Key Points
Inhibition of RAB protein function mediates the anti–acute myeloid leukemia activity of statins.
Statin sensitivity is associated with enhanced vesicle-mediated traffic.
Abstract
Cholesterol homeostasis has been proposed as one mechanism contributing to chemoresistance in AML and hence, inclusion of statins in therapeutic regimens as part of clinical trials in AML has shown encouraging results. Chemical screening of primary human AML specimens by our group led to the identification of lipophilic statins as potent inhibitors of AMLs from a wide range of cytogenetic groups. Genetic screening to identify modulators of the statin response uncovered the role of protein geranylgeranylation and of RAB proteins, coordinating various aspect of vesicular trafficking, in mediating the effects of statins on AML cell viability. We further show that statins can inhibit vesicle-mediated transport in primary human specimens, and that statins sensitive samples show expression signatures reminiscent of enhanced vesicular trafficking. Overall, this study sheds light into the mechanism of action of statins in AML and identifies a novel vulnerability for cytogenetically diverse AML.
Introduction
Despite many advances in acute myeloid leukemia (AML) therapy in recent years, this disease is still associated with poor prognosis. Early preclinical studies suggested that adaptation of cholesterol homeostasis represents 1 of the mechanisms contributing to chemoresistance in AML.1,2 Statins are competitive inhibitors of HMGCR, the rate-limiting enzyme in the mevalonate pathway. This pathway is involved in cholesterol biosynthesis and produces a variety of other bioactive compounds, most notably isoprenoids, required for intracellular signaling and vesicle-mediated trafficking,3 as well as coenzyme Q, which acts as an electron carrier in mitochondrial respiration.4
The effect of statins on AML cell survival has been predominantly studied using AML cell lines that do not completely recapitulate the disease and that poorly reflect the genetic heterogeneity of AML. Studies exploring the impact of statins on primary AML cells show a heterogeneity of responses5,6 and a possible association between poor prognosis and reduced statin sensitivity.5 To improve AML treatment outcomes, addition of pravastatin to standard induction chemotherapy has been explored, and results of a phase 1 study are encouraging.7 A phase 2 study of idarubicin and cytarabine in combination with pravastatin reported an increased complete response rate for relapsed AML8 and improved outcomes for patients with unfavorable prognosis.9
Using our recently developed in vitro culture system that preserves leukemia stem cell activity,10 we report the statin response of a collection of >200 primary human AML specimens. We uncovered vesicular trafficking as a key determinant of statin sensitivity in AML and as a novel vulnerability for this disease.
Methods
Primary human AML specimens and cell viability assays
This study was approved by the Research Ethics Boards of the University of Montréal and Maisonneuve-Rosemont Hospital. All samples were collected with informed consent and were annotated and cryopreserved at the Quebec Leukemia Cell Bank. Viability assays were performed as described.11
Whole-genome CRISPR/Cas9 screening
Screening was performed as described.12,13
Endocytosis/pinocytosis assay
Specimens were treated with atorvastatin (1 μM) with or without geranylgeranyl pyrophosphate (2 μM) for 24 hours. Cells were exposed to dextran–fluorescein isothiocyanate or dextran–Pacific blue at 100 μg/mL for 30 minutes. Data were acquired using FACSCanto and analyzed using Diva software.
