Key Points
The fetal-to-adult transition in HSC states is associated with a global acquisition of megakaryocytic chromatin accessibility signatures.
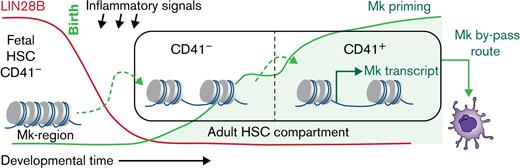
The developmentally restricted expression of LIN28B suppresses establishment of an Mk-biased HSC pool early in life.
Abstract
The fetal-to-adult switch in hematopoietic stem cell (HSC) behavior is characterized by alterations in lineage output and entry into deep quiescence. Here we identify the emergence of megakaryocyte (Mk)-biased HSCs as an event coinciding with this developmental switch. Single-cell chromatin accessibility analysis reveals a ubiquitous acquisition of Mk lineage priming signatures in HSCs during the fetal-to-adult transition. These molecular changes functionally coincide with increased amplitude of early Mk differentiation events after acute inflammatory insult. Importantly, we identify LIN28B, known for its role in promoting fetal-like self-renewal, as an insulator against the establishment of an Mk-biased HSC pool. LIN28B protein is developmentally silenced in the third week of life, and its prolonged expression delays emergency platelet output in young adult mice. We propose that developmental regulation of Mk priming may represent a switch for HSCs to toggle between prioritizing self-renewal in the fetus and increased host protection in postnatal life.
Introduction
In recent years, the use of single-cell transplantations and in situ fate-mapping approaches has cemented the existence of hematopoietic stem cells (HSCs) with preferred megakaryocyte (Mk) lineage output.1-4 This predisposition has inspired the terms Mk-biased or Mk-primed HSCs and has been linked to the expression of CD41, Vwf, cKithigh, and CD9, offering means to prospectively isolate such cells.3,5-8 Functional studies have unveiled a role for CD41-expressing HSCs in mitigating the risk for thrombocytopenia after acute stress by forging a top-down emergency megakaryopoiesis bypass route.9-12 However, when and how this poised HSC state is first established during ontogeny remains unexplored.
Fetal-to-adult changes in HSC behavior are well documented and hallmarked by a transition from rampant self-renewing cell divisions in the fetal liver (FL) into a deeply quiescent state in their bone marrow (BM) niche.13 The switch in HSC behavior was shown to occur precipitously in the 3- to 4-week window following birth and encompasses changes in HSC growth factor responsiveness, repopulation capacity, and lineage output patterns.14-19 Mechanistically, we and others have reported an instructive role for the heterochronic RNA-binding protein LIN28B in controlling this functionally defined switch.20-24 The expression of LIN28B is developmentally restricted and orchestrates a posttranscriptional molecular program that promotes enhanced self-renewal capacity characteristic of fetal HSCs.21 When ectopically expressed during adult hematopoiesis, LIN28B reprograms adult stem cells to assume fetal-like behavior and lineage output patterns.
Here we identify ubiquitous Mk priming as a chromatin-scripted feature associated with the fetal-to-adult transition in HSC states. Mirroring this molecular switch, we show that neonatal HSCs are selectively muted in their ability to induce early Mk differentiation events in response to inflammatory stress. CD41-expressing HSCs and Mk lineage preparedness gradually emerge with the developmental silencing of endogenous LIN28B at around 3 weeks of age. Genetic perturbations show that prolonged LIN28B expression delays the establishment of CD41+ Mk-biased HSCs and thereby blunts the emergency Mk response. Finally, we show that LIN28B protein expression is downregulated by inflammatory stress early in life. Our findings therefore highlight a key developmental intersection for environmental and developmental cues in controlling the postnatal switch in HSC function.
Materials and methods
Mice
Adult (8- to 10-week-old) C57Bl/6 N female and male mice were from Taconic. B6.Cg-Col1a1tm2(tetO-LIN28B)Gqda/J mice (Jax stock #023911) were obtained from the laboratory of Dr. George Daley25 (Harvard Medical School). B6-Cg-Gt(ROSA)26Sortm1(rtTA∗M2)Jae/J mice (Jax stock #006965) were from The Jackson Laboratory.26 The two were intercrossed to obtain trans-heterozygous tet-Lin28b mice used in experiments. In all tet-Lin28b experiments, doxycycline (DOX)-treated littermate control mice harboring the B6-Cg-Gt (ROSA)26Sortm1(rtTA∗M2)Jae/J allele were used. Lin28b−/− (B6.Cg-Lin28btm1.1Gqda/J Jax #023917) mice were from The Jackson Laboratory.27Ifnar−/− mice (B6.129S2-Ifnar1tm1Agt/Mmjax #32045 were from The Jackson Laboratory.28 The Lin28b-eGFP constitutive knock-in strain was generated at Cyagen Biosciences by replacing the endogenous Lin28b TAG stop codon with a 3xGGGGS-eGFP cassette by embryonic stem cell targeting. All animal procedures were performed and mice are kept in accordance with ethical permits approved by the Swedish Board of Agriculture.
In vivo treatment with polyinosinic-polycytidylic acid
To induce acute inflammation, mice were injected intraperitoneally with a single dose of 10 mg/kg polyinosinic-polycytidylic acid (pIpC) (MilliporeSigma) and killed for analysis 16 hours after treatment. Alternatively, 3 consecutive weekly intraperitoneal injections of the same dose were initiated in 10-day-old mice that were ultimately killed at 7 to 9 weeks of age for end point analysis.
