Key Points
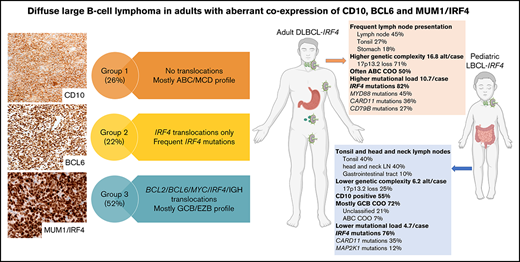
DLBCL in adults aberrantly coexpressing CD10, BCL6, and MUM1 is genetically heterogeneous and enriched in IRF4-rearranged cases.
IRF4-rearranged DLBCL in adults share genetic features with LBCL-IRF4 in children but with higher genomic complexity and often ABC GEP.
Abstract
Diffuse large B-cell lymphoma (DLBCL) with aberrant coexpression of CD10+BCL6+MUM1+ (DLBCL-AE), classified as germinal center B cell (GCB) type by the Hans algorithm (HA), was genetically characterized. To capture the complexity of DLBCL-AE, we used an integrated approach that included gene expression profiling (GEP), fluorescence in situ hybridization, targeted gene sequencing, and copy number (CN) arrays. According to GEP, 32/54 (59%) cases were classified as GCB-DLBCL, 16/54 (30%) as activated B-cell (ABC) DLBCL, and 6/54 (11%) as unclassifiable. The discrepancy between HA and GEP was 41%. Three genetic subgroups were identified. Group 1 included 13/50 (26%) cases without translocations and mainly showing and ABC/MCD molecular profile. Group 2 comprised 11/50 (22%) cases with IRF4 alterations (DLBCL-IRF4), frequent mutations in IRF4 (82%) and NF-κB pathway genes (MYD88, CARD11, and CD79B), and losses of 17p13.2. Five cases each were classified as GCB- or ABC-type. Group 3 included 26/50 (52%) cases with 1 or several translocations in BCL2/BCL6/MYC/IGH, and GCB/EZB molecular profile predominated. Two cases in this latter group showed complex BCL2/BCL6/IRF4 translocations. DLBCL-IRF4 in adults showed a similar copy number profile and shared recurrent CARD11 and CD79B mutations when compared with LBCL-IRF4 in the pediatric population. However, adult cases showed higher genetic complexity, higher mutational load with frequent MYD88 and KMT2D mutations, and more ABC GEP. IRF4 mutations were identified only in IRF4-rearranged cases, indicating its potential use in the diagnostic setting. In conclusion, DLBCL-AE is genetically heterogeneous and enriched in cases with IRF4 alterations. DLBCL-IRF4 in adults has many similarities to the pediatric counterpart.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of non-Hodgkin lymphoma, representing ∼30% to 40% of all newly diagnosed lymphomas.1 DLBCL is clinically, morphologically, and biologically a heterogeneous disease reflected in its highly variable clinical course.2,3 The 2017 World Health Organization (WHO) lymphoma classification recognizes within the group of DLBCL several subtypes characterized by unique clinical, pathological, or genetic features. Nevertheless, most cases of DLBCL fall into the “not otherwise specified” category. Gene expression profiling (GEP) studies have revealed that DLBCL, not otherwise specified, comprises molecular groups that reflect either the stage in B-cell development from which the disease originates or the activity of different biological programs.4-8 This cell-of-origin (COO) classification recognizes 2 broad categories: the germinal center B cell (GCB)-like DLBCL, and the activated B cell (ABC)-like DLBCL; however, ∼15% remains unclassified.5,9
More recently, a genetic classification of DLBCL based on mutational profile, somatic copy-number alterations (CNA), and structural variants was proposed.3,10,11 The similarities of the molecular subtypes identified in 3 different studies indicate that these subgroups reflect true biological differences. The MCD/C5/MYD88 subtypes are part of the ABC group and are enriched for MYD88L265P and CD79B mutations with subsequent activation of the NF-κB pathway. The EZB/C3/BCL2 subtypes are assigned to the GCB group and are characterized by BCL2 translocations and EZH2 mutations associated with other mutations of chromatin modifiers. In contrast, the BN2/C1/NOTCH2 subtypes, mostly assigned to ABC-DLBCL, are characterized by BCL6 translocations and NOTCH2 mutations with a putative origin in marginal zone B cells. The C2 subgroup is defined by TP53 mutations, a high number of CNAs, and increased ploidy. Based on these molecular studies, a predictor model has been developed that dissects and stratifies further the COO classification into 7 genetic subtypes with apparently clinical relevance: 3 mostly related to ABC-type, 3 to GCB-type, and the novel subgroup genetically related to marginal zone lymphoma.12
The increasing defining role of genetic alterations in lymphoma classification is highlighted in the recognition of 2 provisional entities in the 2017 WHO classification, namely, large B-cell lymphoma with IRF4 rearrangement (LBCL-IRF4) and Burkitt-like lymphoma with 11q aberration.13,14 Burkitt-like lymphoma with 11q aberration is a high-grade B-cell lymphoma without MYC translocation and genetic alterations closer to those found in GCB-DLBCL.15,16 LBCL-IRF4 predominates in children and young adults, shows frequent involvement of the Waldeyeŕs ring and gastrointestinal tract, and shows excellent outcome in patients after chemotherapy.13,17-19 A recent study demonstrated that LBCL-IRF4 in children and young adults has a distinct molecular profile different from other LBCL characterized by frequent mutations in IRF4 and NF-κB–related genes (CARD11, CD79B, and MYD88), despite a GCB-transcriptional program, and losses in 17p13 and gains of chromosome 7 and 11q12.3-q25.20 LBCL-IRF4 shows mostly a GCB phenotype according to the Hans algorithm (HA),21 with expression of CD10 (60%) and BCL6 but with strong MUM1/IRF4 expression.13,19,22 An unanswered question is whether all DLBCLs with aberrant coexpression of CD10, BCL6, and MUM1 (DLBCL-AE) correspond to the GCB subtype and/or carry an IRF4 translocation. Furthermore, LBCL-IRF4 has been reported in the adult population13,23,24 ; however, it is not clear whether these cases represent clinically and biologically the same disease with excellent prognosis as in children and young adults.
