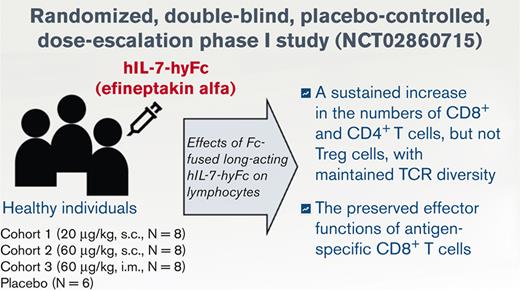
Key Points
A single hIL-7-hyFc administration induced a sustained increase in the numbers of T cells, but not regulatory T cells, in healthy adults.
The effector functions of antigen-specific CD8+ T cells were preserved after hIL-7-hyFc administration.
Abstract
Interleukin-7 (IL-7) is an essential cytokine for T-cell homeostatic proliferation and maintenance. Clinical studies have shown the potential benefits of IL-7 therapy in various diseases associated with lymphopenia. However, the kinetics of the T-cell response to a single administration of IL-7 in humans have not been fully elucidated. Here, we investigated the effects of Fc-fused long-acting recombinant human IL-7 (hIL-7-hyFc, efineptakin alfa) on lymphocytes in healthy adults after a single subcutaneous or intramuscular administration. Administration of hIL-7-hyFc increased the CD8+ and CD4+ T-cell numbers up to 2.5-fold, with corresponding upregulation of Ki-67 and Bcl-2 expression, peaking at day 3 or 7. Regulatory T cells (Tregs) did not expand. Among CD8+ and CD4+ T cells, all T-cell subsets (TN, TEM, TCM, TEMRA, and TSCM) increased for 56 days. The T-cell receptor repertoire diversity of naive CD8+ and CD4+ T cells was increased by hIL-7-hyFc, whereas the memory T-cell subsets did not differ between day 56 and day 0. Transcriptomic analysis revealed that hIL-7-hyFc induced robust T-cell expansion without changes in gene expression profiles associated with T-cell functions or genes related to T-cell exhaustion, senescence, and anergy. The effector functions of antigen-specific CD8+ T cells were preserved after hIL-7-hyFc administration. Our results suggest that hIL-7-hyFc administration induced a sustained increase in the numbers of CD8+ and CD4+ T cells, but not Tregs, without qualitative changes. These results support the potential of hIL-7-hyFc as a treatment for patients with compromised T-cell immunity or as a vaccine adjuvant.
Introduction
Interleukin-7 (IL-7) is a cytokine mainly produced by stromal and epithelial cells of lymphoid organs that plays a critical role in T-cell development, proliferation, and maintenance through thymopoiesis and peripheral T-cell homeostasis.1,2 Furthermore, in the periphery, IL-7 is involved in mature naive T-cell homeostatic expansion and survival, as well as memory T-cell generation and maintenance. IL-7 receptor comprises IL-7Rα (ie, CD127) and common gamma chain (γc; ie, CD132). IL-7 signaling is indispensable during each stage of αβ and γδ T-cell development.3
In mouse and primate models, exogenous IL-7 administration promotes lymphopoiesis and enhances virus- and tumor-specific T-cell responses to subdominant epitopes, supporting improved postvaccination survival.4-9 Clinical trials have been conducted based on preclinical findings in patients with various diseases, including cancers, idiopathic lymphopenia, HIV infection, sepsis, and T-cell–depleted allogeneic hematopoietic stem cell transplantation.1,10-18 Recently, exogenous IL-7 administration to patients with coronavirus disease 2019 (COVID-19) has been studied to promote lymphopoiesis because lymphopenia severity is correlated with mortality from COVID-19.19-22 The results consistently show that exogenously administered IL-7 is well tolerated and dose-dependently induces T-cell expansion. Due to the short half-life of IL-7, previous clinical studies were conducted with repeated administration of IL-7 with 1- to 7-day intervals. Therefore, T-cell expansion kinetics and changes in detailed immunological phenotypes and effector functions of naive and memory T cells after single administration of IL-7 are still unknown.
Exogenous IL-7 has a relatively short half-life (6-23 hours), potentially requiring a short dosing interval to maintain efficacy. Thus, there is a need for a novel long-acting IL-7 that requires less frequent administration. IL-7-hyFc (efineptakin alfa) is a homodimeric IL-7 fused to the immunoglobulin D/G4 (IgD/IgG4) immunoglobulin domain (hyFc). The hyFc region comprises the hinge and N-terminal portion of the heavy chain constant region 2 (hinge-CH2) of human IgD, which is fused to the C-terminal region of CH2 and the entire CH3 region of human IgG4. The hyFc portion of IL-7-hyFc helps IL-7 last longer in the body through neonatal Fc receptor–mediated recycling. We recently reported that hIL-7-hyFc is well tolerated in healthy volunteers after a single subcutaneous (s.c.) or intramuscular (i.m.) administration and has a long half-life with absolute lymphocyte count (ALC)-increasing effects.23
Here, we aimed to extend these findings and gain further insights into the in vivo effects of hIL-7-hyFc on T cells. Using multicolor flow cytometry, RNA sequencing (RNA-seq), and in vitro functional assays, we performed a detailed analysis of peripheral T cells using peripheral blood mononuclear cells (PBMCs) from healthy adults who received a single dose of hIL-7-hyFc. Our results demonstrate that hIL-7-hyFc induced T-cell expansion while maintaining effector functions of memory T cells, and increased T-cell receptor (TCR) repertoire diversity of naive T cells, supporting its potential as an immunomodulator for any indications that could benefit from a boost in T-cell numbers and functionality.
Materials and methods
hIL-7-hyFc
The new formulation of recombinant human IL-7, hIL-7-hyFc, is a fusion of 2 IL-7 molecules with the hybridizing IgD/IgG4 hyFc.23 Because hyFc binds to the neonatal Fc receptor and mediates hIL-7-hyFc recycling, hIL-7-hyFc has a longer half-life in the body compared with previous IL-7 formulations.
Study design and subjects
This randomized, double-blind, placebo-controlled, dose-escalation phase 1 study of hIL-7-hyFc (www.clinicaltrials.gov #NCT02860715) enrolled 31 participants, 30 of whom provided written informed consent before drug administration. Eligible participants were aged 19 to 45 years and healthy based on medical screening 4 weeks before drug administration. In total, 20 men and 10 women received a single dose of hIL-7-hyFc or placebo (ratio, 8:2). We investigated the following doses and administration routes: 20 μg/kg s.c., 60 μg/kg s.c., and 60 μg/kg i.m. The study was approved by the institutional review board of Seoul National University Hospital and complied with the ethical principles of the Declaration of Helsinki and Korean Good Clinical Practice. A detailed description of the eligibility criteria, participants, and clinical results is given in our previous report.23
Flow cytometry
Flow cytometry was performed to assess the expression of cell surface and intracellular molecules on cryopreserved PBMCs collected at specified time points. The surface molecules were stained at room temperature (RT) for 15 minutes with the antibodies. The cells were stained with a tetramer (RT, 30 minutes). MR1-tetramers were prepared using monomeric biotinylated human MR1 5-OPRU provided by the NIH Tetramer Core Facility using streptavidin-APC (Thermo Fisher). Acquisition was performed using an LSR II instrument (BD Biosciences), and data analysis was performed using FlowJo software (FlowJo, LLC).