Flow cytometry analysis and sorting
BM cells were extracted by crushing the bones from forelimbs, hind limbs, and hips with mortar and pestle as previously described.29 BM was subjected to red blood cell lysis with ammonium chloride. FL was dissected and brought into single-cell suspension by pipetting. Red blood cell lysis was not performed for FL. For HSC isolation, BM and FLs were pre-enriched by MACS (Miltenyi Biotec) depletion of lineage-positive cells (BM) or Ter119+ cells (FL) according to manufacturer’s instructions. All steps were performed with Hanks balanced salt solution (Gibco) supplemented with 0.5% bovine serum albumin (BSA) and 2 mM EDTA. This buffer was also used for antibody staining at a density of 10 × 106 cells/100 μL volume for 30 minutes in the dark and on ice. Antibodies are detailed in supplemental Table 1. All fluorescence-activated cell sorting (FACS) experiments were performed at the Lund Stem Cell Center FACS Core Facility (Lund University) on BD FACS Aria III, BD FACS Aria IIu, BD Fortessa, BD Fortessa X20, and BD LSRII instruments. Bulk populations were sorted by using a 70 μm nozzle, 0.32.0 precision mask, and flow rate of 5000 to 6000 events per second. Single-cell sorts were performed with a 85 or 100 μm nozzle, single-cell precision mask, and index sorting at flow rate 2000 to 2500 events per second. Lineage panel used for FACS was Ter119–Gr1–CD11b–CD3–B220–. Note that CD11b was left out of the lineage panel for FL and neonates, including 19-day-old BM samples, for both analysis and sorting (Table 1).30
Gating strategies used for FACS sorting
| FL HSC CD41– | Lineage–Sca1+cKit+ (LSK) CD48–CD150+Flt3– CD41– |
| ABM HSC CD41– | Lineage–Sca1+cKit+ (LSK) CD48–CD150+Flt3– CD41– |
| ABM HSC CD41+ | Lineage–Sca1+cKit+ (LSK) CD48–CD150+Flt3– CD41+ |
| ABM MkP | Lineage–Sca1–cKit+ CD150+CD41+ |
| FL HSC CD41– | Lineage–Sca1+cKit+ (LSK) CD48–CD150+Flt3– CD41– |
| ABM HSC CD41– | Lineage–Sca1+cKit+ (LSK) CD48–CD150+Flt3– CD41– |
| ABM HSC CD41+ | Lineage–Sca1+cKit+ (LSK) CD48–CD150+Flt3– CD41+ |
| ABM MkP | Lineage–Sca1–cKit+ CD150+CD41+ |
FACS analysis was performed by using FlowJo version 10.5.3 (FlowJo LLC). Analysis gate for CD41 in HSCs was set based on fluorescence minus one control and the Mk progenitor (MkP) internal reference population.31 For the Lin28b-eGFP mouse model, wild-type littermate controls from each age were used to set the enhanced green fluorescent protein (eGFP)-positive gate. FlowJo Uniform Manifold Approximation and Projection (UMAP) plug-in32 was used to generate merged UMAP visualizations (Figure 1). First, a concatenated file was generated with equal events from all samples (FlowJo DownSample plugin). The UMAP Euclidean distance function was used with nearest neighbors = 15 and minimum distance = 0.5. Parameters included: forward scatter area, height, and width; side scatter area, height, and width; and all fluorochrome and comp-fluorochrome parameters.
Mk lineage priming of HSCs is developmentally timed. (A) Gene set enrichment analysis comparing bulk RNA-sequencing results from sorted E14.5 FL and ABM HSCs (lineage–Sca1+cKit+ [LSK] CD48–CD150+Flt3–) for the following lineage-restricted progenitor signatures3,37: MkPs, pre-erythrocyte colony-forming units (preCFU-E), common lymphoid progenitors (CLP), and pre-granulocyte/macrophage progenitors (preGM). Normalized enrichment score (NES) and false discovery rate (FDR) are indicated. n = 2 to 3 biological replicates. (B) Normalized RNA-sequencing values for the average abundance of indicated transcripts per kilobase million. (C) Representative FACS analysis of CD41+ HSCs from E14.5 FL and ABM. (D) Mean frequency of CD41+ cells among HSCs from mice of the indicated ages. (E) Representative FACS analysis of MkPs in E14.5 and ABM. (F) Mean frequency of CD150+CD41+MkPs in Lin–cKit+Sca1– HPCs from mice of the indicated ages (n = 4-7). Statistical significance determined by Kruskal-Wallis test followed by Dunn’s multiple comparisons test comparing all ages vs E14.5 FL. (G) Representative images from day 7 single HSC cultures. (H) Analysis of day 7 single HSC cultures from the indicated ages. Wells with dead or no cells were removed before analysis (6%-8% of total wells). d, days; FSC, forward scatter; w, week(s).
Mk lineage priming of HSCs is developmentally timed. (A) Gene set enrichment analysis comparing bulk RNA-sequencing results from sorted E14.5 FL and ABM HSCs (lineage–Sca1+cKit+ [LSK] CD48–CD150+Flt3–) for the following lineage-restricted progenitor signatures3,37: MkPs, pre-erythrocyte colony-forming units (preCFU-E), common lymphoid progenitors (CLP), and pre-granulocyte/macrophage progenitors (preGM). Normalized enrichment score (NES) and false discovery rate (FDR) are indicated. n = 2 to 3 biological replicates. (B) Normalized RNA-sequencing values for the average abundance of indicated transcripts per kilobase million. (C) Representative FACS analysis of CD41+ HSCs from E14.5 FL and ABM. (D) Mean frequency of CD41+ cells among HSCs from mice of the indicated ages. (E) Representative FACS analysis of MkPs in E14.5 and ABM. (F) Mean frequency of CD150+CD41+MkPs in Lin–cKit+Sca1– HPCs from mice of the indicated ages (n = 4-7). Statistical significance determined by Kruskal-Wallis test followed by Dunn’s multiple comparisons test comparing all ages vs E14.5 FL. (G) Representative images from day 7 single HSC cultures. (H) Analysis of day 7 single HSC cultures from the indicated ages. Wells with dead or no cells were removed before analysis (6%-8% of total wells). d, days; FSC, forward scatter; w, week(s).
Platelet recovery after pIpC
Platelet concentrations were assessed by using a Sysmex automated hematology analyzer (Sysmex KX-21N) on whole blood collected by tail vein puncture in EDTA-coated tubes. Starting point blood values were assessed immediately before pIpC treatment.
Single-cell HSC cultures
E14.5 and adult BM (ABM) HSCs (lineage–Sca1+cKit+ CD48–CD150+Flt3–) were index sorted based on CD41 expression with a single-cell precision mask. Cells were cultured in Terasaki plates containing 10 μL Ham’s F-12 nutrient mix media (Gibco) with 10 ng/mL animal-free stem cell factor (PeproTech), 100 ng/mL animal-free thrombopoietin (TPO; PeproTech), 1 mg/mL polyvinyl alcohol (87%-90% hydrolyzed; MilliporeSigma), 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 1× penicillin-streptomycin-glutamine (Gibco), and 1× insulin-transferrin-selenium-ethanolamine (Gibco).33,34 After 7 days, single-cell wells were imaged by using the Olympus CKX53 microscope and scored based on the presence or absence of Mk/MkP clones.