The aim of this study was to unravel the molecular landscape of 55 cases of DLBCL-AE in an adult population. The analysis revealed that DLBCL-AE is genetically a heterogeneous group enriched in IRF4-rearranged (DLBCL-IRF4) cases. The clinical and relevant molecular features of DLBCL-IRF4 in adults were compared with LBCL-IRF4 in the pediatric population.
Material and methods
Patients and samples
Fifty-five cases of de novo DLBCL aberrantly coexpressing CD10, BCL6, and MUM1/IRF4 (DLBCL-AE) in >50% of tumor cells were included in the study. Initially, 210 cases of DLBCL were investigated and 30 cases of DLBCL-AE (14%) were identified. To expand the study, further cases were collected from different institutions. In total, there were 25 cases from the University of Tübingen, Germany, 20 cases from Hospital Clinic of Barcelona, Spain, and 10 cases from the Mayo Clinic, Phoenix, Arizona. As control, 9 cases of DLBCL were included: 5 with GCB phenotype and 4 with non–GCB phenotype according to HA. The cases were classified following the criteria of the 2017 WHO classification.25 The immunohistochemical staining was performed as part of the diagnostic workup. The study was approved by the local Ethics Review Committee and the IRB review panels of the contributing institutions (UKT 199/2020BO2). It was conducted in accordance with the Declaration of Helsinki.
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) analyses for the detection of BCL2, BCL6, MYC, IRF4, IGH, IGK, and IGL translocations were performed using LSI Dual color break-apart probes (Metasystems, Altlußheim, Germany; Agilent Technologies, Santa Clara, CA; Vysis-Abbott Molecular, Wiesbaden, Germany). In some cases, homemade FISH probes were used.13 For the triple-color FISH IGH-BCL2-IRF4, the XL t(14;18) IGH/BCL2 dual fusion probe (Metasystems) was cohybridized with BAC clones spanning the IRF4 loci RP3-416J7, RP5-1077H22, and RP5-856G1 labeled in Spectrum Aqua.
Targeted next generation sequencing (NGS) and mutational analysis
The mutational status of 66 B-cell lymphoma–related genes (supplemental Table 1) was investigated using a SureSelectXT Target Enrichment System Capture NGS strategy library (Agilent Technologies) before sequencing in a MiSeq equipment (Illumina, San Diego, CA) (supplemental Methods). As verification, 25 cases were additionally investigated with a 10-gene Ion AmpliSeq Custom panel (supplemental Table 2) (Ion Torrent S5; Thermo Fisher Scientific). The Custom Panel was designed using the Ion AmpliSeq Designer from Thermo Fisher Scientific (version 3.4). For description of library preparation, sequencing, and raw data analyses see supplemental Methods. The previously published mutational profile of 17 pediatric and young adult cases of LBCL-IRF4 was used for comparison.20 Occurrence of mutations in genes within predefined pathogenic pathways was calculated per each molecular group.26
DNA CN alterations analysis
CN alterations were examined in LBCL-IRF4 and/or IRF-4 mutated using Oncoscan (Thermo Fisher Scientific), according to standard protocols (supplemental Methods). Gains and losses and CN neutral loss of heterozygosity (CNN-LOH) regions were evaluated using Nexus Biodiscovery version 9.0 software (Biodiscovery, Hawthorne, CA). Additional previously published CN data were used for comparison.20
Gene expression analysis (NanoString and HTG)
COO classification was performed using the Lymph2Cx assay (NanoString, Seattle, WA) and/or HTG EdgeSeq System (HTG Molecular Diagnostics Inc., Tucson, AZ) (supplemental Methods). IRF4 mRNA levels were established based on the number of counts of IRF4_NM_002460.1 on the Lymph2Cx assay.
Statistical methods
Differences in the distribution of individual parameters among patient subsets were analyzed by Fisher’s exact test for categorized variables and the Student t-test for continuous variables. The P values for multiple comparisons were adjusted using the Benjamini-Hochberg correction (false discovery rate). A cutoff of P = .05 was considered significant. Statistical analyses were performed using R software version 3.5.0.
Results
Clinical and morphological findings
Clinical information is summarized in Table 1 and supplemental Table 3. A total of 55 patients were included in the study, of whom 22 were male and 33 were female patients, representing a M:F ratio of 1:1.5 with a median age of 70 years (range 37-92 years). Twenty-three patients (42%) showed nodal presentation whereas 32 patients (58%) had extranodal presentation, with 10 cases (18%) involving the tonsils/Waldeyeŕs ring, 6 cases involving the gastrointestinal tract (11%), 3 cases (5%) involving the testis, and 2 cases each (4%) involving bone marrow and breast and others (16%). Of the patients with available clinical information, 19/45 had low-stage disease at presentation (42%) whereas 26/45 patients had high-stage disease (58%). Thirty patients (30/48; 62%) were alive at last follow-up without evidence of disease, with a median follow-up of 36 months (range 7-97 months). Ten patients died; 9 died of disease with a median of 7 months (range 1-21 months), and 1 patient died after 6 months of an unrelated cause. Eight patients were lost to follow-up during or after therapy (range 0-19 months).
Clinical data of 55 patients with DLBCL with aberrant coexpression of CD10, BCL6, and MUM1
| Parameter . | No. of cases . |
|---|---|
| Age | Mean 70 y (range 37-92 y) |
| Male gender | 22 (40%) |
| Female gender | 33 (60%) |
| Stage I/II | 19/45 (42%) |
| Stage III/IV | 26/45 (58%) |
| Localization | |
| Nodal presentation | 23 cases (42%) |
| Extranodal presentation | 32 cases (58%) |
| Tonsil/Waldeyeŕs ring | 10 cases (18%) |
| Gastrointestinal tract | 6 cases (11%) |
| Testis | 3 cases (5%) |
| Breast | 2 cases (4%) |
| Other | 11 cases (20%) |
| Alive, no evidence of disease | 30/48 patients (62%) |
| Median follow-up (range) | 36 mo (7-97 mo) |
| Died of disease | 9/48 patients (19%) |
| Median (range) | 7 mo (1-21 mo) |
| Cell of origin | |
| GCB | 32/54 (59%) |
| ABC | 16/54 (30%) |
| UC | 6/54 (11%) |
| Parameter . | No. of cases . |
|---|---|
| Age | Mean 70 y (range 37-92 y) |
| Male gender | 22 (40%) |
| Female gender | 33 (60%) |
| Stage I/II | 19/45 (42%) |
| Stage III/IV | 26/45 (58%) |
| Localization | |
| Nodal presentation | 23 cases (42%) |
| Extranodal presentation | 32 cases (58%) |
| Tonsil/Waldeyeŕs ring | 10 cases (18%) |
| Gastrointestinal tract | 6 cases (11%) |
| Testis | 3 cases (5%) |
| Breast | 2 cases (4%) |
| Other | 11 cases (20%) |
| Alive, no evidence of disease | 30/48 patients (62%) |
| Median follow-up (range) | 36 mo (7-97 mo) |
| Died of disease | 9/48 patients (19%) |
| Median (range) | 7 mo (1-21 mo) |
| Cell of origin | |
| GCB | 32/54 (59%) |
| ABC | 16/54 (30%) |
| UC | 6/54 (11%) |
UC, unclassifiable.