Results and discussion
Lipophilic statins are potent inhibitors of cytogenetically diverse AML
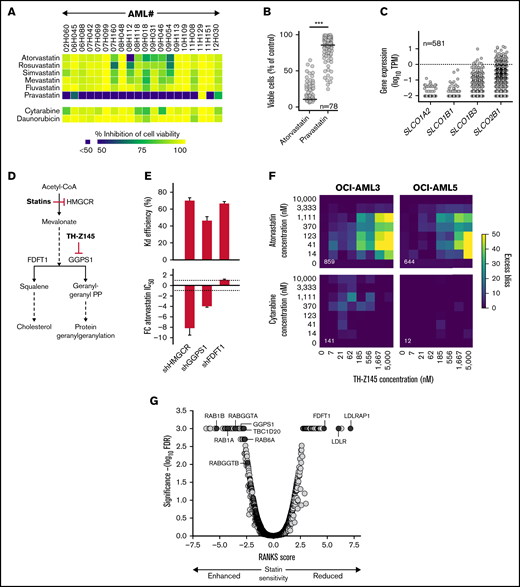
We previously interrogated a collection of 20 cytogenetically diverse primary human AML specimens (supplemental Table 1 screen 1) with a library of 5013 compounds11 and hits included statins (Figure 1A). Screening of an extended cohort of specimens (n = 78; supplemental Table 1 screen 2; supplemental Table 2) highlighted the dramatically reduced ability of pravastatin to inhibit AML cell viability compared with atorvastatin (Figure 1B). Statins differ in their import mechanism; whereas lipophilic statins such as atorvastatin mostly cross cell membranes by passive diffusion, hydrophilic statins like pravastatin mainly enter cells via carrier-mediated mechanisms.14 Interestingly, organic anion transporter proteins, responsible for pravastatin cellular uptake, are poorly transcribed in primary human AML specimens (Figure 1C), likely explaining the low pravastatin efficacy in AML cells in vitro.
Inhibition of RAB protein function mediates the anti-AML activity of statins. (A) Heatmap illustrating sensitivity of 20 primary human AML specimens to 2.5 μM of indicated statins. (B) Viability of primary human AML cells after 6-day incubation in the presence of 2.5 μM of atorvastatin or pravastatin. Horizontal lines represent median inhibition achieved by pravastatin (14%) and atorvastatin (74%). (C) Expression levels of genes implicated in transmembrane transport of pravastatin in primary human AML specimens from the Leucegene cohort. Dotted line, 1 TPM. (D) Schematic representation of the mevalonate pathway. Red bars indicate targets of statins and TH-Z145 compound. (E) Knockdown efficiency achieved by short hairpin RNAs (shRNAs) targeting HMGCR, FDFT1, and GGPS1 (top) and corresponding fold change (FC) in atorvastatin 50% inhibitory concentration (IC50; bottom) in OCI-AML5 cells. Average of 3 shRNAs achieving similar knockdown levels is shown with standard error of the mean. (F) Heatmaps showing excess bliss scores for treatment of OCI-AML3 and OCI-AML5 cells with atorvastatin, cytarabine, and TH-Z145 at indicated concentrations. Numbers in white refer to the sum of all scores >0 (indicative of synergy) for each surface. Representative of 2 independent experiments. Results were analyzed using the R (v3.6.1) SynergyFinder (v2.0.12) package. (G) Results of CRISPR/Cas9 whole-genome screening performed in NALM6 cells treated with 150 nM of cerivastatin. Robust analytics and normalization for knockout screens (RANKS) scores are presented (average of 10 sgRNAs per gene), and statistical assessment was performed by RANKS with false discovery rate (FDR) correction. Genes with FDR values <0.001 were assigned a FDR value of 0.001. ***P < .0001. PP, pyrophosphate; TPM, transcripts per million.
Inhibition of RAB protein function mediates the anti-AML activity of statins. (A) Heatmap illustrating sensitivity of 20 primary human AML specimens to 2.5 μM of indicated statins. (B) Viability of primary human AML cells after 6-day incubation in the presence of 2.5 μM of atorvastatin or pravastatin. Horizontal lines represent median inhibition achieved by pravastatin (14%) and atorvastatin (74%). (C) Expression levels of genes implicated in transmembrane transport of pravastatin in primary human AML specimens from the Leucegene cohort. Dotted line, 1 TPM. (D) Schematic representation of the mevalonate pathway. Red bars indicate targets of statins and TH-Z145 compound. (E) Knockdown efficiency achieved by short hairpin RNAs (shRNAs) targeting HMGCR, FDFT1, and GGPS1 (top) and corresponding fold change (FC) in atorvastatin 50% inhibitory concentration (IC50; bottom) in OCI-AML5 cells. Average of 3 shRNAs achieving similar knockdown levels is shown with standard error of the mean. (F) Heatmaps showing excess bliss scores for treatment of OCI-AML3 and OCI-AML5 cells with atorvastatin, cytarabine, and TH-Z145 at indicated concentrations. Numbers in white refer to the sum of all scores >0 (indicative of synergy) for each surface. Representative of 2 independent experiments. Results were analyzed using the R (v3.6.1) SynergyFinder (v2.0.12) package. (G) Results of CRISPR/Cas9 whole-genome screening performed in NALM6 cells treated with 150 nM of cerivastatin. Robust analytics and normalization for knockout screens (RANKS) scores are presented (average of 10 sgRNAs per gene), and statistical assessment was performed by RANKS with false discovery rate (FDR) correction. Genes with FDR values <0.001 were assigned a FDR value of 0.001. ***P < .0001. PP, pyrophosphate; TPM, transcripts per million.