Single-cell assay for transposase-accessible chromatin sequencing
Isolation, washing, and counting of nuclei suspensions were performed according to the Low Cell Input Nuclei Isolation Demonstrated Protocol: Nuclei Isolation for Single Cell ATAC Sequencing (10x Genomics). Briefly, ∼20 000 FL CD41– HSCs, 20 000 ABM CD41– HSCs, 6800 ABM CD41+ HSCs, and 20 000 adult MkPs were added to separate 2-mL microcentrifuge tubes and centrifuged (300g for 5 minutes at 4°C). The cells were washed once in 45 μL phosphate-buffered saline/0.04% BSA and centrifuged (300g for 5 minutes at 4°C). The supernatant was removed without disrupting the cell pellet, and 45 μL chilled Lysis Buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, 0.1% Nonidet P40 Substitute, 0.01% digitonin, and 1% BSA) was added and pipette-mixed 10 times. The microcentrifuge tube was then incubated on ice for 3 minutes. After lysis, 50 μL of chilled Wash Buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, and 1% BSA) was added, and the resulting solution was mixed by pipetting. Nuclei were centrifuged (500g for 5 minutes at 4°C) and the supernatant removed without disrupting the nuclei pellet. Nuclei were resuspended in chilled Diluted Nuclei Buffer (2000153; 10x Genomics) at approximately 500 to 1700 nuclei per microliter based on the starting number of cells. The resulting nuclei concentration was then manually determined by staining 2 μL of the nuclei Trypan Blue followed by hemocytometer counting. The total number of nuclei used as input for single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) library preparation was as follows: FL CD41– HSCs, 3800; ABM CD41– HSCs, 5400; ABM CD41+ HSCs, 2400; and MkPs, 8700.
scATAC-seq libraries were prepared according to the Chromium Next GEM Single Cell ATAC Reagent Kits User Guide (v1.1 chemistry; 10x Genomics;) by the Center for Translational Genomics, Lund University. Nuclei were sequenced on an Illumina NovaSeq with ∼81 000 to 235 000 read pairs per cell, and reads fastq-files were generated with the cellranger-atac mkfastq command. Data pre-processing and analysis are described in the supplemental Methods.
Statistical test
Statistical analyses were performed in GraphPad Prism 8 (GraphPad Software). Tests for statistical significance are described in the figure legends for the relevant graphs. In all legends, n denotes biological replicates.
Results
Mk lineage priming of HSCs is developmentally timed
To explore changes in lineage priming during the transition from E14.5 FL to ABM HSCs, we performed bulk RNA-sequencing followed by gene set enrichment analysis.35,36 Transcripts associated with Mk, erythroid, lymphoid, and myeloid lineage restriction3,37 were compared between FL and ABM HSCs (supplemental Figure 1A). The analysis identified a selective enrichment for an MkP transcript signature in ABM compared with FL HSCs (Figure 1A) consistent with increased Mk lineage priming. Among Mk transcripts upregulated in ABM HSCs is Itga2b (Figure 1B), encoding for the integrin subunit CD41 that is ubiquitously expressed in the Mk/platelet lineage. Because CD41+ HSCs exhibit an Mk lineage bias both functionally and transcriptionally,38 we measured their abundance in mice ranging from E14.5 to 3 months of age to explore potential changes in lineage priming (Figure 1C-D; supplemental Figure 1B). FACS data show an absence of CD41+ HSCs in fetal and neonatal life. Instead, CD41+ HSCs become detectable first in 3.5-week-old mice and gradually increase in frequency into adulthood. This transition is accompanied by a concomitant increase in MkP frequency among cKit+Sca1– myelo-erythroid progenitors (Figure 1E-F; supplemental Figure 1C). Our results extend previous findings in aged mice5 and suggest that the generation of Mk-biased HSCs is a continuous process first initiated in postnatal life.
To verify that changes in CD41+ HSC frequency translate into Mk output differences, we used a single-cell culture system in which thrombopoietin (TPO) supplementation allows for the parallel assessment of HSC expansion and Mk lineage differentiation.33 Two to three hundred single cells were sorted and individually cultured from FL and ABM HSC populations separated according to CD41 status (supplemental Figure 1D). Seven days later, the cultures were manually scored for Mk output (no Mk, Mk hematopoietic stem and progenitor cell [HSPC] mix, and Mk only) (Figure 1G). Indeed, ABM CD41+ HSC clones produced the highest frequency of “Mk only” cultures consistent with a clear lineage bias (Figure 1H). Upon closer investigation, CD41– ABM clones produced more “Mk HSPC mixed cultures” at the expense of “no Mk” cultures compared with CD41– FL clones. This finding hinted that the basis for Mk lineage priming might be a broader phenomenon among adult HSCs not solely restricted to the CD41+ subset.
Fetal-to-adult HSC transition is associated with global chromatin accessibility signatures for Mk lineage priming
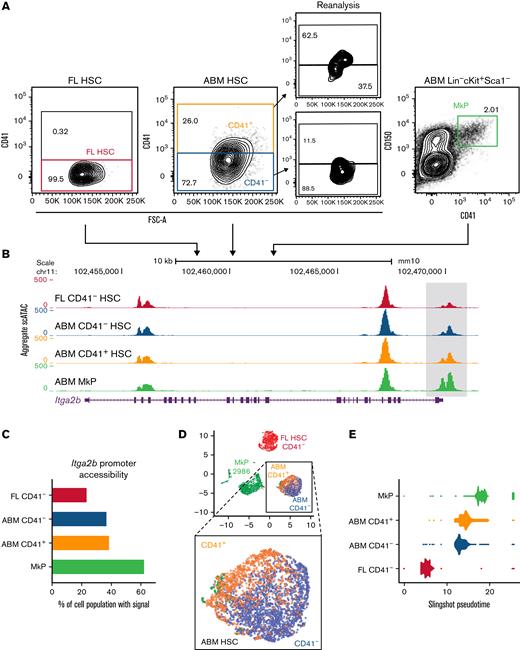
Changes in platelet function and output during ontogeny39 are well documented but have not previously been linked to chromatin-scripted Mk priming in the HSC compartment. Although fetal and adult HSC chromatin status has been compared,40,41 these analyses performed on bulk HSCs did not allow for high-resolution dissection of Mk-priming events. To this end, we performed scATAC-seq on sorted FL CD41– HSCs, ABM CD41– HSCs, ABM CD41+ HSCs, and lineage-committed ABM CD41+ MkPs (Figure 2A). After initial processing,42 the experiment yielded data for a total of 8822 cells with 198 127 unique accessible peaks across the 4 cell types (supplemental Figure 2A). Interestingly, ABM CD41– HSCs exhibit higher chromatin accessibility at the Itga2b promoter region compared with FL CD41– HSCs, at a frequency that is comparable to that of ABM CD41+ HSCs (Figure 2B-C). This example showcases our ability to detect Mk-associated chromatin accessibility changes before CD41 expression.