Materials for flow cytometry
The following anti-human fluorochrome-conjugated antibodies were used: Alexa Fluor 700-conjugated antibody to human CD3 (UCHT1); anti-IL-7Rα-BV421 (A019D5), anti-CD4-PE-Cy5 (OKT4), anti-CD45RA-APC-Cy7 (HI100), anti-Fas-BV650 (DX2), anti-CCR7-BV785 (G043H7), anti-CD31-Alexa Fluor 488 (WM59), anti-TCR γ/δ-Percp-Cy5.5 (B1), anti-CD25-BV650 (BD96; BioLegend), anti-CD19-BV711 (SJ25C1), anti-CD14-BV711 (MφP9), anti-CD3-BV510 (HIT3a), anti-CD8-Alexa Fluor 700 (RPA-T8; all from BD Biosciences); anti-CD19-PE-eFluor610 (HIB19); anti-CD14-PE-eFluor610 (61D3), and anti-CD56-APC (CMSSB; Thermo Fisher). Simultaneously, dead cells were stained using a Live/Dead Fixable Red Dead Cell Stain Kit (Thermo Fisher). The cells were fixed and permeabilized using a Foxp3 Staining Buffer Set (Thermo Fisher), and then intracellular molecules were stained (4°C, 20 minutes) using anti-Bcl-2-Alexa Fluor 488 (100; BioLegend), anti-Ki-67-PE-Cy7 (20Raj1), and anti-Foxp3-PE (236A/E7; Thermo Fisher). Detailed information about the multicolor flow cytometry panel used in this study is shown in supplemental Table 4.
TCR repertoire diversity
RNA-seq and library preparation
The naive and memory subsets of CD8 T cells were isolated from cryopreserved cells using BD FACSAria II (BD Biosciences) after staining with anti-CD8-APC (SK1), anti-CD14-Percp-Cy5.5 (M5E2; BD Biosciences), anti-CD4-Percp-Cy5.5 (RPA-T4; BioLegend), anti-CD19-Percp-Cy5.5 (HIB19), anti-APC-CD45RA (HI100; both from Thermo Fisher), and anti-CCR7-FITC (150503, R&D Systems). Sorted T cells were dissolved in TRIzol reagent (Thermo Fisher), and total RNA was extracted following the manufacturer’s instructions. RNA quality was assessed using an Agilent 2100 Bioanalyzer with an RNA 6000 Nano Chip (Agilent Technologies). Extracted RNAs were processed using the Swift RNA Library Kit (Swift Biosciences) following the manufacturer’s instructions. High-throughput sequencing was performed as paired-end 150 sequencing using a NovaSeq 6000 (Illumina). Raw reads were assembled, and low-quality reads were filtered using Cutadapt (version 2.8). Filtered reads were aligned on a reference genome downloaded from Ensembl (GRCh38.p13, Accession number: GCA_000001405.28) using STAR (version 2.7.1a). The read counts were normalized to sample-specific size factors using DESeq2 (version 1.26.0) (PMID: 25516281). Normalized counts were dimensionally reduced through principal component analysis using the top 2 principal components within the whole sample. Differentially expressed genes (DEGs) were analyzed using the two-sided Wald test according to each condition. DEGs were determined based on cut-offs of an adjusted P value of < .05, and a |log2 fold change (FC)| > 1.0. Gene set enrichment analysis (GSEA) was performed using publicly available gene sets, including Gene Ontology to compare functional changes according to each condition; Biological Process database (GO.BP) and specific public gene sets using GSEA software (version 4.1.0) (ref: PMID: 16199517, 12808457).
Cell proliferation assays
Cryopreserved PBMCs were stained with cell trace violet (CTV; Thermo Fisher) (20 minutes, RT), and then stimulated with 1 μg/mL epitope peptide (NLVPMVATV; JPT Peptide Technologies) for 6 days at 37°C in 500 μL of 10% fetal bovine serum/RPMI 1640 medium (WELGENE). Cells were labeled with HLA-A∗0201-restricted CMVpp65 pentamer (HLA-A2∗0201: CMVpp56495, ProImmune). Next, surface molecule staining, dead cell staining, fixation, permeabilization, and intracellular molecule staining were performed using the following antibodies: anti-CD3-BV510 (HIT3a), anti-CD8-Percp-Cy5.5 (SK1; BD Biosciences), anti-CD19-PE-eFluor610 (HIB19), anti-CD14-PE-eFluor610 (61D3), anti-Ki-67-PE-Cy7 (20Raj1; Thermo Fisher), anti-CD45RA-APC-Cy7 (HI100), and anti-CCR7-BV785 (G043H7; BioLegend), and a Foxp3 Staining Buffer Set. Data were collected using an LSR II instrument.
Cytokine secretion assays
Cryopreserved PBMCs were labeled with HLA-A∗0201-restricted CMVpp65 pentamer (10 minutes, RT) and then stimulated with 1 μg/mL epitope peptide (NLVPMVATV) (4 hours, 37°C) in 200 μL/well of 10% fetal bovine serum/RPMI 1640 medium in 96-well round-bottom plates. Next, the cells were stained with interferon gamma and tumor necrosis factor (TNF) catch reagents (Miltenyi Biotec) (55 minutes, 37°C) on a rotator, and then with FITC-labeled anti-IFN-γ and PE-labeled anti-TNF detection reagents (Miltenyi Biotec) (10 minutes, 4°C). Next, cells were relabeled with pentamer (10 minutes, RT), and stained with anti-CD3-BV510 (HIT3a), anti-CD8-BV711 (SK1; BD Biosciences), anti-CD45RA-APC-Cy7 (HI100), and anti-CCR7-BV785 (G043H7; BioLegend). Data were collected using an LSR II instrument.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 or R software (version 3.6). Analyses included the Wilcoxon matched-pairs signed-rank test, two-way repeated measures (RM) analysis of variance (ANOVA) with the Geisser-Greenhouse correction and Pearson correlation, and the Friedman one-way ANOVA test evaluated P values. Significance was set at P < .05.
Results
hIL-7-hyFc administration induces long-lasting increase of CD8+ and CD4+ T-cell numbers through enhanced proliferation and Bcl-2 upregulation.
Thirty healthy adults were randomly divided into the hIL-7-hyFc–treated and placebo groups. The hIL-7-hyFc–treated group comprised 24 participants divided into 3 cohorts: (1) s.c. injected with low-dose hIL-7-hyFc (20 μg/kg; cohort 1; n = 8), (2) s.c. injected with high-dose hIL-7-hyFc (60 μg/kg; cohort 2; n = 8), and (3) i.m. injected with high-dose hIL-7-hyFc (60 μg/kg; cohort 3; n = 8) (Figure 1A). The remaining 6 participants were included in the placebo group. Using multicolor flow cytometry, we analyzed the changes in the absolute numbers of T (CD3+), NK (CD3–CD56+), and B cells (CD3–CD19+) among cryopreserved PBMCs (gating strategy shown in supplemental Figure 1A). As shown in our previous study,23 a single administration of hIL-7-hyFc robustly and dose-dependently expanded T cells, potentially contributing to the ALC increase (supplemental Figure 1B). In contrast, hIL-7-hyFc had minimal effect on NK cells, B cells, γδ T cells, and mucosal-associated invariant T cells (supplemental Figure 1C-F). Although these cells were increased in the high-dose i.m. group (cohort 3) on day 21 and/or day 56, the increases were not as pronounced as with the conventional T-cell subsets (supplemental Figure 1C-F). Thus, we further investigated the detailed effects of hIL-7-hyFc on the dynamics of various T-cell subpopulations.