Screening of a cohort of 205 primary human AML specimens (supplemental Table 1 screen 3; supplemental Table 3) revealed median statin 50% inhibitory concentrations in the nanomolar range (supplemental Figure 1A) and high correlation between responses to the different lipophilic statins across specimens (supplemental Figure 1B). Statin sensitivity was associated with lower marrow blast counts (P = .015), but not with patient age, sex, or white blood cell counts (data not shown). Core binding factor [inv(16) and t(8;21)] AML and M4Eo AML by French-American-British classification also showed enhanced statin sensitivity (supplemental Figure 1C-D). Normal mobilized peripheral blood– or cord blood–derived CD34+ and more primitive CD34+CD45RA− cells, on the other hand, were characterized by reduced statin sensitivity compared with AML samples (supplemental Figure 1E). Overall, these results demonstrate that lipophilic statins are potent anti-AML agents.
Inhibition of RAB protein function mediates the anti-AML activity of statins
Inhibition of protein geranylgeranylation has been suggested to mediate inhibition of AML cell viability by statins.15,16 In support of this, short hairpin RNA–mediated knockdown of HMGCR and GGPS1 (Figure 1D) in OCI-AML5 cells indeed led to a an increase in statin sensitivity (Figure 1E), likely by rendering cells more dependent on the mevalonate pathway, as reported by others,17 whereas knockdown of the FDFT1 gene did not affect AML cell sensitivity to statins (Figure 1E). Synergy between treatment with atorvastatin and GGPS1 inhibitor TH-Z14518 was also observed for inhibition of AML cell viability (Figure 1F; supplemental Figure 2).
To further dissect the statin response, we performed whole-genome CRISPR/Cas9 genetic screening in the NALM6 acute lymphoblastic leukemia cell line already optimized for CRISPR/Cas9 screening by our group12 and observed an overrepresentation of gene sets related to vesicle-mediated transport among genes showing synthetic lethal interactions with statin treatment (supplemental Figure 3), with RAB family genes (RAB1A, RAB1B, and RAB6A) and RAB activator TBC1D20 found to be among the most significant synthetic lethal interactions (Figure 1G; supplemental Table 4). Short hairpin RNA–mediated knockdown of RAB genes also sensitized AML cells to statins (supplemental Figure 4), suggesting that identified interactions are relevant to AML. Interestingly, RAB proteins are central coordinators of vesicular trafficking,19 requiring geranylgeranylation for membrane association and intrinsic activity, and knockouts of GGPS1 and of the 2 subunits of the RAB geranylgeranyltransferase (RABGGTA and RABGGTB) were found among most significant synthetic lethal interactions (Figure 1G). HMGCR knockout also showed a synthetic lethal interaction with statins (supplemental Table 4). Knockout of genes involved in cholesterol homeostasis (LDLR and LDLRAP1) as well as of the FDFT1 gene, on the other hand, rescued the effects of statins (Figure 1G). This could be explained by the inactivation of negative feedback loops normally induced by high cholesterol levels,20 leading to upregulation of the mevalonate pathway. Altogether, these results, along with the reported ability of statins to inhibit geranylgeranylation and membrane targeting of RAB proteins,21 suggest that inhibition of RAB protein function contributes to the effects of statins on AML cell viability.