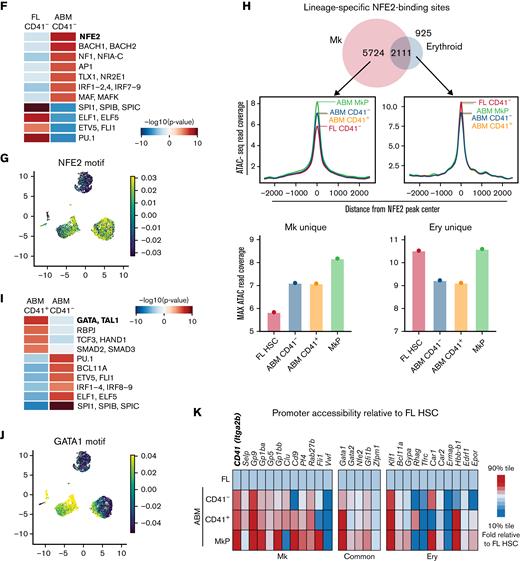
Fetal-to-adult HSC transition is associated with chromatin accessibility signatures for Mk lineage priming. (A) Cell-sorting strategy for populations subjected to scATAC-seq. Post-sort reanalysis is shown for CD41– and CD41+ABM HSCs to indicate CD41 separation. (B) Aggregate scATAC-seq tracks show chromatin accessibility at the Itga2b locus of the sorted populations. Box highlights promoter region. (C) Itga2b promoter accessibility represented as percentage of single cells with detectable ATAC-seq signal within the promoter region. (D) UMAP visualization of single FL HSC CD41– (red = 2403 HSCs), ABM CD41– (blue = 2310 HSCs), ABM CD41+ (orange = 1123 HSCs), and MkPs (green = 2986 ABM MkPs). Magnified section shows combined ABM HSC cluster. (E) Slingshot pseudotime analysis44 based on PCA trajectory of the 10 000 most variable peaks. Each cell is represented as one dot. Accessibility analysis in panels F and I was performed on aggregate scATAC-seq data for the indicated samples. (F) Top enriched transcription factor–binding motifs for differentially accessible regions between ABM CD41– HSCs and FL CD41– HSCs. (G) UMAP visualization NFE2 motif accessibility enrichment across all samples at the single-cell level. Blue to yellow hues show relative accessibility. (H) Accessibility read coverage for previously published erythroid- and Mk-specific NFE2-binding sites defined per chromatin immunoprecipitation sequencing.47 Bar plots show maximum ATAC read coverage values for the indicated samples. (I) Top enriched transcription factor–binding motifs for differentially accessible regions between ABM CD41+ HSCs and ABM CD41– HSCs. (J) UMAP visualization of GATA1 motif accessibility enrichment across all samples at the single-cell level. Blue to yellow hues show relative accessibility. (K) Relative promoter accessibility of select Mk, erythroid, and shared lineage genes represented as the fraction of single cells with detectable scATAC-seq signal relative to the value of FL CD41– HSCs. FSC-A, forward scatter area.
Fetal-to-adult HSC transition is associated with chromatin accessibility signatures for Mk lineage priming. (A) Cell-sorting strategy for populations subjected to scATAC-seq. Post-sort reanalysis is shown for CD41– and CD41+ABM HSCs to indicate CD41 separation. (B) Aggregate scATAC-seq tracks show chromatin accessibility at the Itga2b locus of the sorted populations. Box highlights promoter region. (C) Itga2b promoter accessibility represented as percentage of single cells with detectable ATAC-seq signal within the promoter region. (D) UMAP visualization of single FL HSC CD41– (red = 2403 HSCs), ABM CD41– (blue = 2310 HSCs), ABM CD41+ (orange = 1123 HSCs), and MkPs (green = 2986 ABM MkPs). Magnified section shows combined ABM HSC cluster. (E) Slingshot pseudotime analysis44 based on PCA trajectory of the 10 000 most variable peaks. Each cell is represented as one dot. Accessibility analysis in panels F and I was performed on aggregate scATAC-seq data for the indicated samples. (F) Top enriched transcription factor–binding motifs for differentially accessible regions between ABM CD41– HSCs and FL CD41– HSCs. (G) UMAP visualization NFE2 motif accessibility enrichment across all samples at the single-cell level. Blue to yellow hues show relative accessibility. (H) Accessibility read coverage for previously published erythroid- and Mk-specific NFE2-binding sites defined per chromatin immunoprecipitation sequencing.47 Bar plots show maximum ATAC read coverage values for the indicated samples. (I) Top enriched transcription factor–binding motifs for differentially accessible regions between ABM CD41+ HSCs and ABM CD41– HSCs. (J) UMAP visualization of GATA1 motif accessibility enrichment across all samples at the single-cell level. Blue to yellow hues show relative accessibility. (K) Relative promoter accessibility of select Mk, erythroid, and shared lineage genes represented as the fraction of single cells with detectable scATAC-seq signal relative to the value of FL CD41– HSCs. FSC-A, forward scatter area.
UMAP visualization of global chromatin accessibility showed cells positioning in clear sample-wise clusters except for ABM CD41– and CD41+ HSCs, which localized to opposite poles of the same cluster (Figure 2D). Thus, despite differential lineage bias, ABM CD41+ and CD41– HSCs possess highly analogous chromatin states consistent with their shared stem cell identity established by transplantation studies.5 Conversely, FL and ABM CD41– HSCs fell into highly distinct clusters, highlighting age-specific differences in chromatin accessibility independent of CD41 status. These global developmental changes in chromatin accessibility cannot be accounted for by changes in cell cycle status, as reviewed by Ma et al43 (supplemental Figure 2B), and are consistent with the observed increase in Mk lineage output by ABM compared with FL CD41– HSCs (Figure 1G). Supporting this, slingshot pseudotime analysis44 positioned ABM CD41– HSCs closer to the CD41+ ABM HSCs than the naive CD41– FL HSCs (Figure 2E; supplemental Figure 2C), suggesting that initial steps toward Mk priming is established during the fetal-to-adult CD41– HSC transition.