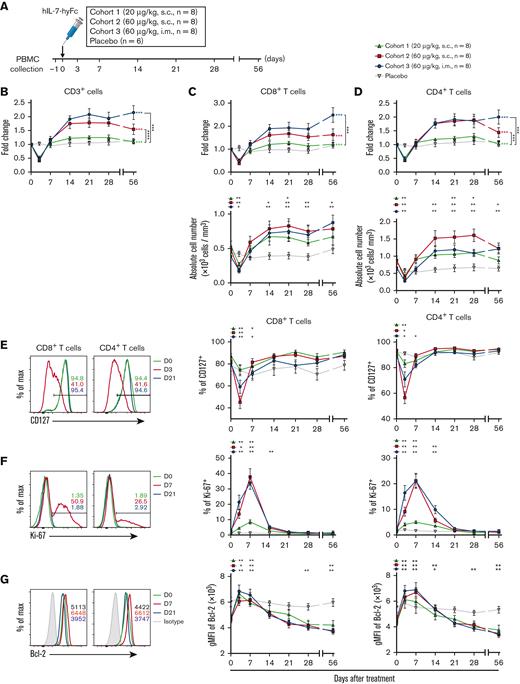
hIL-7-hyFc induces the expansion of circulating CD8+and CD4+T cells with cell proliferation and upregulated Bcl-2 expression. (A) Study design, showing the injection route and dose, and the PBMC collection schedule. The absolute counts of lymphocyte subsets were obtained from the complete blood counts using the ALC and flow cytometry–based frequencies. (B) FC in absolute counts of CD3+ cells over baseline, which was an average value on a day before and the day of administration. The data set for this graph was adapted from a previous report.23 (C) Absolute cell number of CD8+ T cells (bottom) and FC over baseline in CD8+ T-cell number (top). (D) Absolute cell number of CD4+ T cells (bottom) and FC over baseline in CD4+ T-cell number (top). (E-G) Expression changes of CD127, Ki-67, and Bcl-2 among CD8+ and CD4+ T cells after hIL-7-hyFc administration, as analyzed by flow cytometry. Plots show expression levels on days 0 and 21 and the maximum change time point. (E) Frequencies of CD127+ cells among CD8+ and CD4+ T cells. (F) Frequencies of Ki-67+ cells among CD8+ and CD4+ T cells. (G) Bcl-2 expression as gMFI among CD8+ and CD4+ T cells. Plot shows the mean values (± standard error of the mean) for each cohort at the indicated time points. P values of FC over baseline in panels B to D represent comparisons between cohorts by two-way RM ANOVA with Geisser-Greenhouse correction. Green, red, and blue asterisks indicate P values between an indicated group and the placebo group. Black asterisks next to the graphs indicate P values between 2 groups indicated. Black asterisks above the graphs indicate P values between the value of each time point to that of baseline (day 0) by a Wilcoxon matched-pairs signed-rank test. ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
hIL-7-hyFc induces the expansion of circulating CD8+and CD4+T cells with cell proliferation and upregulated Bcl-2 expression. (A) Study design, showing the injection route and dose, and the PBMC collection schedule. The absolute counts of lymphocyte subsets were obtained from the complete blood counts using the ALC and flow cytometry–based frequencies. (B) FC in absolute counts of CD3+ cells over baseline, which was an average value on a day before and the day of administration. The data set for this graph was adapted from a previous report.23 (C) Absolute cell number of CD8+ T cells (bottom) and FC over baseline in CD8+ T-cell number (top). (D) Absolute cell number of CD4+ T cells (bottom) and FC over baseline in CD4+ T-cell number (top). (E-G) Expression changes of CD127, Ki-67, and Bcl-2 among CD8+ and CD4+ T cells after hIL-7-hyFc administration, as analyzed by flow cytometry. Plots show expression levels on days 0 and 21 and the maximum change time point. (E) Frequencies of CD127+ cells among CD8+ and CD4+ T cells. (F) Frequencies of Ki-67+ cells among CD8+ and CD4+ T cells. (G) Bcl-2 expression as gMFI among CD8+ and CD4+ T cells. Plot shows the mean values (± standard error of the mean) for each cohort at the indicated time points. P values of FC over baseline in panels B to D represent comparisons between cohorts by two-way RM ANOVA with Geisser-Greenhouse correction. Green, red, and blue asterisks indicate P values between an indicated group and the placebo group. Black asterisks next to the graphs indicate P values between 2 groups indicated. Black asterisks above the graphs indicate P values between the value of each time point to that of baseline (day 0) by a Wilcoxon matched-pairs signed-rank test. ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
After hIL-7-hyFc administration, T cells transiently decreased on day 3 and returned to baseline on day 7 (Figure 1B). In cohorts 2 (60 μg/kg, s.c.) and 3 (60 μg/kg, i.m.), T cells increased significantly and rapidly, which was maintained at least up to day 56, particularly in cohort 3. Moreover, hIL-7-hyFc administration markedly increased both CD8+ and CD4+ T cells in cohorts 2 and 3, and these increases in cohort 3 were maintained up to day 56 (Figure 1C-D).
IL-7 signaling is important for cell cycling and T-cell survival; therefore, we evaluated the cellular response of T cells to hIL-7-hyFc by multicolor flow cytometry. CD127 surface expression on both CD8+ and CD4+ T cells was temporarily and dose-dependently downregulated on day 3 and restored to basal levels by day 14 (Figure 1E). CD127 transcription and protein levels are downregulated by IL-7 signaling25; thus, our results supported that hIL-7-hyFc directly increased CD8+ and CD4+ T cells. Moreover, the frequency of Ki-67+ (a proliferation marker) CD8+ and CD4+ T cells was significantly and dose-dependently increased in all cohorts, peaked on day 7, and then gradually decreased to basal levels by day 21 (Figure 1F). Ki-67 expression correlated strongly with an increased number of CD8+ and CD4+ T cells (supplemental Figure 1G-H). In addition, hIL-7-hyFc administration significantly upregulated the expression of the antiapoptotic protein Bcl-2 in CD8+ and CD4+ T cells during the first week (Figure 1G). Subsequently, Bcl-2 expression in both subsets declined to basal levels by day 56, consistent with a previous report of rhIL-7 administration in patients with HIV infection.13 In addition, when we examined the expression of activation (CD38 and HLA-DR) and exhaustion (PD-1 and CTLA-4) markers on CD8+ and CD4+ T cells, the frequencies of CD38+ HLA-DR+ cells, PD-1+ cells, and CTLA+ cells among CD8+ and CD4+ T cells tended to be transiently increased on day 3 and then restored to basal levels after day 7, although the differences did not reach statistical significance (supplemental Figure 2A-C).