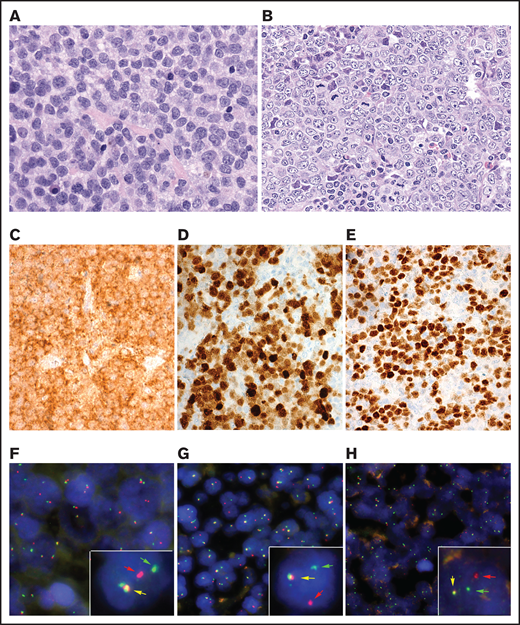
Morphologically, 6 cases revealed a blastoid cytology (Figure 1A), whereas 49 cases showed a characteristic centroblastic cytology (Figure 1B). All cases showed expression of CD10, BCL6, and MUM1/IRF4 in more than 50% of tumor cells (Figure 1C-E).
Morphological, immunophenotypic, and genetic features of triple-positive DLBCLs. (A) B-cell lymphoma with blastic morphology characterized by medium- to large-sized cells with irregular nuclei, fine chromatin, inconspicuous nucleolus, and scant cytoplasm (case 10), (original magnification ×400; hematoxylin and eosin stain). (B-G) Case 5 shows a characteristic centroblastic morphology (original magnification ×400; hematoxylin and eosin stain). The tumor cells are CD10+ (C), MUM1/IRF4+ (D), and BCL6+ (E) (original magnification ×400; immunostaining). FISH demonstrates an IRF4 (F) and IGL (G) break with 1 colocalized signal (yellow arrow) and 1 split signal (green and red arrows) consistent with gene rearrangement. (H) In case 2, an IGK break was demonstrated. FISH shows a signal constellation of 1 colocalized signal (yellow arrow) and 1 split signal (green and red arrows) consistent with gene rearrangement.
Morphological, immunophenotypic, and genetic features of triple-positive DLBCLs. (A) B-cell lymphoma with blastic morphology characterized by medium- to large-sized cells with irregular nuclei, fine chromatin, inconspicuous nucleolus, and scant cytoplasm (case 10), (original magnification ×400; hematoxylin and eosin stain). (B-G) Case 5 shows a characteristic centroblastic morphology (original magnification ×400; hematoxylin and eosin stain). The tumor cells are CD10+ (C), MUM1/IRF4+ (D), and BCL6+ (E) (original magnification ×400; immunostaining). FISH demonstrates an IRF4 (F) and IGL (G) break with 1 colocalized signal (yellow arrow) and 1 split signal (green and red arrows) consistent with gene rearrangement. (H) In case 2, an IGK break was demonstrated. FISH shows a signal constellation of 1 colocalized signal (yellow arrow) and 1 split signal (green and red arrows) consistent with gene rearrangement.
FISH results
The results of FISH analysis for BCL2, BCL6, MYC, IRF4, and IG genes are summarized in supplemental Table 4. Of the 55 cases, 15 cases (27%) showed no rearrangements whereas 40 cases (73%) showed 1 or more rearrangements (Figure 1F-H). The most frequently rearranged genes were BCL2 and BCL6 (12 cases each) followed by IRF4 and MYC (11 cases each). Two cases were double-hit (MYC/BCL6), 2 cases were triple-hit (MYC/BCL2/BCL6), and 4 cases showed rearrangements of both BCL2 and BCL6. IGH was detected as the translocation partner in 23 cases whereas in 2 cases IGL was demonstrated. In 6 cases, an IG break was detected (4 IGH, 1 IGL, and 1 IGK) without identifiable partner. Of the 11 cases with IRF4 rearrangement, 2 cases (cases 25 and 26) also had a BCL2 and/or a BCL6 translocation. Because of the complex rearrangements, these 2 cases were further analyzed.
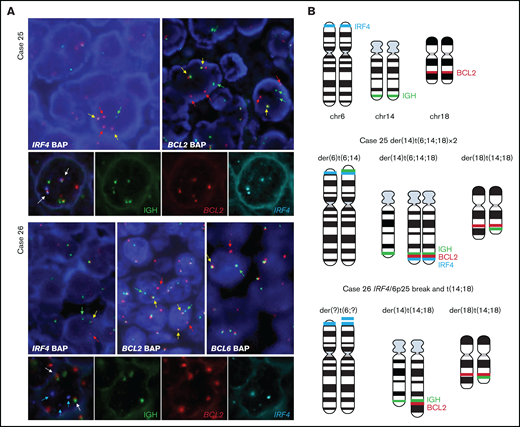
Triple-color FISH IGH-BCL2-IRF4 on case 25 showed that all cells had rearrangements of BCL2 and IRF4 and colocalized with IGH locus in the same derivative chromosome (Figure 2A). This derivative chromosome containing these 3 loci was duplicated. These results suggest that in case 25, a t(6;14;18) translocation occurred as 1 hit.