To explore upstream transcription factors that might orchestrate the FL to ABM CD41– HSC changes in chromatin accessibility, we performed binding motif enrichment analysis within differentially accessible regions between the 2 samples (Figure 2F; supplemental Figure 2A,D). Interestingly, the binding motif for NFE2, an essential Mk and erythroid lineage transcription factor,45,46 was most enriched in ABM over FL CD41– HSCs (Figure 2G). To distinguish whether the increased accessibility at NFE2-binding sites reflects differences in erythroid or Mk lineage priming, we took advantage of previously published Mk- and erythroid-specific chromatin immunoprecipitation–sequencing peak coordinates for NFE2.47 Differential accessibility was queried at lineage-specific NFE2-binding sites. Interestingly, only binding sites unique to the Mk lineage showed higher accessibility in adult compared with FL CD41– HSCs (Figure 2H). Thus, transcription factor motif accessibility analysis supports a scenario in which the main fetal-to-adult shift in HSC lineage priming is megakaryocytic.
Among the chromatin regions more open in adult CD41+ HSCs compared with CD41– HSCs (Figure 2I; supplemental Figure 2E), the motif for GATA was most enriched (Figure 2J). As with NFE2, the GATA family of transcription factors plays an essential role in both Mk and erythroid lineage differentiation.47-49 In contrast, the binding motif for the erythroid lineage-specific transcription factor BCL11A was progressively less open during the FL to ABM and CD41– to CD41+ ABM HSC transitions (Figure 2I; supplemental Figure 2F). Thus, increased GATA-binding site accessibility likely reflects advancing Mk priming in CD41+ compared with CD41– ABM HSCs.
To directly query epigenetic Mk priming in the inferred population hierarchy, we analyzed the accessibility of a panel of known Mk- and erythroid-associated gene loci. From FL to ABM CD41– HSCs, we observed increased promoter accessibility of genes associated with the Mk but not the erythroid lineage (Figure 2K). As expected, the observed selective increase in Mk-accessible promoters is further augmented in the CD41+ ABM HSCs, consistent with a stepwise progression toward the Mk lineage. Taken together, a level of early Mk priming in adult CD41– HSCs was identified that is further specified within the CD41+ subset, and a component of ontogenic control in the chromatin scripted program for Mk output is shown.
Neonatal HSCs are resistant to inflammation-induced acute Mk lineage differentiation
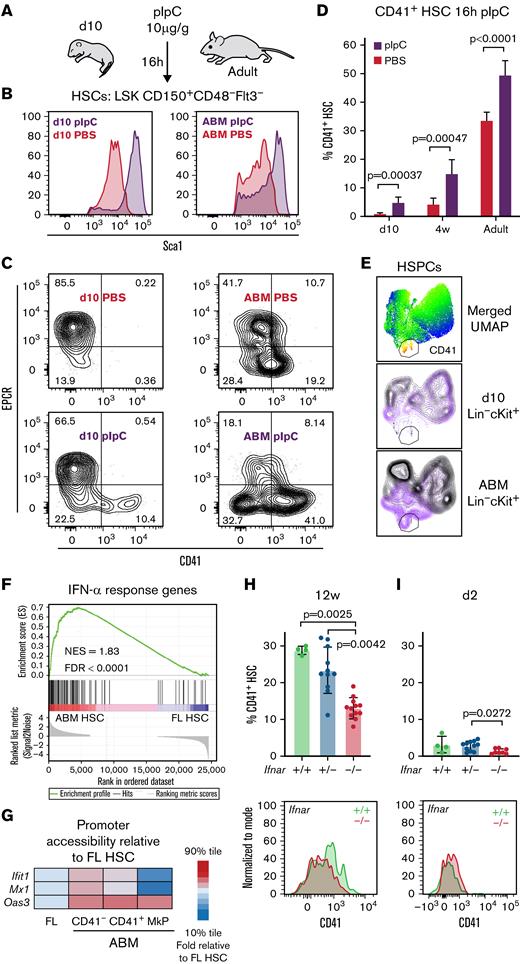
To test whether the paucity in Mk lineage priming observed in early-life HSCs correlates with altered emergency Mk lineage differentiation, we treated adult and 10-day-old neonatal mice with a single dose of pIpC (10 μg/g) to mimic an acute viral infection50 (Figure 3A). All immunophenotypic BM HSCs regardless of age responded 16 hours later by upregulating Sca-1 and type I interferon response genes (Figure 3B; supplemental Figure 3A-B). Despite the apparent ability of neonatal HSCs to acutely respond to inflammatory insult, surface CD41 levels remained unchanged in the majority of cells (Figure 3C-D). This finding is in contrast to adult HSCs in which a marked increase was observed in both the percentage and median fluorescence intensity of CD41+ cells, consistent with a previous report.9 Furthermore, this CD41 upregulation was accompanied by an increase in Mk-associated transcripts at the single-cell level (supplemental Figure 3A-B) and a collapse in the expression of the stem cell marker endothelial protein C receptor (EPCR), which was sustained in treated neonatal HSCs (Figure 3C). These age-specific HSC responses to pIpC treatment are mirrored by a downstream increase in the output of CD41+ Mk progenitors (Figure 3E). We conclude that neonatal HSCs are better able to uphold their undifferentiated stem cell status in the face of pIpC insult compared with adult HSCs that undergo early Mk differentiation.
Neonatal HSCs are resistant to inflammation-induced acute Mk lineage differentiation. (A) pIpC challenge in 10-day-old (d10) and adult mice. (B-C) Representative FACS data showing BM HSCs for experiment outlined in panel A. (D) CD41+ HSC frequency in PBS- or pIpC-treated mice of the indicated ages (n = 6-9 for each treatment/age). Error bars indicate standard deviation of the mean. Statistical significance determined by multiple t tests using the Holm-Šídák method. (E) Representative merged UMAP visualization of concatenated FACS data from d10 and ABM HSPCs 16 hours after pIpC (purple) or PBS (black) treatment. (F) Gene set enrichment analysis of the interferon-α (IFN-α) response hallmark gene set.66 (G) Relative accessibility for select IFN-inducible genes represented as the fraction of single cells with detectable ATAC-seq signal. (H) FACS analysis of 12-week-old (12w) Ifnar-deficient mice. Upper panel: CD41+ frequency in HSCs of the indicated genotypes (n = 4-12). Lower panel: representative histogram overlay of CD41 expression. (I) FACS analysis of 2-day-old (d2) Ifnar-deficient pups. Upper panel: CD41+ frequency in HSCs of the indicated genotypes (n = 4-11). Lower panel: representative histogram overlay of CD41 expression. Statistical significance was determined by using the Kruskal-Wallis test followed by Dunn’s multiple comparisons test. EPCR, endothelial protein C receptor; FDR, false discovery rate; LSK, lineage–Sca1+cKit+; NES, normalized enrichment score.