Next, we analyzed the different effects of IL-7 according to the differences in the participants’ age and base T-cell counts at day 0. We subdivided individuals treated with hIL-7-hyFc into 2 subgroups based on the median age, and then we compared the maximal FC of cell number between these 2 subgroups. There were no significant differences in the maximal fold increases of the numbers of total, CD8+, and CD4+ T cells (supplemental Figure 3A-B). Moreover, when individuals treated with hIL-7-hyFc were subdivided into 2 groups based on median base T-cell counts at day 0, we observed a significantly higher increase of CD4+ T-cell numbers in individuals with lower CD4+ T-cell counts compared with those with higher T-cell counts (supplemental Figure 3C). In addition, there was a significant correlation of base total and CD4+ T-cell counts with the maximal fold increases of T cells (supplemental Figure 3D). Overall, these results demonstrate that even a single administration of hIL-7-hyFc induced long-lasting enhancement of CD8+ and CD4+ T cells in humans, which could be due to enhanced proliferation and Bcl-2 upregulation via IL-7 signaling.
Both naive and memory T cells are durably increased by hIL-7-hyFc
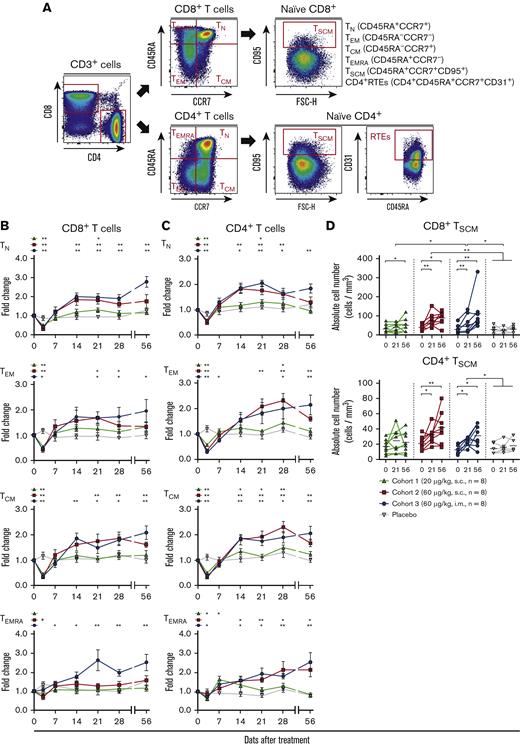
Next, we examined the effects of hIL-7-hyFc administration according to the differentiation status of CD8+ and CD4+ T cells; for example, naive (TN; CCR7+CD45RA+), stem cell–like memory T cells (TSCM; CCR7+CD45RA+CD95+), central memory (TCM; CCR7+CD45RA–), effector memory (TEM; CCR7–CD45RA–), and effector memory CD45RA+ (TEMRA; CCR7–CD45RA+) (Figure 2A). Among CD8+ T cells, all subsets in cohorts 2 and 3 were significantly increased after day 21 and preserved up to day 56 (during the 56 days observation period), whereas only the TN and TSCM subsets in cohort 1 were increased (Figure 2B,D). In addition, Ki-67 expression of all subsets was significantly increased, peaking on day 3 or 7, consistent with the results among total CD8+ T cells (supplemental Figure 4A). Notably, the Ki-67 peak was observed earlier in the memory CD8+ and CD4+ T-cell subsets than naive subsets, whereas Bcl-2 expression showed a greater increase in naive T cells than in the memory T-cell subsets (supplemental Figure 4A-D). This result suggests that IL-7 regulates the homeostasis of naive and memory subsets via different molecular mechanisms. Interestingly, the highest Ki-67 expression was observed in TSCM cells, which have self-renewal capacity and multipotency for differentiation into diverse T-cell subsets (supplemental Figure 4A).
hIL-7-hyFc increases the absolute numbers of CD8+and CD4+T-cell subsets. (A) Gating strategy for obtaining flow cytometry-based frequencies of the TN, TEM, TCM, TEMRA, TSCM subsets, and CD4+ RTEs. (B-C) FC over baseline for the absolute counts of the TN, TEM, TCM, and TEMRA subsets among CD8+ T cells (B) and CD4+ T cells (C). TN, TEM, TCM, and TEMRA subsets are shown in order from top to bottom. Black asterisks above the graphs indicate P values comparing the value of each time point with that of baseline (day 0) by a Wilcoxon matched-pairs signed-rank test. (D) Absolute numbers of the TSCM subsets among CD8+ T cells (top) and CD4+ T cells (bottom) on days 0, 21, and 56. Values shown are the mean (± standard error of the mean) for cohorts and placebo at the specified times. P values between the value of each time point in each cohort were calculated with a Wilcoxon matched-pairs signed-rank test. The statistical significance between the cohorts were calculated with a two-way RM ANOVA with Geisser-Greenhouse correction. ∗∗P < .01, ∗P < .05.
hIL-7-hyFc increases the absolute numbers of CD8+and CD4+T-cell subsets. (A) Gating strategy for obtaining flow cytometry-based frequencies of the TN, TEM, TCM, TEMRA, TSCM subsets, and CD4+ RTEs. (B-C) FC over baseline for the absolute counts of the TN, TEM, TCM, and TEMRA subsets among CD8+ T cells (B) and CD4+ T cells (C). TN, TEM, TCM, and TEMRA subsets are shown in order from top to bottom. Black asterisks above the graphs indicate P values comparing the value of each time point with that of baseline (day 0) by a Wilcoxon matched-pairs signed-rank test. (D) Absolute numbers of the TSCM subsets among CD8+ T cells (top) and CD4+ T cells (bottom) on days 0, 21, and 56. Values shown are the mean (± standard error of the mean) for cohorts and placebo at the specified times. P values between the value of each time point in each cohort were calculated with a Wilcoxon matched-pairs signed-rank test. The statistical significance between the cohorts were calculated with a two-way RM ANOVA with Geisser-Greenhouse correction. ∗∗P < .01, ∗P < .05.
Similar to the results for CD8+ T cells, all CD4+ T-cell subsets were significantly increased in cohorts 2 and 3, whereas in cohort 1, only the TN, TEM, and TCM subsets were increased (Figure 2C-D). In cohort 3, the TN, TEM, and TCM cells doubled at the apex of the cells, and TEMRA cells were increased by approximately 2.5 fold at the peak. Ki-67 and Bcl-2 expression was significantly increased in each CD4+ T-cell subset (supplemental Figure 4B,D), with the highest Ki-67 expression in TSCM cells (supplemental Figure 4B). In addition, we examined the expressions of activation (CD38 and HLA-DR) and exhaustion (PD-1) markers on each T-cell subset (supplemental Figure 5A-D). The frequency of CD38+ HLA-DR+ cells was increased in memory CD8+ and CD4+ T-cell subsets during the first week after IL-7 administration (supplemental Figure 5A-B). PD-1 expressions were transiently upregulated during the first week in memory CD8+ and CD4+ T-cell subsets except for the CD8+ TEMRA subset (supplemental Figure 5C-D). Notably, the upregulated levels of activation and exhaustion markers in the CD4+ and CD8+ TN subsets was not as pronounced as in memory T-cell subsets.
Like the naive CD4+ subset, the CD4+ recent thymic emigrants (RTEs) transiently decreased on day 3 in all cohorts and then gradually increased until day 14 in cohort 2 or day 21 in cohort 3 (supplemental Figure 6A-B). In addition, the frequency of RTEs among naive CD4+ T cells remained constant during the IL-7–induced expansion of naive CD4+ T cells (supplemental Figure 6C).
Collectively, our results indicate that hIL7-hyFc administration increased naive CD8+, CD4+ T cells, and CD4+ RTEs, as well as memory T-cell subsets, including TSCM, TCM, TEM, and TEMRA.