FISH analysis demonstrates complex rearrangement with IRF4 and BCL2/BCL6 genes. (A) FISH analyses using IRF4, BCL2, and BCL6 break-apart probes and triple-color FISH for IGH, BCL2, and IRF4 loci in the 2 cases with IRF4 rearrangement together with BCL2 in case 25 and BCL2/BCL6 rearrangement in case 26. These hybridizations were performed using t(14;18) dual-color fusion (Metasystems) and BAC clones spanning IRF4 locus (BAC clones 5' RP3-416J7 and 3' RP5-1077H22 and RP5-856G1) labeled in spectrum aqua. Red signals correspond to BCL2, green signals correspond to IGH, and aqua/blue signals represent IRF4. Case 25. FISH demonstrates IRF4 and BCL2 breaks (upper panel) with 1 or 2 colocalized signals (yellow arrows) and 1 or 2 split signals (green and red arrows) consistent with BCL2 and IRF4 gene rearrangement. The triple rearrangement (IGH-IRF4-BCL2) (lower panel) is demonstrated by colocalization of the 3 colors (white arrows). Case 26. FISH analysis for IRF4, BCL2, and BCL6 with break-apart probes demonstrates 1 colocalized signal (yellow arrow) and 1 split signal (green and red arrows), indicating a triple rearrangement. FISH analyses of concomitant IGH, BCL2 and IRF4 rearrangements (lower panel) show 2 colocalized signals (white arrow) of the derivative chromosomes 14 and 18 resulting in t(14;18) translocation. IRF4 locus (blue arrows) did not colocalize with either the IGH locus or the t(14;18) translocation, indicating that this is an independent rearrangement with unknown partner. (B) Schematic representation of IRF4 (aqua blue), IGH (green), and BCL2 (red) breaks/rearrangements in cases 25 and 26.
FISH analysis demonstrates complex rearrangement with IRF4 and BCL2/BCL6 genes. (A) FISH analyses using IRF4, BCL2, and BCL6 break-apart probes and triple-color FISH for IGH, BCL2, and IRF4 loci in the 2 cases with IRF4 rearrangement together with BCL2 in case 25 and BCL2/BCL6 rearrangement in case 26. These hybridizations were performed using t(14;18) dual-color fusion (Metasystems) and BAC clones spanning IRF4 locus (BAC clones 5' RP3-416J7 and 3' RP5-1077H22 and RP5-856G1) labeled in spectrum aqua. Red signals correspond to BCL2, green signals correspond to IGH, and aqua/blue signals represent IRF4. Case 25. FISH demonstrates IRF4 and BCL2 breaks (upper panel) with 1 or 2 colocalized signals (yellow arrows) and 1 or 2 split signals (green and red arrows) consistent with BCL2 and IRF4 gene rearrangement. The triple rearrangement (IGH-IRF4-BCL2) (lower panel) is demonstrated by colocalization of the 3 colors (white arrows). Case 26. FISH analysis for IRF4, BCL2, and BCL6 with break-apart probes demonstrates 1 colocalized signal (yellow arrow) and 1 split signal (green and red arrows), indicating a triple rearrangement. FISH analyses of concomitant IGH, BCL2 and IRF4 rearrangements (lower panel) show 2 colocalized signals (white arrow) of the derivative chromosomes 14 and 18 resulting in t(14;18) translocation. IRF4 locus (blue arrows) did not colocalize with either the IGH locus or the t(14;18) translocation, indicating that this is an independent rearrangement with unknown partner. (B) Schematic representation of IRF4 (aqua blue), IGH (green), and BCL2 (red) breaks/rearrangements in cases 25 and 26.
FISH constellation in case 26 showed typical patterns of break for IGH and IRF4 loci, whereas BCL2 break-apart probes revealed 3 clones with different BCL2 rearrangements: the first clone with a typical BCL2 break, the second clone with a gain of normal chromosome 18 to the break, and the third clone with loss of 3' signal (red signal) in derivative chromosome 18 (Figure 2B). Using triple-color FISH IGH-BCL2-IRF4, IRF4 locus did not colocalize with either the IGH locus or the t(14;18) derivative chromosomes, indicating that IRF4 rearrangement was an independent, clonal alteration observed in more than 80% of the cells.
Gene expression patterns
GEP was performed in 54 cases: 48 cases with the Lymph2Cx, and 24 cases with HTG EdgeSeq DLBCL COO assay (supplemental Table 4). In 18 cases, both techniques were performed. According to GEP, 32/54 (59%) cases were classified as GCB, 16/54 (30%) were classified as ABC, and 6/54 (11%) were unclassifiable. The correlation between the 2 different assays was very good (13/18; 72%). No discordant cases were classified as GCB or ABC; however, in 5 cases assigned as unclassifiable by the Lymph2Cx, HTG EdgeSeq classified 3 as ABC and 2 as GCB. The discrepancy between the HA and GEP in the DLBCL-AE was 41%. In contrast, in the control cases there was a 100% agreement between HA and GEP (supplemental Figure 1).
Identification of recurrent mutations by NGS
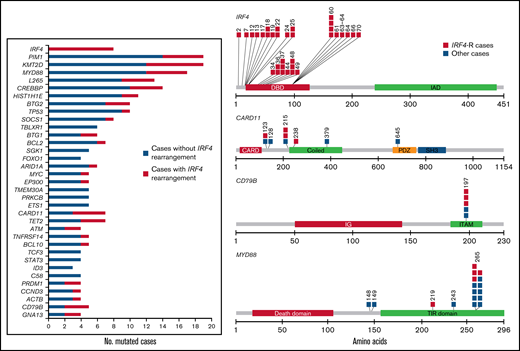
All cases were analyzed by next generation sequencing (NGS). Fifty cases were informative, of which 46 were analyzed with the 66-gene SureSelectXT panel (mean coverage 873; range 26-2693) and 24 were analyzed with the 10-gene Ampliseq panel (mean coverage 1650; range 100-65 280). Twenty cases were analyzed with both panels and 4 cases only with the Ampliseq panel (supplemental Table 5). In the 50 evaluable cases, 531 mutations were identified in 47 (94%) whereas in 3 cases (6%) no mutations were identified (mean 10.62 mutations/case). A total of 392 variants (74%) were predicted as potential driver mutations with a mean of 7.84 mutations per case. The most recurrently mutated genes were KMT2D and PIM1 (19/50; 38%) followed by MYD88 (17/50; 34%); CREBBP (14/50; 28%); HIST1H1E (11/50; 22%); BTG2, TP53 (10/50; 20%); IRF4, SOCS1 (8/50; 16%); BCL2, CARD11, TET2 (7/50;14%); BTG1, ARID1A, TBLXR1 (6/50; 12%); MYC, EP300, TNFRSF14, TMEM30A, PRKCB, BCL10, CD79B, SGK1, ETS1 (5/50%; 10%); and others (Figure 3).