Neonatal HSCs are resistant to inflammation-induced acute Mk lineage differentiation. (A) pIpC challenge in 10-day-old (d10) and adult mice. (B-C) Representative FACS data showing BM HSCs for experiment outlined in panel A. (D) CD41+ HSC frequency in PBS- or pIpC-treated mice of the indicated ages (n = 6-9 for each treatment/age). Error bars indicate standard deviation of the mean. Statistical significance determined by multiple t tests using the Holm-Šídák method. (E) Representative merged UMAP visualization of concatenated FACS data from d10 and ABM HSPCs 16 hours after pIpC (purple) or PBS (black) treatment. (F) Gene set enrichment analysis of the interferon-α (IFN-α) response hallmark gene set.66 (G) Relative accessibility for select IFN-inducible genes represented as the fraction of single cells with detectable ATAC-seq signal. (H) FACS analysis of 12-week-old (12w) Ifnar-deficient mice. Upper panel: CD41+ frequency in HSCs of the indicated genotypes (n = 4-12). Lower panel: representative histogram overlay of CD41 expression. (I) FACS analysis of 2-day-old (d2) Ifnar-deficient pups. Upper panel: CD41+ frequency in HSCs of the indicated genotypes (n = 4-11). Lower panel: representative histogram overlay of CD41 expression. Statistical significance was determined by using the Kruskal-Wallis test followed by Dunn’s multiple comparisons test. EPCR, endothelial protein C receptor; FDR, false discovery rate; LSK, lineage–Sca1+cKit+; NES, normalized enrichment score.
Considering the reported instructive role of inflammatory signaling in the transition from a fetal to an adult HSC molecular landscape,41,51 we hypothesized that Mk lineage priming might be similarly instructed. In line with this notion, we observed a clear interferon-related transcript and chromatin accessibility signature enriched in ABM over FL HSCs (Figure 3F-G). To test if the emergence of Mk-biased HSCs is dependent on type I interferon signaling, we analyzed BM HSCs from Ifnar−/− mice at 12 weeks of age (Figure 3H; supplemental Figure 3C). Our results show a 50% decrease in CD41+ HSC frequency in Ifnar−/− compared with wild-type littermates. Considering the role of sterile inflammatory signaling at multiple stages of prenatal hematopoiesis,41,52 and the surface expression of CD41 in emerging definitive HSPCs in the embryo,53,54 we wanted to assess the degree of Ifnar-dependent accumulation of CD41 in neonates. Our results show low levels of Ifnar-dependent CD41 expression at day 2 (Figure 3I), suggesting that the accumulation of CD41+ HSCs during the fetal-to-adult transition occurs predominantly but not exclusively because of postnatal inflammatory signals.
Sustained LIN28B expression limits functional emergency megakaryopoiesis
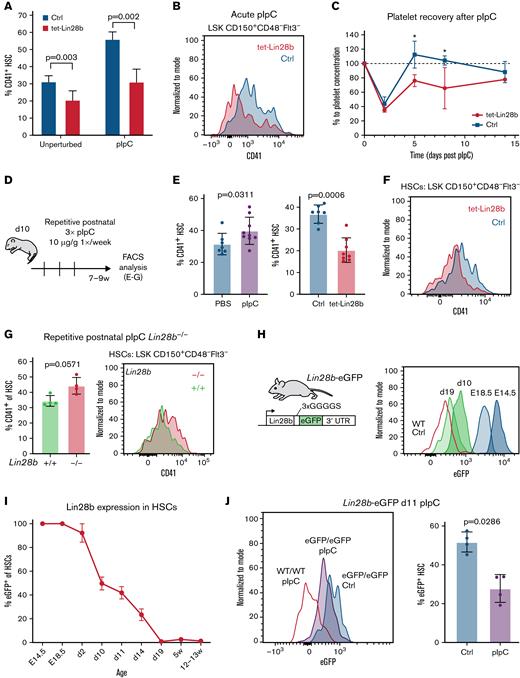
In addition to cell extrinsic inflammatory cues, we hypothesized that early Mk lineage priming of HSCs might be regulated by their endogenous levels of the fetal stemness factor LIN28B. scATAC-seq showed higher chromatin accessibility at the promoter region of the Lin28b locus in FL (supplemental Figure 4A-B), consistent with its developmentally restricted expression pattern in HSCs.21,22 To test whether sustained LIN28B protein alters the generation of Mk-biased HSCs, we analyzed tet-Lin28b–inducible mice kept on continuous DOX food following birth (Figure 4A, unperturbed; supplemental Figure 4C). Our results show a significant decrease in CD41+ HSC frequency by 8 weeks of age, indicating that sustained LIN28B expression is sufficient to diminish the spontaneous emergence of Mk-biased HSCs. To test whether LIN28B affects emergency megakaryopoiesis, DOX-fed tet-Lin28b mice were subjected to acute pIpC challenge at 8 weeks of age. LIN28B-induced mice displayed less surface CD41 upregulation at 16 hours’ posttreatment (Figure 4A-B; supplemental Figure 4D), reminiscent of the behavior of neonatal HSCs (Figure 3C-D). Importantly, peripheral blood platelet recovery monitored across 15 days after pIpC insult was notably delayed in LIN28B-induced mice (Figure 4C). These results show that sustained LIN28B insulates the HSC compartment against inflammation-induced emergency megakaryopoiesis.
