TO THE EDITOR:
Clinical features of pediatric del(5q) acute myeloid leukemia (AML) overlap with FAB M0, and its prognosis is dismal that worsens with the addition of complex karyotypes.1-5 However, few studies have examined the molecular features beyond cytogenetics of childhood del(5q) AML due to the low prevalence of this disease. Therefore, it is important to better understand their molecular mechanisms leading to novel treatment strategy.
The MLLT10 gene, located at 10p12, can fuse with numerous partner genes in both AML and T-cell acute lymphoblastic leukemia (T-ALL).6,7 The MLLT10 encodes AF10 protein, of which OM-LZ domain is crucial for leukemogenesis and is retained in all fusions that lead to leukemia. HNRNPH1-MLLT10 fusion has been reported in T-ALL and shares the gene expression profile signatures of the HOXA subgroup,7 but no such fusion has ever been reported in AML.
Here, we are the first, to our knowledge, to report a case of pediatric AML with del(5q) and HNRNPH1-MLLT10 fusion.
A 9-year-old boy was admitted to our hospital with enlargement of the cervical lymph nodes and gingival swelling, along with a history of a continuous fever for 2 months. A peripheral blood examination revealed leukopenia without circulating blasts. Bone marrow analysis revealed myeloperoxidase− blasts accounting for 70% of the cells. Immunophenotyping of the leukemic cells showed positivity for CD5, CD7, CD11b, CD13, CD33, CD34, CD41, CD45, CD117, and HLA-DR, which was consistent with AML (FAB M0). Conventional cytogenetic analysis revealed a complex karyotype, characterized by abnormalities in both chromosomes 5, additions and deletions in multiple chromosomes, and a marker chromosome (supplemental Table 1; supplemental Figure 3). Multicolor fluorescence in situ hybridization (Figure 1A) enabled us to further characterize some of the abnormalities noted by G-banding. One abnormal chromosome 5 was a derivative from a translocation with chromosome 10, and the second abnormal chromosome 5 carried a deletion. Abnormalities of other chromosomes initially described as additions were subsequently found to be deletions, and the marker chromosome was found to be derived from chromosome 11. We diagnosed the patient with del(5q) AML with a complex karyotype, and he was enrolled in the Japanese Pediatric Leukemia/Lymphoma Study Group AML-12 protocol with complete remission after 2 cycles of induction therapy.8 Considering the high relapse rate in previous reports of del(5q) AML with complex karyotype,1-3 he received unrelated cord blood cell transplantation during the first complete remission after 2 subsequent cycles of consolidation therapy. His first stem cell transplantation from an unrelated donor was rejected due to preengraftment immune reactions and BK viral cystitis, and therefore he received a second graft from his haploidentical mother. The patient has been in complete remission for >2 years since his last stem cell transplantation.
Molecular characterization of the HNRNPH1-MLLT10 fusion. (A) Multicolor fluorescence in situ hybridization indicating a complex karyotype including del(5q). (B) HNRNPH1-MLLT10 splicing isoform identified in the present case is shown. GenBank accession numbers referred to are NM_001364227.2 for HNRNPH1 and NM_004641.4 for MLLT10. Hypothetical fusion protein is shown in which HNRNPH1 maintained all 3 RNA-recognition motifs at the N-terminus and MLLT10 maintained the critical OM-LZ domain at the C-terminus. Sanger sequencing of complementary DNA revealed that exon 10 of HNRNPH1 (rather than exon 12, which was reported previously) fused in frame with exon 15 of MLLT10 (as reported previously). (C) Sanger sequencing indicates genomic breakpoints at position Chr5:179 042,232 within HNRNPH1 gene and Chr10: 21 999,262 in MLLT10 gene with a single nucleotide insertion.
Molecular characterization of the HNRNPH1-MLLT10 fusion. (A) Multicolor fluorescence in situ hybridization indicating a complex karyotype including del(5q). (B) HNRNPH1-MLLT10 splicing isoform identified in the present case is shown. GenBank accession numbers referred to are NM_001364227.2 for HNRNPH1 and NM_004641.4 for MLLT10. Hypothetical fusion protein is shown in which HNRNPH1 maintained all 3 RNA-recognition motifs at the N-terminus and MLLT10 maintained the critical OM-LZ domain at the C-terminus. Sanger sequencing of complementary DNA revealed that exon 10 of HNRNPH1 (rather than exon 12, which was reported previously) fused in frame with exon 15 of MLLT10 (as reported previously). (C) Sanger sequencing indicates genomic breakpoints at position Chr5:179 042,232 within HNRNPH1 gene and Chr10: 21 999,262 in MLLT10 gene with a single nucleotide insertion.
