statistically different between high and low ABCA3 expressers who received GO (7.5% vs 13.7%, P = .50 and 18.5% vs 23.7%, P = .79, Fisher’s exact test, respectively). Complete response rates were not affected by the ABCA3 expression level. In multivariate analyses of all samples (Table 1), high ABCA3 expression retained its prognostic impact on EFS and LFS but not on OS. Subgroup analyses based on risk stratification used in the ALFA0701 trial revealed that high ABCA3 expression had a negative impact on LFS in standard-risk and GO-treated SR patients (supplemental Figure 3A-B). However, in the latter case, this correlation did not reach statistical significance, likely due to the limited number of patients. Similar results were obtained when censoring patients receiving HSCT at the date of transplantation (not shown). Using LSC17 scores previously described for 160 (72%) adult patients,16 we also evidenced a positive correlation between LSC17 and ABCA3 levels (R2 = 0.09; P < .0001, Spearman rank). ABCA3 expression allowed stratification of patients with low LSC17 values, for whom high ABCA3 expression was associated with as low survival as patients with high LSC17 scores in the overall LSC17-tested cohort as well as in the GO-treated subgroup (n = 82) (supplemental Figure 3C-D). Regarding the GO target, no correlation was noticed between CD33 and the expression levels of ABCA3 in ALFA0701 samples (supplemental Figure 3E). However, the frequency of AML cells expressing CD33 at the cell membrane level was negatively correlated with ABCA3 expression, suggesting that ABCA3 might affect cell response to GO (supplemental Figure 3F). As already described for rituximab resistance in B-cell lymphoma,7 the role of ABCA3 in the humoral evasion of AML cells through extravesicular sequestration of the CD33 antibody remains to be investigated.
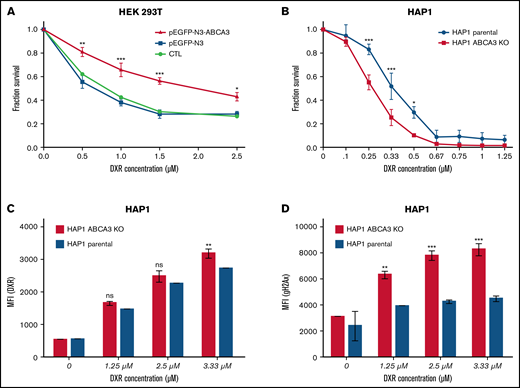
To assess the impact of ABCA3 expression on cell viability upon anthracycline exposure, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assays were carried out with HEK293T cells overexpressing an ectopic ABCA3 (or not) that were exposed to increasing concentrations of doxorubicin (DXR). Figure 2A shows that ABCA3-overexpressing cells were less sensitive to DXR-induced cell death than control cells. To eliminate nonspecific effects that could arise from the protein overexpression system and HEK293 phenotype, MTT experiments were repeated with parental and ABCA3-KO HAP1 cells, which were knocked out for ABCA3 by a CRISPR-Cas9 approach. No change in cell viability was noticed between parental and HAP1-ABCA3-KO cells under normal growth conditions. In contrast, DXR-exposed parental cells displayed a survival advantage compared with their counterpart HAP1-ABCA3-KO cells (Figure 2B). This coincided with a lower cellular uptake of DXR (Figure 2C) as well as a lower number of phospho-H2AX–positive cells than that exhibited by HAP1-ABCA3-KO cells (Figure 2D).
ABCA3 expression induces resistance to DXR and GO in myeloid cells. (A) MTT assays (Cell Growth Determination kit, Sigma-Aldrich) determining the survival proportions of HEK 293T cells nontransfected and those transfected (using JetPrime) with the plasmids pEGFP-N3 and pEGFP-N3-ABCA3 encoding ABCA3 fused to GFP. (B) MTT assays of HAP1 parental cells and HAP1 cells knocked out for ABCA3 (HAP-ABCA3-KO) using CRISPR-Cas9–induced indels (product ID HZGHC006065c005, Horizon Discovery) upon increasing concentrations of DXR (48-hour incubation). (C) Uptake of DXR in HAP1 parental cells and HAP1-ABCA3-KO cells. The mean fluorescence intensity (MFI) was measured by flow cytometry analysis HAP1 cells after 6-hour exposure to increasing concentrations of DXR. (D) gammaH2AX expression in HAP1 parental cells and HAP1-ABCA3-KO cells exposed to DXR. The primary antiphospho-gammaH2AX antibody (Ser139; Cell Signaling Technology) and secondary anti-rabbit APC-linked antibody (Cell Signaling Technology) was used. (A-D) The mean +/− SEM (n = 3) and P values from Sidak’s multiple analyses are shown (*P < .05; **P < .01; ***P < .001).