Sustained postnatal LIN28B expression limits inflammation-induced Mk lineage output. (A) CD41+ frequency in phenotypic HSCs from 8-week-old tet-Lin28b and littermate (ctrl) mice induced with DOX from birth. Mice were either kept unperturbed or treated with 10 μg/g pIpC and analyzed by using FACS 16 hours later. Statistical significance determined by multiple t tests using the Holm-Šídák method. n = 3 to 8. (B) Representative FACS histograms showing CD41+ HSC frequency in pIpC-treated mice. (C) Relative platelet frequency in peripheral blood from tet-Lin28b and ctrl mice after pIpC treatment measured by FACS. Statistical significance for each time point was determined by multiple t tests assuming equal standard deviation. Day 5, P = .003; day 8, P = .002. (D) Experimental outline for repetitive postnatal pIpC treatments of juvenile mice. Three consecutive weekly treatments of intraperitoneal injection of 10 μg/g pIpC started in 10-day-old (d10) wild-type (WT) pups followed by end point analysis between 7 and 9 weeks of age. (E) FACS analysis of CD41+ HSC frequency (left) in WT mice after repetitive pIpC or PBS injections and (right) tet-Lin28b and ctrl mice after repetitive pIpC treatment (n = 7-9). P values were determined by using the Mann-Whitney test. (F) Representative FACS histograms of CD41+ HSCs from repetitive pIpC-treated tet-Lin28b and control mice. (G) FACS analysis of CD41+ frequency in HSCs from 8-week-old mice of the indicated genotypes treated with the repetitive postnatal pIpC regimen. P value was determined by using the Mann-Whitney test. (H) LIN28B-eGFP reporter mouse model design and FACS histograms showing decreasing LIN28B protein levels across developmental time. (I) Compiled frequency of eGFP+ HSCs at the indicated ages. eGFP+ gating is based on WT littermate controls (n = 2-7 per time point). (J) FACS analysis of eGFP+ levels and frequency in HSCs from 11-day-old (d11) mice 16 hours after 10 μg/g pIpC challenge. ∗P < .05. P value was determined by using the Mann-Whitney test. 3′UTR, 3′ untranslated region; LSK, lineage–Sca1+cKit+.
Sustained postnatal LIN28B expression limits inflammation-induced Mk lineage output. (A) CD41+ frequency in phenotypic HSCs from 8-week-old tet-Lin28b and littermate (ctrl) mice induced with DOX from birth. Mice were either kept unperturbed or treated with 10 μg/g pIpC and analyzed by using FACS 16 hours later. Statistical significance determined by multiple t tests using the Holm-Šídák method. n = 3 to 8. (B) Representative FACS histograms showing CD41+ HSC frequency in pIpC-treated mice. (C) Relative platelet frequency in peripheral blood from tet-Lin28b and ctrl mice after pIpC treatment measured by FACS. Statistical significance for each time point was determined by multiple t tests assuming equal standard deviation. Day 5, P = .003; day 8, P = .002. (D) Experimental outline for repetitive postnatal pIpC treatments of juvenile mice. Three consecutive weekly treatments of intraperitoneal injection of 10 μg/g pIpC started in 10-day-old (d10) wild-type (WT) pups followed by end point analysis between 7 and 9 weeks of age. (E) FACS analysis of CD41+ HSC frequency (left) in WT mice after repetitive pIpC or PBS injections and (right) tet-Lin28b and ctrl mice after repetitive pIpC treatment (n = 7-9). P values were determined by using the Mann-Whitney test. (F) Representative FACS histograms of CD41+ HSCs from repetitive pIpC-treated tet-Lin28b and control mice. (G) FACS analysis of CD41+ frequency in HSCs from 8-week-old mice of the indicated genotypes treated with the repetitive postnatal pIpC regimen. P value was determined by using the Mann-Whitney test. (H) LIN28B-eGFP reporter mouse model design and FACS histograms showing decreasing LIN28B protein levels across developmental time. (I) Compiled frequency of eGFP+ HSCs at the indicated ages. eGFP+ gating is based on WT littermate controls (n = 2-7 per time point). (J) FACS analysis of eGFP+ levels and frequency in HSCs from 11-day-old (d11) mice 16 hours after 10 μg/g pIpC challenge. ∗P < .05. P value was determined by using the Mann-Whitney test. 3′UTR, 3′ untranslated region; LSK, lineage–Sca1+cKit+.
The postnatal period is associated with a naturally occurring inflammatory signaling caused by de novo exposure to environmental and microbial antigens.55 To further challenge the regulatory capacity of LIN28B, and mimic the cumulative effects of postnatal infections, we administered weekly pIpC treatments to DOX-fed tet-Lin28b mice for 3 consecutive weeks starting on postnatal day 10 (Figure 4D). Interestingly, this challenge enhanced the accumulation of Mk-biased CD41+ HSCs in WT but not tet-Lin28b–induced mice (Figure 4E-F). Instead, the latter retained a higher fraction of CD41– HSCs marked by EPCR (supplemental Figure 4E-F). Conversely, 8-week-old Lin28b−/− mice27 subjected to the same treatment exhibited a trend toward accelerated CD41+ HSC accumulation (Figure 4G; supplemental Figure 4G). Collectively, these results show that LIN28B actively suppresses the establishment of the emergency Mk bypass route, providing a possible explanation for the paucity in Mk lineage priming of HSCs early in life.
To position these developmental changes in the context of the attenuation of endogenous LIN28B protein expression during ontogeny, we generated a reporter mouse line in which LIN28B-eGFP fusion protein is expressed from the endogenous Lin28b locus (Figure 4H). FACS analysis revealed a gradual and uniform dilution of eGFP signal on a per-cell basis within immunophenotypic HSCs from E14.5 to adulthood, reaching near background levels around day 19 (Figure 4I). Thus, the attenuation of LIN28B protein coincides with the previously established timing for the fetal-to-adult switch in HSC function17 and precedes the postnatal accumulation of CD41+ HSCs. Interestingly, we observed that pIpC treatment results in a significant decrease in eGFP levels (Figure 4J), showing that inflammatory signaling triggers an acute decline in LIN28B protein levels. Thus, we propose that early-life inflammatory signaling promotes the establishment of Mk-biased HSCs, at least in part, by the downregulation of LIN28B.
Discussion
In this study, we dissect the establishment of Mk-biased HSCs into 2 progressive stages discernable at the chromatin level. Importantly, our findings reveal a layer of developmental control and identify LIN28B as a negative regulator of emergency Mk output during ontogeny. These findings position the Mk-poised HSC state as an outcome of the previously established developmental switch in HSC function.17 Considering the critical role played by platelets in immune protection of the host,56 we propose that the postnatal attenuation of LIN28B may represent a licensing cue to sensitize the hematopoietic system for inflammatory responsiveness aimed at mitigating the increased threats of infections following birth.