TCR repertoire diversity of naive CD8+ and CD4+ T cells was not decreased after hIL-7-hyFc–mediated T-cell expansions
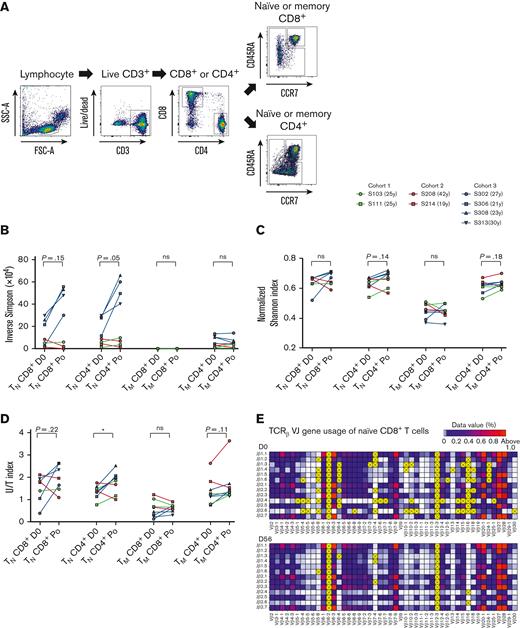
Several studies have reported that repeated rhIL-7 administration increased the TCR repertoire diversity of T cells in patients with refractory cancer or HIV infection that are immunocompromised and patients who received an allogeneic stem cell transplant.1,15,18 Therefore, we investigated whether CD8+ and CD4+ T-cell expansions induced by a single hIL-7-hyFc administration altered the TCR profile of naive and memory T-cell subsets in individuals that were immunocompetent and healthy. TCR repertoire diversity mainly depends on that of the naive population, including RTEs. Thus, we separately analyzed the TCR repertoires after FACS sorting of naive and memory subsets of CD8+ or CD4+ T cells from PBMCs obtained from 4 individuals in cohort 3 on day 0 and on day 56 after hIL-7-hyFc administration (post), and 4 individuals in cohort 1 and 2 (2 individuals from each cohort) on day 0 and post–hIL-7-hyFc administration (day 21, 28, or 56) (Figure 3A). TCRβ CDR3 sequencing revealed an average of 7 462 715 ± 6 087 046 (mean ± standard deviation) total reads per sample, of which 96 154 ± 96 971 were unique productive nucleotide (nt) sequences in each repertoire (supplemental Table 1). We assessed changes in the TCR repertoire of each T-cell subset based on diversity parameters, including the normalized Shannon, inverse Simpson, and U/T indices. Overall, normalized Shannon and inverse Simpson indices of naive CD8+ and CD4+ T-cell subsets showed that there were no significant differences in TCR repertoire diversity between day 0 and post, although there was a trend of TCR repertoire diversity increase especially in naive CD4+ T cells that was further confirmed by the U/T index (the number of productive unique nt sequences/the number of total productive nt sequences) (Figure 3B-D). Indeed, representative TCRβ V(D)J gene usages of naive CD8+ T cells from 1 individual in cohort 3 showed that dominant VJ gene usage (shown as red boxes) was maintained, and undetected VJ gene usage (shown as yellow boxes) on day 0 were newly detected on day 56 (Figure 3E). TCR repertoire diversity of memory CD8+ and CD4+ T-cell subsets did not differ between day 0 and day 56 post–hIL-7-hyFc administration (Figure 3B-C).
TCR repertoire diversities of naive CD8+and CD4+T cells are increased after the T-cells expansion induced by hIL-7-hyFc. (A) Gating strategy for isolating the naive and memory subsets of CD8+ and CD4+ T cells by flow cytometry. Each subset was isolated on days 0 and 56 from 4 individuals in cohort 3 (n = 4) or was isolated on day 0 and post (day 21-56) from 4 individuals in cohort 1 and 2 (n = 2 from each cohort, respectively). The TCR repertoire diversity of each isolated subset was analyzed by sequencing the TCRβ CDR3 regions. (B) Inverse Simpson index, (C) normalized Shannon index, and (D) U/T index are shown. (E) V(D)J gene usage of CDR3 regions of naive CD8+ T cells on day 0 (top) and day 56 (bottom), shown as VJ gene combinations. Yellow boxes with “X” indicate VJ gene combinations with undetected use. Bright red boxes indicate VJ gene combinations with >1% usage of total combinations. V or J genes not shown on the horizontal or vertical axis were excluded because they were not detected in CDR3 sequencing. P values were calculated with a Wilcoxon matched-pairs signed-rank test. ∗P < .05.
TCR repertoire diversities of naive CD8+and CD4+T cells are increased after the T-cells expansion induced by hIL-7-hyFc. (A) Gating strategy for isolating the naive and memory subsets of CD8+ and CD4+ T cells by flow cytometry. Each subset was isolated on days 0 and 56 from 4 individuals in cohort 3 (n = 4) or was isolated on day 0 and post (day 21-56) from 4 individuals in cohort 1 and 2 (n = 2 from each cohort, respectively). The TCR repertoire diversity of each isolated subset was analyzed by sequencing the TCRβ CDR3 regions. (B) Inverse Simpson index, (C) normalized Shannon index, and (D) U/T index are shown. (E) V(D)J gene usage of CDR3 regions of naive CD8+ T cells on day 0 (top) and day 56 (bottom), shown as VJ gene combinations. Yellow boxes with “X” indicate VJ gene combinations with undetected use. Bright red boxes indicate VJ gene combinations with >1% usage of total combinations. V or J genes not shown on the horizontal or vertical axis were excluded because they were not detected in CDR3 sequencing. P values were calculated with a Wilcoxon matched-pairs signed-rank test. ∗P < .05.
Collectively, these results indicate that the long-term increase of CD8+ and CD4+ T-cell numbers by a single administration of hIL-7 was not accompanied by a decrease in TCR repertoire diversity.
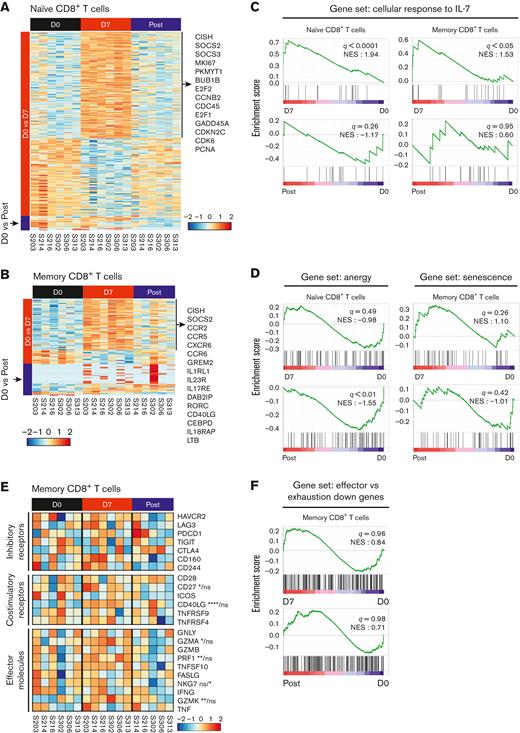
hIL-7-hyFc upregulates genes related to the cell cycle and IL-7 signaling pathway, without altering genes related to T-cell exhaustion, senescence, and anergy
T-cell cellular responses to hIL-7-hyFc peaked on day 7 and returned to the basal state after day 21 (Figure 1D-E). Thus, we performed RNA-seq to comprehensively investigate the differences in gene expression profiles between sorted naive and memory CD8+ and CD4+ T cells on day 0, day 7, and post (day 21-56). Direct comparison of the naive CD8+ T cells on day 7 and day 0 revealed 286 DEGs (adjusted P < .05, |log2 FC| > 1.0) (Figure 4A). Of these 286 DEGs, 166 were upregulated on day 7, including suppressor of cytokine signaling (SOCS) family genes (CISH, SOCS2, and SOCS3) that regulate the IL-7 signaling pathway, and cell cycle–related genes (MKI67, PKMYT1, BUB1B, E2F2, etc) (supplemental Table 2). However, the gene expression profiles were similar between day 0 and post, with only 21 DEGs. Memory CD8+ T cells on day 7 and day 0 exhibited distinct transcriptomic differences (94 DEGs), whereas post and day 0 showed 45 DEGs (Figure 4B). Among memory CD8+ T cells, 74 genes were upregulated on day 7 compared with day 0, which included SOCS family genes (CISH and SOCS2), chemokine receptors (CCR2, CCR5, CXCR6, and CCR6), and genes associated with response to cytokine stimulus (GREM2, IL1RL1, IL23R, etc) (supplemental Table 2).