Vesicular trafficking is a key determinant of the statin response in AML
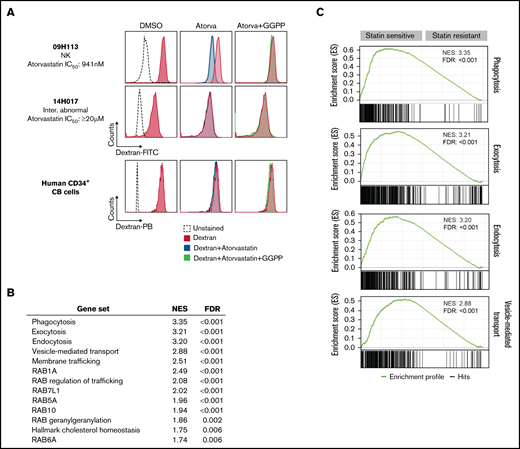
We next assessed the impact of statin treatment on vesicular trafficking in AML cells. Atorvastatin was indeed able to inhibit endocytosis/pinocytosis in statin-sensitive primary human AML specimens (Figure 2A; supplemental Figure 5), but not in statin-resistant AML or CD34+ cord blood cells (Figure 2A). Addition of geranylgeranyl pyrophosphate (Figure 1D) rescued the effect of atorvastatin on endocytosis/pinocytosis (Figure 2A), suggesting that statins inhibit vesicular transport by interfering with protein geranylgeranylation. Vesicle-mediated transport has been shown to be essential for cell viability,12 suggesting that inhibition of vesicular trafficking by statins can account for their effects on cell viability. We next performed differential transcriptome analysis of statin-sensitive and -resistant specimens (Figure 2B), and among gene sets enriched in statin-sensitive specimens compared with statin-resistant ones were found several related to vesicular trafficking (Figure 2B-C). These results suggest that statin-sensitive leukemias show enhanced vesicle-mediated trafficking, likely rendering them more vulnerable to inhibition of this process by statins and providing a possible explanation for the observed impact of vesicular trafficking regulator downregulation on statin sensitivity in AML.
Vesicular trafficking is a key determinant of statin sensitivity in AML. (A) Impact of atorvastatin treatment on endocytosis as assessed by uptake of dextran–fluorescein isothiocyanate (FITC) or dextran–Pacific blue (PB) determined by flow cytometry in primary human AML specimens and CD34+ cord blood (CB) cells. Results for statin-sensitive specimen 09H113 are representative of 12 primary human AML specimens (supplemental Figure 5). Results for CD34+ CB cells are representative of 2 independent experiments performed with CD34+ cells isolated from 2 different CB units. Specimen 14H017 was the only statin-resistant specimen among samples tested as part of this study. (B) Enrichment scores of gene sets related to vesicular transport in top 25% statin-sensitive vs -resistant primary human AML specimens of the Leucegene cohort (n = 204). Gene set enrichment analysis (GSEA) was performed with GSEA 4.1.0 software from the Broad Institute using a list of differentially expressed genes between sensitive and resistant specimens ordered based on fold change of expression. (C) Enrichment profiles of top 4 enriched gene sets related to vesicular transport in statin-sensitive compared with statin-resistant specimens. FDR, false discovery rate; GGPP, geranylgeranyl pyrophosphate; IC50, 50% inhibitory concentration; NES, normalized enrichment score; NK, natural killer.
Vesicular trafficking is a key determinant of statin sensitivity in AML. (A) Impact of atorvastatin treatment on endocytosis as assessed by uptake of dextran–fluorescein isothiocyanate (FITC) or dextran–Pacific blue (PB) determined by flow cytometry in primary human AML specimens and CD34+ cord blood (CB) cells. Results for statin-sensitive specimen 09H113 are representative of 12 primary human AML specimens (supplemental Figure 5). Results for CD34+ CB cells are representative of 2 independent experiments performed with CD34+ cells isolated from 2 different CB units. Specimen 14H017 was the only statin-resistant specimen among samples tested as part of this study. (B) Enrichment scores of gene sets related to vesicular transport in top 25% statin-sensitive vs -resistant primary human AML specimens of the Leucegene cohort (n = 204). Gene set enrichment analysis (GSEA) was performed with GSEA 4.1.0 software from the Broad Institute using a list of differentially expressed genes between sensitive and resistant specimens ordered based on fold change of expression. (C) Enrichment profiles of top 4 enriched gene sets related to vesicular transport in statin-sensitive compared with statin-resistant specimens. FDR, false discovery rate; GGPP, geranylgeranyl pyrophosphate; IC50, 50% inhibitory concentration; NES, normalized enrichment score; NK, natural killer.