Mutational landscape of 50 cases of DLBCL-AE. Bar graph shows mutated genes in 4 or more cases. The results are given in number of mutated cases per gene. The colored bars indicate the presence (orange) or absence (blue) of IRF4 rearrangement. A diagram of the relative positions of driver mutations is shown for IRF4, CARD11, CD79B, and MYD88. Multiple passenger mutations for IRF4 also are depicted. The x-axis indicates amino acid positions. The approximate location of somatic mutations identified in each gene is indicated. IRF4 mutations are mainly in the DNA binding domain. All CD79B mutations are located in a hotspot Y197 in the immunoreceptor tyrosine-based activation motif (ITAM). Domains of the protein are represented according to the Uniprot database (www.uniprot.org).
Mutational landscape of 50 cases of DLBCL-AE. Bar graph shows mutated genes in 4 or more cases. The results are given in number of mutated cases per gene. The colored bars indicate the presence (orange) or absence (blue) of IRF4 rearrangement. A diagram of the relative positions of driver mutations is shown for IRF4, CARD11, CD79B, and MYD88. Multiple passenger mutations for IRF4 also are depicted. The x-axis indicates amino acid positions. The approximate location of somatic mutations identified in each gene is indicated. IRF4 mutations are mainly in the DNA binding domain. All CD79B mutations are located in a hotspot Y197 in the immunoreceptor tyrosine-based activation motif (ITAM). Domains of the protein are represented according to the Uniprot database (www.uniprot.org).
MYD88 gene mutations were mostly identified in the hotspot p.L265P; however, in 4/17 cases, variant mutations were observed (p.S219C, p.D148V, p.S243N, p.S149N), all of which were predicted as driver mutations. Multiple IRF4 mutations were identified in 4 of 8 cases, including synonymous variants. These mutations were exclusively found in the highly conserved N-terminal DNA-binding domain. In 2 additional cases (cases 26 and 44), only 1 IRF4 mutation was identified but predicted as passenger. All IRF4 mutations were exclusively identified in IRF4-rearranged cases. Multiple mutations in >3 cases were identified in IRF4 and PIM1. The pattern of these mutations is concordant with aberrant somatic hypermutation (aSHM) (supplemental Table 6). In the 20 cases analyzed with both panels, 67 mutations were verified (89%), whereas 8 mutations (5 PIM1, 1 BCL2, and 2 IRF4) were only identified with the SureSelectXT panel.
Molecular subgroup prediction
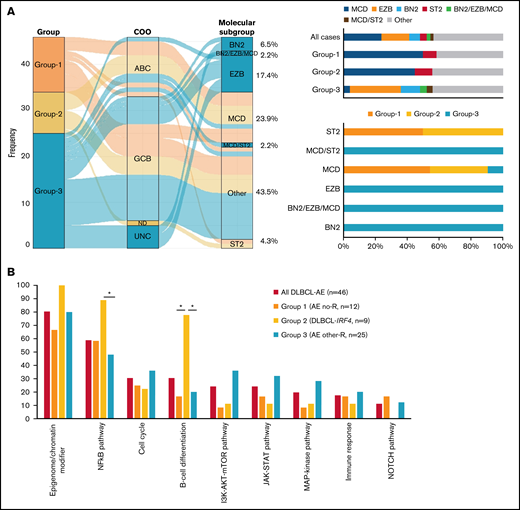
Molecular subgroup prediction of 46 DLBCL-AE cases was performed using the LymphGen tool.12 Only information on 42/114 genes used by the algorithm was available. This algorithm predicted 57% of the cases to belong to 1 of the molecular subgroups. Most cases were predicted as belonging to either the MCD group (24%) or the EZB group (17%), with few cases belonging to the BN2 (7%), ST2 (4%), MCD/ST2 (2%), and BN2/EZB/MCD (2%) groups (Figure 4A).
Molecular subgroup prediction and recurrent mutated pathways in DLBCL-AE. (A) Alluvial plot shows the frequency and relationship between groups, COO classification according to LYMPH2Cx, and/or HTG and molecular subgroup prediction according to LymphGen tool12 of 46 cases of DLBCL-AE. The bars indicate the molecular predicted subgroup in all cases and in each genetic group identified; 57% of all cases were assigned to a specific molecular subgroup. (B) Recurrent mutated pathways22 in 46 cases of DLBCL-AE. Genes included in each pathway are indicated in supplemental Table 1. Bar graph shows the total number of mutated cases for each pathway. Asterisk represents significant mutated pathways. AE, aberrant expressor CD10+BCL6+MUM1+; R, rearrangement.
Molecular subgroup prediction and recurrent mutated pathways in DLBCL-AE. (A) Alluvial plot shows the frequency and relationship between groups, COO classification according to LYMPH2Cx, and/or HTG and molecular subgroup prediction according to LymphGen tool12 of 46 cases of DLBCL-AE. The bars indicate the molecular predicted subgroup in all cases and in each genetic group identified; 57% of all cases were assigned to a specific molecular subgroup. (B) Recurrent mutated pathways22 in 46 cases of DLBCL-AE. Genes included in each pathway are indicated in supplemental Table 1. Bar graph shows the total number of mutated cases for each pathway. Asterisk represents significant mutated pathways. AE, aberrant expressor CD10+BCL6+MUM1+; R, rearrangement.