hIL-7-hyFc upregulates genes related to the cell cycle and IL-7 signaling pathway. Naive and memory CD8+ T cells at days 0 and 7, and after day 21 (post) were isolated with 6 individuals receiving the high-dose of hIL-7-hyFc (60 μg/kg). Gene expression profiles of isolated naive and memory CD8+ T cells were analyzed by RNA-seq. (A-B) Heatmap shows DEGs (adjusted P < .05, |log2 FC| > 1.0) identified from (A) naive and (B) memory CD8+ T cells at day 0 vs day 7 and at day 0 and post. (C, D, and G) GSEA of gene sets related to response to IL-7, anergy, senescence, and exhaustion in the transcriptomes of naive and memory CD8+ T cells at each time point. Results are shown as normalized enrichment scores. (C) GSEA of a gene set associated with the cellular response to IL-7 obtained from GO BP in naive and memory CD8+ T cells. (D) GSEA of genes upregulated in anergic T cells in naive CD8+ T cells (left) and senescent cells in memory CD8+ T cells (right). (E) Expression of selected genes related to T-cell functions, including inhibitory, costimulatory, and effector molecules in memory CD8+ T cells. (F) GSEA using genes related to T-cell exhaustion in memory CD8+ T cells. Statistical analyses were calculated with a Wald test. ns, not significant; ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
hIL-7-hyFc upregulates genes related to the cell cycle and IL-7 signaling pathway. Naive and memory CD8+ T cells at days 0 and 7, and after day 21 (post) were isolated with 6 individuals receiving the high-dose of hIL-7-hyFc (60 μg/kg). Gene expression profiles of isolated naive and memory CD8+ T cells were analyzed by RNA-seq. (A-B) Heatmap shows DEGs (adjusted P < .05, |log2 FC| > 1.0) identified from (A) naive and (B) memory CD8+ T cells at day 0 vs day 7 and at day 0 and post. (C, D, and G) GSEA of gene sets related to response to IL-7, anergy, senescence, and exhaustion in the transcriptomes of naive and memory CD8+ T cells at each time point. Results are shown as normalized enrichment scores. (C) GSEA of a gene set associated with the cellular response to IL-7 obtained from GO BP in naive and memory CD8+ T cells. (D) GSEA of genes upregulated in anergic T cells in naive CD8+ T cells (left) and senescent cells in memory CD8+ T cells (right). (E) Expression of selected genes related to T-cell functions, including inhibitory, costimulatory, and effector molecules in memory CD8+ T cells. (F) GSEA using genes related to T-cell exhaustion in memory CD8+ T cells. Statistical analyses were calculated with a Wald test. ns, not significant; ∗∗∗∗P < .0001, ∗∗∗P < .001, ∗∗P < .01, ∗P < .05.
We also compared naive and memory CD8+ T cells on day 0, day 7, and post with regard to the expression of gene sets associated with the cellular response to IL-7 (GO:0098761),26-28 anergy,29 and senescence.30 GSEA confirmed that naive and memory CD8+ T cells exhibited significant enrichment of the gene set associated with cellular response to IL-7 on day 7 compared with day 0, whereas day 0 and post did not significantly differ (Figure 4C). Gene sets associated with anergy and senescence were also not enriched in the naive and memory subsets (Figure 4D).
Next, we analyzed the expression of genes associated with T-cell function in memory CD8+ T cells on day 0, day 7, and post. Among the memory CD8+ T cells, costimulatory molecules (CD27 and CD40LG), effector molecules (GZMA, PRF1, and GZMK), and chemokine receptors (CCR5 and CXCR6) were significantly upregulated on day 7 compared with day 0 (Figure 4E and supplemental Figure 7B). GSEA confirmed that hIL-7-hyFc did not upregulate genes associated with T-cell exhaustion31-33 among expanded memory or naive CD8+ T cells (Figure 4F).
Moreover, 180 DEGs (adjusted P < .05, |log2 FC| > 1.0) were revealed by direct comparison of the naive CD4+ T cells between day 7 and day 0, and 270 DEGs were identified between post and day 0 (supplemental Figure 8A). Among memory CD4+ T cells, 148 DEGs were identified between day 0 vs day 7, and 187 were found between day 0 vs post (supplemental Figure 8B). The DEGs identified through each comparison were presented in supplemental Table 3. Consistent with the results from CD8+ T cells, GSEA confirmed the cellular response to IL-7 on day 7, by showing significant enrichment of genes related to cellular response to IL-7 (GO: 0098761) in both naive and memory CD4+ T cells on day 7, not day 0 nor post (supplemental Figure 8C). Furthermore, we found that gene sets associated with anergy and senescence were not enriched in the naive and memory CD4+ T-cell subsets, respectively (supplemental Figure 8D). A heatmap of selected genes related to inhibitory receptor, costimulatory, and effector molecules revealed no significant change in those genes among memory CD4+ T cells after IL-7 treatment (supplemental Figure 8E). Finally, GSEA confirmed that the IL-7-induced expansion of memory T cells did not lead to upregulation of genes related to T-cell exhaustion (supplemental Figure 8F).
Overall, these results suggest that hIL-7-hyFc upregulates genes associated with the cell cycle and IL-7 signaling pathway without causing significant changes in genes associated with T-cell exhaustion, senescence, or anergy, and expanded CD8+ T cells do not downregulate genes associated with T-cell effector functions.