ABCA3 expression induces resistance to DXR and GO in myeloid cells. (A) MTT assays (Cell Growth Determination kit, Sigma-Aldrich) determining the survival proportions of HEK 293T cells nontransfected and those transfected (using JetPrime) with the plasmids pEGFP-N3 and pEGFP-N3-ABCA3 encoding ABCA3 fused to GFP. (B) MTT assays of HAP1 parental cells and HAP1 cells knocked out for ABCA3 (HAP-ABCA3-KO) using CRISPR-Cas9–induced indels (product ID HZGHC006065c005, Horizon Discovery) upon increasing concentrations of DXR (48-hour incubation). (C) Uptake of DXR in HAP1 parental cells and HAP1-ABCA3-KO cells. The mean fluorescence intensity (MFI) was measured by flow cytometry analysis HAP1 cells after 6-hour exposure to increasing concentrations of DXR. (D) gammaH2AX expression in HAP1 parental cells and HAP1-ABCA3-KO cells exposed to DXR. The primary antiphospho-gammaH2AX antibody (Ser139; Cell Signaling Technology) and secondary anti-rabbit APC-linked antibody (Cell Signaling Technology) was used. (A-D) The mean +/− SEM (n = 3) and P values from Sidak’s multiple analyses are shown (*P < .05; **P < .01; ***P < .001).
In conclusion, our data show that ABCA3 expression has a prognostic impact on pediatric AML and helps to predict response to GO in adult AML. This prognostic impact may improve current risk stratification of patients with AML and help predict which patients may benefit from GO therapy. The prognostic impact of ABCA3 expression in pediatric patients with AML treated with GO remains to be explored. Furthermore, we identified a positive correlation between ABCA3 expression and LSC17 scores. This finding is in line with high ABCA3 expression levels in leukemic stem cells.3 ABCA3 expression demonstrated prognostic value in pediatric and adult patients with AML displaying intermediate and low LSC17 scores, respectively. Given the positive effects of ABCA3 on cell survival upon drug exposure, future studies should determine whether targeting ABCA3 could be a strategy for sensitizing leukemic stem cells to therapy.
Acknowledgments: The authors gratefully thank patient and family associations (Association Philanthropique de Parents d'Enfants atteints de Leucémie [APPEL] and association Arche) for funding PCR supplies as well as patients and their families who participated in the ELAM02 and the ALFA0701 trials. The authors also thank Pr. Raphael Itzykson for reviewing the manuscript.
This work was was supported by ARC (Association de Lutte pour la Recherche Contre le Cancer) (A.C.), INSERM (F.M.), and Hospices Civils de Lyon and Lyon I University (E.W.).
Authorship: A.C., F.M., and E.W. conceptualized the study; H.L., C.P., C.T., G.L., H.D., Y.B., and E.W. provided the resources; A.C., H.L., and M.H.C. conducted the experiments; F.M. and E.W. supervised the study; A.C. and F.M. wrote the original draft; and A.C., H.L., M.H.C., C.P., H.D., C.T., J.L., G.L., Y.B., F.M., and E.W. reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Antony Ceraulo, Department of Pediatric Oncology and Hematology, Institut d’Hématologie et d’Oncologie Pédiatrique (IHOPe), and University Lyon I, Lyon, France; e-mail: antony.ceraulo@lyon.unicancer.fr; Franck Mortreux, Laboratory of Biology and Modelling of the Cell, University of Lyon, ENS de Lyon, University Claude Bernard, CNRS UMR 5239, INSERM U1210, Lyon, France; e-mail: franck.mortreux@ens-lyon.fr; and Eric Wattel, Hospices Civils de Lyon, Department of Hematology, Lyon-Sud Hospital, Pierre Bénite, France; email: eric.wattel@chu-lyon.fr.
References
Author notes
F.M. and E.W. contributed equally to this study.
Requests for data sharing may be submitted to Franck Mortreux (franck.mortreux@ens-lyon.fr).
The full-text version of this article contains a data supplement.