Correlation of clinical features, FISH, GEP, and mutational analysis
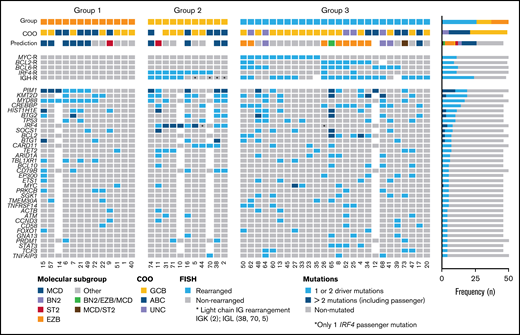
Based on primary events detected by FISH, 3 groups were recognized (Figure 5). Five cases without mutational analysis were excluded. Group 1 included 13/50 cases (26%) without translocations. There were 7 male and 6 female patients with a median age of 70 years (range 38-84 years). Eight cases (54%) were GCB-type whereas 7 cases were ABC-type (46%). In total, 66 driver mutations were identified with a mean of 5.5 mutations/case. The MCD mutational profile predominated in this group (6/12; 50%). Interestingly, 3 cases assigned to the GCB group by GEP (case 7 with HTG, case 15 with HTG and NanoString, and case 57 with NanoString) carried MYD88 and CD79B mutations and were predicted to belong to the MCD molecular subgroup. Summing the molecular subgroup and GEP, 9 cases (60%) showed ABC/MCD profile. Group 2 included 11/50 (22%) cases of DLBCL with IRF4 rearrangements (DLBCL-IRF4). In 2 cases, no IRF4 break was demonstrated; however, these 2 cases had a light chain IG break and IRF4 mutations indicating a cryptic IRF4 rearrangement. The group consisted of 3 male and 8 female patients with a median age of 67 years (range 37-85 years). From the 10 cases analyzed for GEP, 5 cases were subclassified as GCB and 5 as ABC. In total, 96 driver mutations (mean 10.7 mutations/case) were identified. Four cases were predicted as MCD mutational profile and 1 was assigned to the ST2 group. This group was characterized by 1 or multiple IRF4 mutations (82%) and frequent MYD88 (45%), CARD11 (36%), and CD79B (27%) mutations. Finally, group 3 included 26/50 cases (52%) with 1 or several translocations in BCL2/BCL6/MYC/IGH. The 2 cases with complex BCL2 and/or BCL6 translocations together with IRF4 were included in this group. There were 9 male and 17 female patients with a median age of 68 years (range 44-87 years). In this group, the GCB-type (16/26; 62%) and the EZB-type (8/25; 32%) mutational profile predominated. In total, 219 driver mutations were identified, with a mean of 8.8 mutations/case.
Overview of 50 cases of DLBCL with aberrant coexpression of CD10, BCL6, and MUM1 clustered in 3 groups. Oncoprint includes FISH results, molecular subgroup prediction according to LymphGen tool, COO classification according to Lymph2Cx, and/or HTG and frequently mutated genes (>3 cases). Each column of the plot represents 1 TP case and each line is a specific analysis. On the right side of the figure, the frequency of the particular result of the analysis is shown.
Overview of 50 cases of DLBCL with aberrant coexpression of CD10, BCL6, and MUM1 clustered in 3 groups. Oncoprint includes FISH results, molecular subgroup prediction according to LymphGen tool, COO classification according to Lymph2Cx, and/or HTG and frequently mutated genes (>3 cases). Each column of the plot represents 1 TP case and each line is a specific analysis. On the right side of the figure, the frequency of the particular result of the analysis is shown.
The presence of recurrent mutated pathways was evaluated in the 3 groups (Figure 4B). This analysis showed significantly more mutations in NF-κB pathway and B-cell differentiation genes in group 2 (DLBCL-IRF4) when compared with the other 2 groups (P < .05). Group 2 also carried more driver mutations per case when compared with group 1 (10.7 vs 5.5 driver/case; Wilcoxon, P = .01) and group 3 (10.7 vs 8.84 driver/case; Wilcoxon, P = .33) with significantly more IRF4, CARD11, and CD79B mutations (P < .05).
The clinical parameters including sex, age, and overall survival did not differ in the 3 groups (supplemental Table 7; supplemental Figure 2). These results were also observed when a correlation analysis was performed, including the clinical features and molecular characteristics of the 3 groups (mutations >4 cases) (supplemental Figure 3).
CN results of DLBCL-IRF4 cases in adults
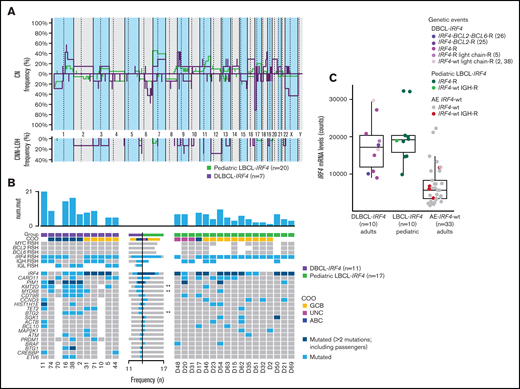
CN analysis of 7 DLBCL-IRF4 revealed a total of 118 genetic alterations (mean 17 alterations per case; range, 0-63 alterations) (Figure 6A; supplemental Table 8). Specifically, 62 CN losses, 43 CN gains, 8 homozygous deletions, and 5 high-copy gains were detected. The most recurrent alterations were 17p13.2 losses detected in 71% of cases (5/7 cases), including TP53 locus in all 5 cases. Additional frequent alterations observed were CN gain of 18q22.3-q23 and CN loss of 6q23.3-q24.1. Twenty-one regions of CNN-LOH were detected in 6 cases, with 11p11.2-p11.12 being the most recurrently affected region. Case 74 presented a complex genetic profile, with 63 regions of CNA and a chromothripsis-like pattern in chromosome 13. Interestingly, it harbored homozygous deletion of chromosome 6q23.3, encompassing TNFAIP3 gene.
Comparison of CN profile and genetic features of adult and pediatric LBCL-IRF4 cases. (A) Comparative plot of CN and CNN-LOH between 7 LBCL-IRF4 cases in adults and 20 LBCL-IRF4 cases in the pediatric population. No significantly different regions were identified. (B) GEP and mutational comparison between LBCL-IRF4 in adults (n = 11) and previously published profiles in children (n = 17) showed ABC COO more often (P = .05) in adults, with higher mutational load (10.6 vs 4.7 mutations/case; Wilcoxon test, P = .004) and higher frequency of KMT2D, MYD88, and BTG2 (P < .05, marked with asterisk). (C) IRF4 mRNA expression levels obtained from the IRF4_NM_002460.1 probe on the Lymph2Cx assay. IRF4 mRNA levels in LBCL-IRF4 in children and adults was similar but higher when compared with DLBCL-AE without IRF4 rearrangement (17 570 vs 6948; Wilcoxon test, P ≤ .01). AE, aberrant expressor CD10+BCL6+MUM1+; R, rearrangement; wt, wild-type.