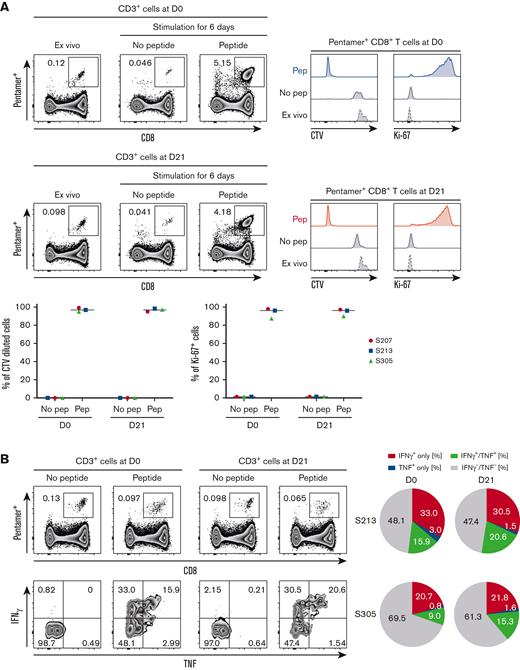
Effector functions of antigen-specific CD8+ T cells are maintained after hIL-7-hyFc administration
Transcriptomic analysis suggested that the effector functions of CD8+ T cells were maintained after a robust cellular response to hIL-7-hyFc. For hIL-7-hyFc to be useful as a therapeutic agent, antigen-specific memory CD8+ T cells should maintain their effector functions after hIL-7-hyFc treatment. To investigate this, we investigated the effector functions of antigen-specific CD8+ T cells after hIL-7-hyFc administration by evaluating the proliferation and cytokine production of CMVpp65495-503-specific memory CD8+ T cells from PBMCs on day 0 and day 21. There was no significant change in the frequency of CMVpp56495-503-specific CD8+ T cells among total CD8+ T cells up to 56 days (supplemental Figure 9A). The proliferative capacity of CMVpp65495-503-specific CD8+ T cells before and after hIL-7-hyFc administration was compared using a CTV dilution assay and Ki-67 expression (Figure 5A). The proliferative capacity of CMVpp65495-503-specific CD8+ T cells did not differ significantly between day 0 and day 21 (Figure 5A). We further examined IFN-γ and/or TNF production using a cytokine secretion assay after stimulation of PBMCs with CMVpp65495-503 peptide (Figure 5B and supplemental Figure 9B). In consequence, the frequency of cytokine-producing cells among pentamer+ CD8+ T cells were not decreased on day 21 compared with day 0 (Figure 5B), indicating that hIL-7-hyFc treatment did not reduce the ability of cytomegalovirus (CMV)-specific CD8+ T cells to produce cytokines upon antigen stimulation.
Effector functions of CMV-specific CD8+T cells are maintained after expansion by hIL-7-hyFc. CMVpp65495-503-specific CD8+ T cells on days 0 and 21 were stimulated with epitope peptide (NLVPMVATV). (A) The percentage of CTV-diluted cells and Ki-67+ cells, respectively, among pentamer+ CD8+ T cells after in vitro stimulation for 6 days. (B) The percentage of IFN-γ- and/or TNF-secreting cells among pentamer+ CD8+ T cells.
Effector functions of CMV-specific CD8+T cells are maintained after expansion by hIL-7-hyFc. CMVpp65495-503-specific CD8+ T cells on days 0 and 21 were stimulated with epitope peptide (NLVPMVATV). (A) The percentage of CTV-diluted cells and Ki-67+ cells, respectively, among pentamer+ CD8+ T cells after in vitro stimulation for 6 days. (B) The percentage of IFN-γ- and/or TNF-secreting cells among pentamer+ CD8+ T cells.
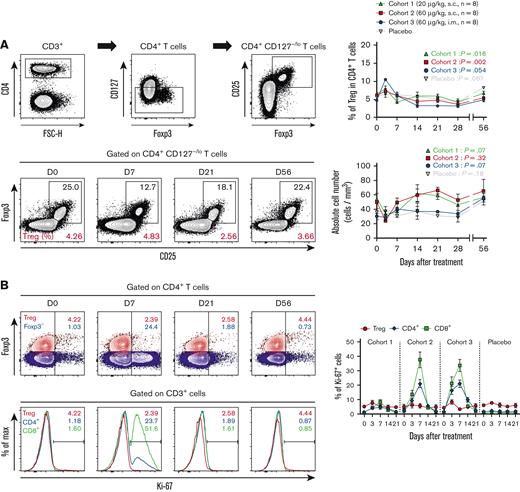
Regulatory T cells (Tregs) are not selectively expanded by hIL-7-hyFc
Previous clinical studies of rhIL-7 in patients infected with HIV or with refractory cancer revealed that the relative frequency of Tregs expressing low-level CD127 was relatively reduced due to IL-7-induced expansion of conventional T cells.1,10,14 Thus, we examined whether exogenous hIL-7-hyFc administration affected Tregs in healthy adults. The frequency of Tregs among CD4+ T cells transiently increased on day 3 (possibly caused by the transient decrease of non-Treg CD4+ T cells on day 3) but then decreased until day 28 (Figure 6A). We assessed the absolute Treg numbers in the periphery after hIL-7-hyFc administration and found no overall increase in Treg numbers in any of the cohorts for at least 56 days (Figure 6A). Notably, Ki-67 expression in Treg cells remained constant during the first week after hIL-7-hyFc administration, whereas we observed a remarkable increase in Ki-67 expression in CD8+ T cells and Foxp3– non-Treg CD4+ T cells (Figure 6B). These results show that hIL-7-hyFc administration in healthy individuals does not induce the selective expansion of Treg cells.
Single administration of hIL-7-hyFc has no significant effect on Treg cell expansion. (A) Gating strategy for Treg cells, frequency of Treg cells among CD4+ T cells, and absolute counts of Tregs. Each value of cohorts and placebo are shown as mean (± standard error of the mean) at the indicated time points. (B) Upper plots display the difference in Ki-67 expression among Tregs (Foxp3+CD127−/loCD25+ CD4+; red) and Foxp3−CD4+ (blue) T cells. Bottom plots display the difference in Ki-67 expression among Treg cells (red), CD4+ T cells (blue), and CD8+ T cells (green). For each cohort, the frequency of Ki-67+ cells in each T-cell subset is shown. P values were calculated with a Friedman one-way ANOVA test. ∗∗P < .01, ∗P < .05; ns, not significant.
Single administration of hIL-7-hyFc has no significant effect on Treg cell expansion. (A) Gating strategy for Treg cells, frequency of Treg cells among CD4+ T cells, and absolute counts of Tregs. Each value of cohorts and placebo are shown as mean (± standard error of the mean) at the indicated time points. (B) Upper plots display the difference in Ki-67 expression among Tregs (Foxp3+CD127−/loCD25+ CD4+; red) and Foxp3−CD4+ (blue) T cells. Bottom plots display the difference in Ki-67 expression among Treg cells (red), CD4+ T cells (blue), and CD8+ T cells (green). For each cohort, the frequency of Ki-67+ cells in each T-cell subset is shown. P values were calculated with a Friedman one-way ANOVA test. ∗∗P < .01, ∗P < .05; ns, not significant.
Discussion
In this study, we demonstrated long-lasting T-cell expansion by a single administration of hIL-7-hyFc (efineptakin alfa) with maintained effector functions and TCR repertoire diversity. IL-7 is a potential modulator of peripheral T-cell homeostasis that induces the expansion of CD8+ and CD4+ T cells. Previous clinical studies used IL-7 drugs with low stability in vivo that requires frequent administration. hIL-7-hyFc is a novel formulation that overcomes these limitations. We recently reported that hIL-7-hyFc is long acting and well tolerated.23
Previously published studies repeatedly administered nonglycosylated rhIL-7 at 2- or 3-day intervals, whereas later studies repeatedly administered glycosylated IL-7 at 7-day intervals. The results rarely indicated that repeated dosing every 7 days generated additional effects on cell proliferation. The present insights into the kinetics of T-cell responses to a single administration of exogenous hIL-7-hyFc can help determine an optimal dosing schedule. The restoration of CD127 surface expression levels and cell proliferation suggests that subsequent hIL-7-hyFc administration should be done at least at 3-week intervals after the termination of the cellular response to hIL-7-hyFc. Moreover, the number of CD8+ and CD4+ T cells reached a plateau after the upsurge between days 7 and 14 on day 21 after administration. Based on results from this study, we have designed a clinical trial for hIL-7-hyFc administration in patients with solid tumors with a 3-week dosing interval (www.clinicaltrials.gov #NCT02860715). Clinical trial is ongoing and the results have not yet been reported.