To determine the genetic alteration of the patient’s AML, we extracted the total RNA from the bone marrow sample at diagnosis and performed transcriptome analysis using TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA). The fusion gene HNRNPH1-MLLT10 was identified and then validated by reverse transcription polymerase chain reaction and direct sequencing. Normalized count data obtained by the variance-stabilizing transformation function of the R package DESeq2 1.16.1 were used for the expression analysis. Prior to the clustering analysis, the data were filtered to remove genes deemed to be unrelated to disease, such as those from the sex chromosomes, as well as genes from contaminating normal erythrocytes (supplemental Table 2). For the clustering analysis, we used Ward’s hierarchical clustering method and included 500 of the top 2% of differentially expressed genes (supplemental Table 3) that were extracted using DESeq2. To compare the patient’s sample with samples from other children with leukemia, we also used the open data set of expression data for pediatric AML and T-ALL.9,10 To analyze the expression status of the patient’s sample, we performed hierarchical clustering based on genes specific for T-ALL (supplemental Table 4) and KMT2A-rearranged leukemia with activation of HOXA9 pathway genes (supplemental Table 5). Fusion transcripts were detected using Genomon version 2.6.1 and filtered by excluding fusions: (1) mapping to repetitive regions, (2) with fewer than 4 spanning reads, (3) that occurred out of frame, or (4) that had junctions not located at known exon-intron boundaries.9 The study was approved by the ethics committee of the Hirosaki University Graduate School of Medicine and was conducted in accordance with the Declaration of Helsinki.
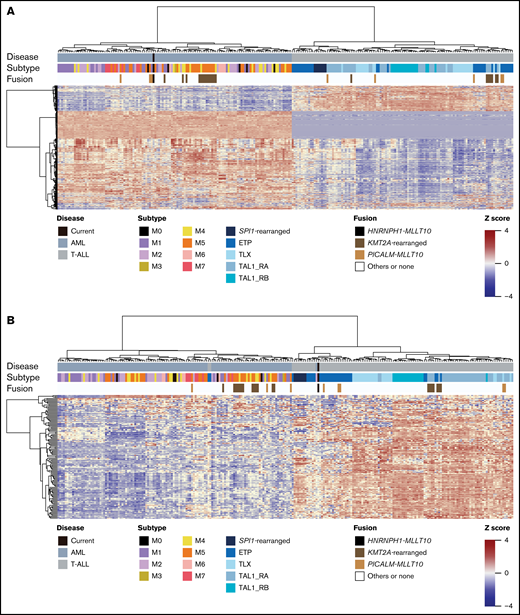
Here, we report the first known case of pediatric AML with del(5q) and HNRNPH1-MLLT10 fusion and reveal similarities in gene expression profiles in AML (FAB M0) and T-ALL by transcriptome analysis. The immunophenotype and gene expression profile based on unsupervised clustering analysis confirmed the diagnosis of AML, and the gene expression patterns of this HNRNPH1-MLLT10 fusion case were similar to those of the PICALM-MLLT10 fusion, which has previously been identified in both AML and T-ALL (Figure 2A).6,7
Gene expression pattern of the HNRNPH1-MLLT10 fusion in AML. (A) All samples were clustered by unsupervised hierarchical clustering by differentially expressed genes (supplemental Table 3). Samples were divided into 2 clusters: the AML cluster and the T-ALL cluster. This unsupervised clustering was based on the biological nature of samples. Our current case was included in the AML cluster. (B) All samples were clustered by supervised hierarchical clustering by a gene set specific for T-ALL genes (supplemental Table 4). The samples were classified into 2 clusters according to the presence or absence of T-ALL genes expression. Our current sample was clustered into the ETP branch of the T-ALL cluster.
Gene expression pattern of the HNRNPH1-MLLT10 fusion in AML. (A) All samples were clustered by unsupervised hierarchical clustering by differentially expressed genes (supplemental Table 3). Samples were divided into 2 clusters: the AML cluster and the T-ALL cluster. This unsupervised clustering was based on the biological nature of samples. Our current case was included in the AML cluster. (B) All samples were clustered by supervised hierarchical clustering by a gene set specific for T-ALL genes (supplemental Table 4). The samples were classified into 2 clusters according to the presence or absence of T-ALL genes expression. Our current sample was clustered into the ETP branch of the T-ALL cluster.