Comparison of CN profile and genetic features of adult and pediatric LBCL-IRF4 cases. (A) Comparative plot of CN and CNN-LOH between 7 LBCL-IRF4 cases in adults and 20 LBCL-IRF4 cases in the pediatric population. No significantly different regions were identified. (B) GEP and mutational comparison between LBCL-IRF4 in adults (n = 11) and previously published profiles in children (n = 17) showed ABC COO more often (P = .05) in adults, with higher mutational load (10.6 vs 4.7 mutations/case; Wilcoxon test, P = .004) and higher frequency of KMT2D, MYD88, and BTG2 (P < .05, marked with asterisk). (C) IRF4 mRNA expression levels obtained from the IRF4_NM_002460.1 probe on the Lymph2Cx assay. IRF4 mRNA levels in LBCL-IRF4 in children and adults was similar but higher when compared with DLBCL-AE without IRF4 rearrangement (17 570 vs 6948; Wilcoxon test, P ≤ .01). AE, aberrant expressor CD10+BCL6+MUM1+; R, rearrangement; wt, wild-type.
Cases with IRF4 rearrangement together with BCL2 (case 25) and BCL2/BCL6 (case 26) rearrangements were also analyzed. They displayed a complex CN profile (22 and 24 regions of CNA) but were not higher than the cases with only IRF4 rearrangement. Interestingly, case 25 presented a CN gain in chromosome 18q21.33, with BCL2 locus in the breakpoint where the gain starts, supporting the presence of the rearrangement. Case 26 harbored CN loss of 17p13 but TP53 was not included in that alteration; instead, it was encompassed in a region of CN gain.
Comparison between pediatric and adult IRF4-rearranged cases
The comparison of CN profile between DLBCL-IRF4 in adults and previously published LBCL-IRF4 data in pediatric population (20 cases) (Figure 6A) showed no differences in terms of recurrent CN-altered regions; however, adult cases showed higher genetic complexity (16.85 alt/case in adults vs 6.25 alt/case in pediatric cases; P = .33), and more ABC COO (P = .05). Although there is a considerable difference in the genetic complexity based on CN alterations, it was not statistically significant, probably because of the small size of the compared groups.
Similarly, mutational comparison between DLBCL-IRF4 in adults (n = 9) and previously published profiles in children (n = 17) (Figure 6B) showed higher mutational load in adult cases (10.7 vs 4.7 mutations/case; Wilcoxon test, P = .004) with higher frequency of KMT2D, MYD88, and BTG2 mutations (Fisher’s exact test; P < .05).
IRF4 mRNA expression levels were obtained from the IRF4_NM_002460.1 probe on Lymph2Cx assay and compared between DLBCL-IRF4 in adults (n = 10), DLBCL-AE without IRF4 rearrangement (n = 33), and pediatric LBCL-IRF4 (n = 10) (Figure 6C). IRF4-rearranged cases in adults and children had similar IRF4 mRNA levels (number of counts; 17 570 vs 20 155; Wilcoxon test, P < .44), which was higher than those observed in DLBCL-AE without IRF4 rearrangement (number of counts; 17 570 vs 6948; Wilcoxon test, P ≤ .01).
Discussion
In this study, we comprehensively characterized the molecular features of 55 DLBCL cases with aberrant coexpression of CD10, BCL6, and IRF4/MUM1 that, according to the HA, belong to the GCB-type of DLBCL. We now demonstrate that DLBCL-AE is genetically heterogeneous and 41% (22/54) of the cases show either an ABC-type GEP or are unclassifiable, contributing to the discordant results between GEP and HA.21,27 These findings clearly indicate that DLBCL-AE should not be classified with the HA. An important aspect of the study was to demonstrate that DLBCL-AE is enriched in cases carrying IRF4 alterations (13/55; 24%).
Recent studies have shown that to capture the complexity of DLBCL, an integrative multilayer analysis is needed that includes GEP, mutational profile, chromosomal translocations, and CNAs.3,10-12 Accordingly, this multiplatform approach in DLBCL-AE identified 3 genetic subgroups. Group 1 included cases without chromosomal translocations and, despite expressing CD10, often showed GEP and/or mutational profile of ABC/MCD subgroup. Although MCD usually lacks GCB signature expression,12 we identified 3 cases with GCB-type GEP and MCD mutational landscape, indicating a clear discrepancy between genotype and phenotype of the tumor cells. Cases similar to these were not captured in the recently described genetic classification12 highlighting the challenges that present the extreme genetic and phenotypic heterogeneity of DLBCL for the development of precision therapy.
Group 2 was composed of cases with only IRF4 chromosomal translocation, including 2 cases with putative cryptic IRF4 translocations.20,22 In 5 cases, IGH was the partner of the translocation, whereas in 4 cases a break in the IG light chain was identified. A hallmark of LBCL-IRF4 in pediatric cases20 not observed in other cases of DLBCL28 is the association of GCB GEP and the presence of 1 or multiple IRF4 mutations, identified in this study in 82% of the cases. Because IRF4 mutations show the pattern of aSHM and predominant AID mutational signature,20,29 its presence is indicative of an IRF4 translocation and of potential use in the diagnostic setting. An important question is whether cases in adults represent the same disease as in children and young adults (≤25 years).19,20 The genetic comparison demonstrated that although pediatric and adults cases share the same recurrent altered CN regions and frequent mutations in IRF4- and NF-kB–related genes (MYD88, CARD11, and CD79B), as a group, adult cases showed higher genetic complexity, had a higher mutational load (10.7 vs 4.7 mutations/case; Wilcoxon test, P = .004) with more KMT2D, MYD88, CD79B, and BTG2 mutations (P < .05), and more often had an ABC-type GEP (P = .05).