Genomic, chemical, and molecular approaches thus identified vesicle-mediated trafficking as a determinant of the statin response in AML and as a novel vulnerability in this disease. Targeting this process therefore represents a promising therapeutic avenue in AML, and these results suggest that clinical development of statins, so far mostly limited to pravastatin, and more particularly of potent lipophilic statins, might demonstrate benefits for AML treatment.
Acknowledgments
The authors thank Muriel Draoui for project coordination, Sophie Corneau for sample coordination, and Jean Duchaine and Karine Audette from the Institute for Research in Immunology and Cancer (IRIC) high-throughput screening platform for managing compound additions and data acquisition. The authors also acknowledge the contribution of Banque de cellules leucémiques du Québec members Giovanni d’Angelo, Claude Rondeau, and Sylvie Lavallée.
This work was supported by the Government of Canada through Genome Canada and the Ministère de l’économie et de l’innovation du Québec through Génome Québec and was also partly funded by the IRIC philanthropic fund for strategic projects. Support from Canadian Cancer Society Research Institute to G.S. is also acknowledged. V.-P.L. was supported by a fellowship from the Cole Foundation and a Vanier scholarship awarded by the Canadian Institutes of Health Research. N.N. was supported by a scholarship from the Cole Foundation. I. Baccelli was supported by a fellowship from the Human Frontier Science Program. C.P. was supported by fellowships from the German Cancer Aid and the Cole Foundation. We dedicate this article to Jana Krosl, who died during the writing of the manuscript.
Authorship
Contribution: J.K. designed the project and performed and analyzed screens and experiments; M.-E.B. contributed to project design, analyzed data, and wrote the paper; C.M. contributed to project design and data analysis; T.M. cloned the short hairpin RNAs used in functional studies and performed dextran uptake experiments; I. Boivin performed chemical screening as well as synergy experiments; N.M. and D.G. contributed to project design; I. Baccelli performed viability screening; V.-P.L., R.B., and B.L. contributed to data analysis; R.M.-S. and R.R. synthesized TH-Z145; T.B. and J.C.-H. performed and analyzed the CRISPR/Cas9 genetic screening under the supervision of M.T.; G.B. performed analysis of synergy experiments; N.N. contributed to functional studies assessing the impact of HMGCR knockdown on statin sensitivity; C.P. and F.B. contributed to project design; P.G., G.B., and S.L. developed the Leucegene database that integrates clinical data and sequencing results; A.M. is responsible for the chemistry team as part of the Leucegene Project and contributed to project design and compound selection; J.H. contributed to project design, selected all the AML samples, and revised their cytogenetic and clinical annotations; and G.S. contributed to project design and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Dedication: We dedicate this article to Jana Krosl, who died during the writing of the manuscript.
Correspondence: Guy Sauvageau, Institute for Research in Immunology and Cancer (IRIC), P.O. Box 6128, Station Centre-Ville, Montreal, QC H3C 3J7, Canada; e-mail: guy.sauvageau@umontreal.ca; and Josée Hébert, Division of Hematology-Oncology and Quebec Leukemia Cell Bank, Maisonneuve-Rosemont Hospital, 5415 L’Assomption Blvd, Montreal, QC H1T 2M4, Canada; e-mail: josee.hebert@umontreal.ca.
References
Author notes
For data sharing, please contact the corresponding authors at guy.sauvageau@umontreal.ca or josee.hebert@umontreal.ca.
The full-text version of this article contains a data supplement.