The immunophenotype of the leukemic cells of the patient did not show staining of the cytoplasmic CD3 and immunoglobulin M, allowing us to exclude early T-cell precursor acute lymphoblastic leukemia (ETP-ALL). However, his leukemic cells expressed immature markers downstream of the early T-cell progenitors.10-12 Notably, hierarchical clustering based on genes specific for T-ALL9 (supplemental Table 4) classified the fusion gene–positive AML into a subgroup of T-ALL, specifically the same branch as SP1 fusion–positive T-ALL and ETP-ALL (Figure 2B). Thus, our data suggested that AML with del(5q) and HNRNPH1-MLLT10 fusion has the gene expression profile signature of both immature myeloid precursors and early T-cell precursors.
We confirmed the presence of the HNRNPH1-MLLT10 fusion transcript and identified the breakpoint (Figure 1B-C). The OM-LZ domain of MLLT10 has been retained in all reported cases of MLLT10 fusion gene–positive leukemia, indicating that the domain is crucial for leukemogenesis through its interaction with chromatin-modifying proteins.6,13-16 Indeed, the OM-LZ domain drives the abnormal enzyme activity of disruptor of telomeric silencing 1-like (DOT1L), which maintains a gene activation state, resulting in an upregulation of the HOXA9 group of genes.13-18
We performed unsupervised hierarchical clustering by differentially expressed genes. Based on the biological characteristics of the samples, they were divided into 2 clusters: an AML cluster and a T-ALL cluster, and this case was included in the AML cluster (Figure 2A). Then, all samples were clustered by supervised hierarchical clustering of all samples with gene sets specific to T-ALL genes (supplemental Table 4). The samples were classified into 2 clusters according to the presence or absence of T-ALL gene expression, and the present case was classified into the ETP-ALL branch of the T-ALL cluster (Figure 2B).
As DOT1L-mediated activation of the HOXA9 pathway has been reported in leukemia that are positive for KMT2A or MLLT10 fusion genes,6,13-18 we performed a supervised clustering analysis of HOXA9 pathway–specific genes to confirm that the HOXA9 pathway was activated in this case (supplemental Table 5)19 and showed the expression level of several representative genes. In the patient, MEIS1 was highly expressed, combined with PBX3 silencing similar to the PICALM-MLLT10 fusion gene,20 suggesting that the HOXA9 pathway was also activated (supplemental Figure 1).
This case study found similarities between the gene expression patterns of AML (FAB M0) and T-ALL. The gene expression signature profile of FAB M0 is heterogeneous (supplemental Figure 2). There have been reports of FAB M0 AML with del(5q) and a few cases in which the fusion genes expressed T-cell antigen, suggesting that the combination of del(5q) and fusion gene might be involved in leukemogenesis in the subgroup of AML (FAB M0).21,22
In conclusion, we report the first case of pediatric AML with del(5q) and HNRNPH1-MLLT10 fusion, in which the gene expression profiling revealed similarities between immature AML and the subgroup of early T-cell precursor ALL. Further studies on pediatric AML (FAB M0) are warranted to investigate whether it shares a gene expression profile signature with T-ALL and to explore new treatment strategies for this subgroup.
Acknowledgments: The authors would like to thank Hisae Kudo and Miwa Hashimoto for their technical assistance.
This work was supported in part by the Japan Society for the Promotion of Science (JSPS) through a Grant-in-Aid for Scientific Research (KAKENHI, grant number 21K07813).
Contribution: K.K., T.T., Y.K., and E.I. designed the study; K.K., Y.K., T.T., and R.K. performed the experiments; K.K., Y.K., T.T., and R.K. analyzed and interpreted the data; K.K. and Y.K. wrote the manuscript; K.K., A.K., T.S., T.K., S.S., E.I., and K.T. evaluated the patient and collected and interpreted the clinical data; N.S., D.T., S.A., K.Y., S.O., M.S., and J.T. provided data sets; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kiminori Terui, Department of Pediatrics, Hirosaki University Graduate School of Medicine, 5 Zaifucho, Hirosaki, Aomori 036-8562, Japan; e-mail: teru@hirosaki-u.ac.jp.
References
Author notes
Requests for data sharing may be submitted to Kiminori Terui (teru@hirosaki-u.ac.jp).
The full-text version of this article contains a data supplement.