Our findings position the establishment of an Mk-poised HSC state as a manifestation of the fetal-to-adult developmental switch on the basis of its developmental onset and Lin28B-imposed control. Interestingly, the notion of a precipitous postnatal switch in HSC states has recently been challenged by a study showing a gradual and asynchronous shift of the HSC transcriptional landscape already in utero.41 To this end, our finding that LIN28B protein levels are detectable until the third week of life to promote fetal-like HSC behavior highlights the possibility that posttranscriptional regulation may intercept transcriptional and functional changes in HSC behavior.
Many questions remain regarding the mechanisms by which LIN28B as a posttranscriptional regulator preserves an indolent stem cell state and prevents the accumulation of Mk-biased HSCs during early life. Previous reports have shown that inflammation-induced emergency megakaryopoiesis can be controlled at the level of protein translation in HSCs.9 It is thus interesting to note that Lin28A/B controls protein translation independent of its repression of the Let-7 family of microRNAs,57-59 raising the possibility that Lin28B may restrict lineage-specific protein expression. Indeed, Lin28B can bind to Bcl11a transcripts in human HSPCs to repress protein synthesis and developmental hemoglobin switching.60 Thus, analogous investigations into the ability of LIN28B to control key Mk factors are critical to dissecting the hierarchical relationship between the LIN28B action and the Mk-primed state in HSCs. A parallel clue to the mode of LIN28B action is its ability to regulate the quality and requirements for growth factor signaling in normal development and cancer.20,25,61,62 More recently, its downregulation was implicated in the developmental acquisition of TPO dependence in HSCs during late fetal stages.19 Considering the supportive role of this cytokine in HSC expansion and Mk differentiation, it is possible that changes in the response to TPO may contribute to the ability of LIN28B to dampen emergency Mk lineage output.
Taken together, our study shows an early-life re-wiring of the path to platelet biogenesis that is controlled at the transcriptional as well as the posttranscriptional level of gene expression. Although our findings are limited to mice, analogous downregulation of LIN28B takes place from fetal to neonatal human HSPCs with undetectable expression in the adult.20,60 This downregulation has been implicated in both human hemoglobin switching63 and ontogenic changes in regulatory T-cell differentiation,62 suggesting that its role in controlling a developmental switch might be conserved during human hematopoiesis. Indeed, LIN28b is overexpressed in specific subtypes of human pediatric leukemias,64 and further investigations into a potential link between changes in inflammation-induced differentiation and leukemogenic potential are of critical future interest.
Despite the questions remaining, our findings highlight how the hematopoietic system adapts to changing needs for host protection in postnatal life. These findings have potential implications for the well-documented risks of thrombocytopenia in infants65 as well as the importance of platelet-mediated innate immune functions during postnatal life.
Acknowledgments
The authors thank J.A. Daniel, G. Karlsson, and P. Dhapola for their critical input. They also thank B. Johansson Lindbom for sharing of reagents.
J.Y. was supported by the European Research Council (715313), the Swedish Research Council (SRC) and Swedish Cancer Society (SCS), and the Knut and Alice Wallenberg Foundation (KAW). J.U. was supported by SCS. D.B. was supported by SRC, SCS, KAW, and the Tobias Foundation. M.S. was supported by SRC, SCS, and KAW.
Authorship
Contribution: T.A.K., J.Y., and J.U. designed experiments; T.A.K., J.U., Q.Z., S.V., E.B., and N.K. performed the experiments; J.U. performed bioinformatics analysis; D.B., M.S., P.N., and O.A. provided critical expertise and project input; and J.Y. and T.A.K. conceived of the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joan Yuan, Developmental Immunology Unit, Division of Molecular Hematology, Department of Laboratory Medicine, Lund Stem Cell Center, Lund University, 22242 Lund, Sweden; e-mail: joan.yuan@med.lu.se.
References
Author notes
The RNA-sequencing and single-cell assay for transposase-accessible chromatin–sequencing data in this study have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus database and are accessible through accession number GSE161725 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE161725). Custom code and analysis are available at https://github.com/jonasungerback/yuan_atac_seq_project. The corresponding author may be contacted for other forms of data sharing (e-mail: joan.yuan@med.lu.se).
The full-text version of this article contains a data supplement.

![Mk lineage priming of HSCs is developmentally timed. (A) Gene set enrichment analysis comparing bulk RNA-sequencing results from sorted E14.5 FL and ABM HSCs (lineage–Sca1+cKit+ [LSK] CD48–CD150+Flt3–) for the following lineage-restricted progenitor signatures3,37: MkPs, pre-erythrocyte colony-forming units (preCFU-E), common lymphoid progenitors (CLP), and pre-granulocyte/macrophage progenitors (preGM). Normalized enrichment score (NES) and false discovery rate (FDR) are indicated. n = 2 to 3 biological replicates. (B) Normalized RNA-sequencing values for the average abundance of indicated transcripts per kilobase million. (C) Representative FACS analysis of CD41+ HSCs from E14.5 FL and ABM. (D) Mean frequency of CD41+ cells among HSCs from mice of the indicated ages. (E) Representative FACS analysis of MkPs in E14.5 and ABM. (F) Mean frequency of CD150+CD41+MkPs in Lin–cKit+Sca1– HPCs from mice of the indicated ages (n = 4-7). Statistical significance determined by Kruskal-Wallis test followed by Dunn’s multiple comparisons test comparing all ages vs E14.5 FL. (G) Representative images from day 7 single HSC cultures. (H) Analysis of day 7 single HSC cultures from the indicated ages. Wells with dead or no cells were removed before analysis (6%-8% of total wells). d, days; FSC, forward scatter; w, week(s).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/6/24/10.1182_bloodadvances.2021006861/4/m_blooda_adv-2021-006861-gr1.jpeg?Expires=1770324863&Signature=ubqbKJWSrnJ9XFisCOkn2lmY3fqdqLPfdF7~tCXVIv8j8IzWyhSexQoUOnKnie4Bknyr7CT5sQ0CE13taYUwI--e1uRSIVTEouuXtnqeccNn85emBLohgzY~QjOxaZKcB3ay558Zl1bxMvloDQtfUZwHljO8McPHCbqokYRyis7a18-OJol0iCoq1Bq5WaRJYcb4MdFAB~VJtK2hzAy3DFmk9GBQ35yhbLAz20ky7qIM2v~Hce0~5gUBfM~ZSEHUJFjmOsL73Zq9szNdpbDZpZBc84wRZnnx0QARxbnqIBLgA9WcNVpevy~1IPx7MzbIMKjA7atxqhGghBy9Tz0UGA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)