Interestingly, by dissecting the IRF4 altered cases into GCB- vs ABC-type, important differences were observed. The 5 cases with GCB GEP showed lower genomic complexity and multiple IRF4 mutations, confirming that AID-driven aSHM is a GCB hallmark. The molecular prediction did not assign these cases into any of the 7 well-defined molecular subgroups,12 indicating that these cases have molecular features that distinguish them from other DLBCL. Furthermore, 3 cases (cases 5, 10, and 44) were in patients younger than age 40. Two cases presented in the tonsil and 1 in a submandibular lymph node. All 3 had excellent response to conventional chemotherapy and achieved complete remission. Nevertheless, similar cases can also occur in older patients (cases 31 and 71). In contrast, most elderly patients with IRF4 alterations showed an ABC GEP (5 cases) and carried 1 or no IRF4 mutations but frequent KMT2D (4/5), MYD88 (4/5), and BTG2 (3/5) mutations, which are acquired only sporadically as byproducts of aSHM.30 Recently, it has been shown that ABC-type lymphomas most probably derive from memory B cells that infrequently reenter the GC,31 explaining why these cases have no or only 1 IRF4 mutation. However, KMT2D mutations are not characteristic of ABC/MCD lymphomas; furthermore, KMT2D and MYD88 mutations are genetic events that increase with age and are rarely observed in the pediatric population.10 Importantly, in previous studies on DLBCL in adults,3,10-12 IRF4-rearranged cases have not been recognized or evaluated as a group, most probably precluding the correct identification of these cases with available algorithms. Although the number of cases of DLBCL-AE with IRF4 alterations in the study is relatively low, these findings suggest that patients under 40 years of age and/or with GCB GEP have the same clinical and genetic disease as patients with LBCL-IRF4 in the pediatric population. This finding supports the contention that molecular characteristics in “pediatric” DLBCL are associated with age and represent a continuum that appears to end close to 40 years of age rather than 18 years of age.32 In contrast, cases in elderly patients with DLBCL-IRF4 and IRF4 mutations, although belonging to the same group, show additional genetic alterations associated with age and are assigned to ABC GEP. Further analysis is warranted to confirm the molecular features of DLBCL-IRF4 in adults, which might influence treatment strategies in the future. Another important issue is that not all LBCL-IRF4 cases aberrantly coexpressed CD10, BCL6, and MUM1. Approximately 40% to 50% of the cases are CD10−, making the identification of these cases difficult.13,19,20,22,33
Group 3 was composed of DLBCL carrying 1 or several translocations in BCL2/BCL6/MYC/IGH and assigned mainly to the GCB/EZB group. The 3 cases predicted to be in the BN2 molecular subgroup carried BCL6 translocations. The 2 cases with complex chromosomal translocations affecting IRF4 and BCL2/BCL6 genes were included in this group because in addition to GCB GEP, they showed genetic alterations closer to those found in GCB-DLBCL and were predicted to be in the EZB molecular group.
In conclusion, we have demonstrated that cases of DLBCL-AE are genetically heterogeneous. An important novel finding was that 24% of these cases carried an IRF4 alteration. Furthermore, DLBCL-IRF4 in adults has some similarities but also important differences from its pediatric counterpart. Our data suggest that within the group of IRF4-rearranged cases in adults, there are genetic features similar to LBCL-IRF4 disease in children and young adults, with some differences inherent to the aging process. Cases with complex IRF4 translocations with BCL2 or BCL2/BCL6 genes have the molecular characteristics of GCB/BCL2/EZB-type DLBCL and should not be included in the group of DLBCL-IRF4 in adults. Cases of DLBCL-AE show different genetic features that may impact therapeutic strategies, emphasizing the value of molecular subtyping in DLBCL.
Acknowledgments
The authors thank Claudia Hermann, Esther Kohler, Noelia García, Elena Gonzalvo, and Ingrid López for the excellent technical assistance.
This work was supported in part by the Deutsche Forschungsgemeinschaft (DFG) under Germany's Excellence Strategy (iFIT) EXC2180-390900677 (L.Q.-M.); the Ministerio de Ciencia e Innovación (MCI) grant No. RTI2018-094274-B-I00 (E.C.); EDER: European Regional Development Fund “Una manera de hacer Europa” and Generalitat de Catalunya Suport Grups de Recerca AGAUR grant Nos. 2017-SGR-1142 (E.C.) and 2017-SGR-1107 (I.S.); Acció instrumental d’incorporació de científics i tecnòlegs PERIS 2016 (SLT002/16/00336) and Asociación Española Contra el Cáncer (AECC CICPFI6025SALA) (I.S.). E.C. is an academia researcher of the Institució Catalana de Recerca I Estudis Avançats of the Generalitat de Catalunya (ICREA). I.S. is a Miguel Servet researcher (CPII18/00015) from Instituto de Salud Carlos III.
Authorship
Contribution: L.Q.-M., E.C., and O.B. conceived and designed the study; S.S., B.M., I.S., J.S.-V., and L.F. performed the FISH analysis; J.E.R.-Z., F.O., A.K.M., J.S., and I.B. performed NGS mutational analysis; M.P., F.O., S.S., and L.F. performed the GEP analysis; J.S.-V. and I.S. performed the CNA analysis; L.M.R. and C.R. contributed material; L.Q.-M., E.C., O.B., F.F., L.F., and N.C.-d.-A. reviewed the cases; I.B., F.O., and I.S. supervised the experimental work; L.Q.-M., O.B., and I.S. wrote the manuscript; and L.F., N.C.-d.-A., E.C., and F.F. helped write the manuscript.
Conflict-of-interest disclosure: E.C. and L.M.R. are co-inventors of the Lymph2Cx gene expression profiling assay used in this study. The remaining authors declare no competing financial interests.
Correspondence: Leticia Quintanilla-Martinez, Institute of Pathology, University Hospital Tübingen, Liebermeisterstr 8, 72076 Tübingen, Germany; e-mail: leticia.quintanilla-fend@med.uni-tuebingen.de; and Olga Balagué, Pathology Department, Hematopathology Unit, Hospital Clinic de Barcelona, Calle de Villarroel 170, 08036 Barcelona, Spain; e-mail: OBALAGUE@clinic.cat.
References
Author notes
L.F. and N.C.-d.-A. contributed equally to this study.
E.C., O.B., and L.Q.-M. supervised this study equally.
The full-text version of this article contains a data supplement.