Several clinical studies have suggested that the response to IL-7 is also affected by disease conditions, which induces changes in the proportion of subsets. For example, it has been reported that IL-7 administration increased the number of TCM in patients with HIV infection, TN in patients with refractory cancer, and TEM in patients with hematopoietic stem cell transplantation.1,10,13,14,18 In contrast, we found that hIL-7-hyFc did not increase specific subsets (TN, TEM, TCM, and TEMRA) in healthy individuals. Because TN constitutes the largest proportion, the increased absolute number of the TN subset was remarkable. However, the increases in each subset were similar in both CD8+ and CD4+ T cells, and thus the proportion of each subset was maintained throughout the observation period. This suggests that exogenous IL-7 can potentially induce homeostatic proliferation in all subsets, when not under disease conditions, even though each subset exhibits distinct population dynamics.
Administration of hIL-7-hyFc augmented the absolute cell number of naive subsets among both CD8+ and CD4+ T cells, and TCR diversity did not decrease but rather tended to increase in naive CD4+ T cells. Although there is a limitation that this study was conducted on very young healthy subjects, the results suggest that hIL-7-hyFc may contribute to a broad spectrum of naive T cells and that hIL-7-hyFc may be a useful vaccine adjuvant for the elderly. With aging, thymic output decreases, and the existing naive T cells encounter their antigens and differentiate into memory T cells. As total naive T cells decrease, elderly hosts have reduced ability to fight new antigens. If hIL-7-hyFc administration to the elderly increases their circulating naive T cells, it would enhance the possibility that the naive T cells would encounter a specific antigen originating from a vaccine. In viral infections such as severe acute respiratory syndrome coronavirus 2, antigen-specific T cells have been suggested to play an important role in virus clearance and protection from developing severe COVID-19.34-37 Therefore, hIL-7-hyFc is a promising candidate as an adjuvant for severe acute respiratory syndrome coronavirus 2 vaccines and/or a therapeutic option for preventing the development of severe COVID-19.
For hIL-7-hyFc to be useful as a therapeutic agent, hIL-7-hyFc–expanded T cells should maintain their function. The T-cell function was reportedly preserved after rhIL-7 administration to patients with cancer when PBMCs were stimulated with anti-CD3 antibodies ex vivo.1 In this study, analysis of naive and memory CD8+ T cells suggested that T cells expanded 21 days after administration compared with those collected before administration, exhibiting similar transcriptional profiles and gene expression patterns related to the functionality of T cells. We also confirmed that the effector functions of CMV-specific T cells were maintained by in vitro stimulation. These results imply that the increase in ALC or T-cell numbers by hIL-7-hyFc administration to patients with cancer might improve patient survival because it has been suggested that the ALC before treatment or lymphocyte recovery after chemoradiotherapy significantly correlates with overall and progression-free survival in cancer.38-40 In addition, these results suggest that hIL-7-hyFc can expand tumor-antigen–specific T cells with effector functions without features of T-cell exhaustion and senescence. In a nonhuman primate study, exogenous IL-7 administration induced the homing of circulating T-cells to various organs.41 It was also reported that rhIL-7 administration to patients with HIV infection augmented T-cell gut homing with the expansion of circulating CD8+ and CD4+ T cells.16 Collectively, these studies suggest that circulating T cells expanded by hIL-7-hyFc administration can migrate into tumor tissues and may enhance antitumor immunity, which may have a synergistic effect with immune checkpoint blockades such as anti-PD-1/PD-L1. In addition, results this study suggest that Tregs are not selectively expanded by hIL-7-hyFc, which was consistent with previous reports.1,10 Recently, it was reported that Tregs were amplified by PD-1 blockade,42 supporting the value of utilizing hIL-7-hyFc in cancer immunotherapy. However, considering that Tregs are relevant for immune tolerance and that a robust expansion of effector T cells without expansion of Tregs might lead to autoimmune diseases in some individuals, further studies will be needed to evaluate the effects of hIL-7-hyFc administration on development of autoimmune diseases.
In summary, we administered a new IL-7 formulation to healthy adults and demonstrated the therapeutic potential of hIL-7-hyFc as an effective hematopoietic growth factor. hIL-7-hyFc induces robust expansion of both naive and memory T cells while maintaining TCR diversity and T-cell effector function. Mechanistically, T-cell expansion was attributed to cell proliferation and enhanced antiapoptotic signaling. We further revealed the T-cell population dynamics and cellular response kinetics of T cells to a single administration of exogenous IL-7, suggesting a means of optimizing multidose IL-7 administration. However, this study has a limitation that this phase 1 clinical study was designed to collect blood samples only up to day 56 after administration. Therefore, further clinical studies will be needed to evaluate the long-term effects of hIL-7-hyFc on T cells. Another limitation of the current study is that this clinical trial was conducted only in a very young healthy volunteer cohort. Therefore, based on our results, further studies are warranted to verify the effectiveness of hIL-7-hyFc in aged individuals, vaccinated individuals, or patients with various diseases accompanied by lymphopenia.
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (Ministry of Science and ICT) (NRF-2022R1A2C3007292) and by the 2020 Joint Research Project of Institutes of Science and Technology.
Authorship
Contribution: S.K., Y.C.S., E.-C.S., and S.-H.P. conceptualized and designed the study; S.K., S.W.L., D.C., M.H., J.-Y.C., and H.L. collected and assembled the data; S.K., S.W.L., M.H., Y.C.S., E.-C.S., and S.-H.P. analyzed and interpreted the data; S.K. and S.-H.P. wrote the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: D.C., B.H.L., and S.H.Y. are employees of NeoImmuneTech, Inc. M.H. and Y.C.S. are employees of Genexine, Inc. The remaining authors declare no competing financial interests.
Correspondence: Su-Hyung Park, Laboratory of Translational Immunology and Vaccinology, Graduate School of Medical Science and Engineering, Korea Advanced Institute of Science and Technology, 291 Daehak-ro, Daejeon 34141, Republic of Korea; e-mail: park3@kaist.ac.kr; Eui-Cheol Shin, Laboratory of Immunology and Infectious Diseases, Graduate School of Medical Science and Engineering, Korea Advanced Institute of Science and Technology, 291 Daehak-ro, Daejeon 34141, Republic of Korea; e-mail: ecshin@kaist.ac.kr; and Howard Lee, Department of Clinical Pharmacology and Therapeutics, Seoul National University College of Medicine and Hospital; Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, 101 Daehak-ro, Jongno-gu, Seoul 03080, Republic of Korea; e-mail: howardlee@snu.ac.kr.
References
Author notes
Sequencing data can be found in the Gene Expression (accession number GSE214137).
Data are available on request from the corresponding author, Su-Hyung Park (park3@kaist.ac.kr).
The full-text version of this article contains a data supplement.