Key Points
SH2B3 mutations in JMML are associated with more aggressive disease in humans and Nf1 Sh2b3-mutant mice.
This novel mouse model provides a platform for studying the biology and therapy of high-risk JMML.
Abstract
Juvenile myelomonocytic leukemia (JMML) is initiated in early childhood by somatic mutations that activate Ras signaling. Although some patients have only a single identifiable oncogenic mutation, others have 1 or more additional alterations. Such secondary mutations, as a group, are associated with an increased risk of relapse after hematopoietic stem cell transplantation or transformation to acute myeloid leukemia. These clinical observations suggest a cooperative effect between initiating and secondary mutations. However, the roles of specific genes in the prognosis or clinical presentation of JMML have not been described. In this study, we investigate the impact of secondary SH2B3 mutations in JMML. We find that patients with SH2B3 mutations have adverse outcomes, as well as higher white blood cell counts and hemoglobin F levels in the peripheral blood. We further demonstrate this interaction in genetically engineered mice. Deletion of Sh2b3 cooperates with conditional Nf1 deletion in a dose-dependent fashion. These studies illustrate that haploinsufficiency for Sh2b3 contributes to the severity of myeloproliferative disease and provide an experimental system for testing treatments for a high-risk cohort of JMML patients.
Introduction
Juvenile myelomonocytic leukemia (JMML) is an aggressive myeloid malignancy of early childhood initiated by germline or somatic mutations in NF1, CBL, NRAS, KRAS, PTPN11, and other Ras-pathway genes.1 In patients predisposed to JMML due to a heterozygous mutation of NF1 or CBL in the germline, JMML occurs after somatic loss of the normal allele in the hematopoietic compartment. Because of its low-grade histologic appearance and association with refractory anemia and thrombocytopenia, JMML is categorized as a mixed myelodysplastic syndrome/myeloproliferative neoplasm (MPN). Hematopoietic stem cell (HSC) transplantation is the only curative therapy; however, 40% to 50% of JMML patients subsequently relapse.
Genome-wide sequencing studies have recently demonstrated, in some JMML patients, secondary mutations in cooperating genes that are associated with adverse outcomes, even when they are present in minor subclones at diagnosis.2-5 Importantly, coexistence of these mutations with the initiating Ras-pathway lesions in JMML cells supports a role in disease progression.3,4,6 Together, these observations suggest that secondary mutations may contribute to treatment failure.
SH2B3 (also known as LNK) alterations are among the most common secondary mutations observed in JMML. SH2B3 is also somatically mutated in other myeloid diseases such as polycythemia vera, essential thrombocythemia, primary myelofibrosis, and acute myeloid leukemia.7 The SH2B3 alterations found in JMML and other hematologic disorders are typically heterozygous point mutations that reduce SH2B3 function, although biallelic mutations have also been described in some patients. SH2B3, an adapter protein that attenuates signaling downstream of multiple cytokines,7 was recently shown to promote degradation of signaling molecules by recruiting the ubiquitin ligase CBL, which is encoded by one of the canonical JMML Ras-pathway genes.8 SH2B3 targets the tyrosine kinase JAK2 to dampen signals induced by cytokines such as thrombopoietin (TPO), erythropoietin, and granulocyte-macrophage colony-stimulating factor (GM-CSF). In addition, SH2B3 can suppress responses to stem cell factor via the receptor c-kit, and to interleukin 7 via JAK3.
In mice, homozygous Sh2b3 deficiency causes an increase in both quantity and repopulating ability of HSCs, mediated by excessive TPO signaling.9-12 B-cell and myeloid precursors, megakaryocytes, and platelets are also expanded through deregulation of interleukin 7, GM-CSF, stem cell factor, and TPO.13-15 MPNs developed in Sh2b3−/− mice by age 12 to 18 months, and these mice were more sensitive to the proliferative effects of transduced Jak2 or BCR/ABL oncogenes.16 No published studies have described a phenotype associated with heterozygous Sh2b3 mutations, as typically occur in leukemia patients.
We hypothesized that Sh2b3 heterozygosity cooperates with Ras-pathway driver mutations to deregulate myeloid growth in vivo. To test this and to model the underlying molecular genetics observed in patients with JMML, we generated mice with homozygous Nf1 inactivation in the context of heterozygous or homozygous ablation of Sh2b3. Here, we show that Nf1 and Sh2b3 interact to exacerbate multiple aspects of the myelodysplastic syndrome/MPN phenotype.
Methods
Mice
C57BL/6 mice received a single intraperitoneal injection of 250 μg of polyinosinic-polycytidylic acid (Sigma Aldrich USA, St. Louis, MO) at 1 to 3 days of age. Animals were maintained in the specific pathogen-free rodent facility at the University of California, San Francisco (UCSF) and the experimental procedures were approved by the UCSF Committee on Animal Research. Flow cytometry was performed as described.17 Statistical methods are described in figure legends. Log transformations were applied to leukocyte counts and spleen weights to correct heteroscedasticity; if data contained zeroes, square root transformation was used instead.
Human subjects
Studies were approved by the UCSF Committee on Human Research. All participants/guardians provided informed consent in accordance with the Declaration of Helsinki. Data used for this study were obtained as part of the clinical care of the patients. Mutations were identified using a targeted sequencing panel as described.3
Results and discussion
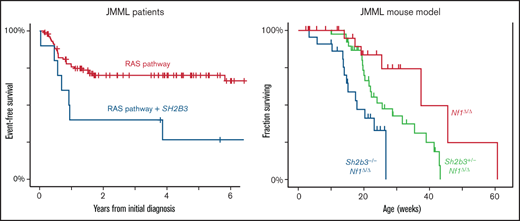
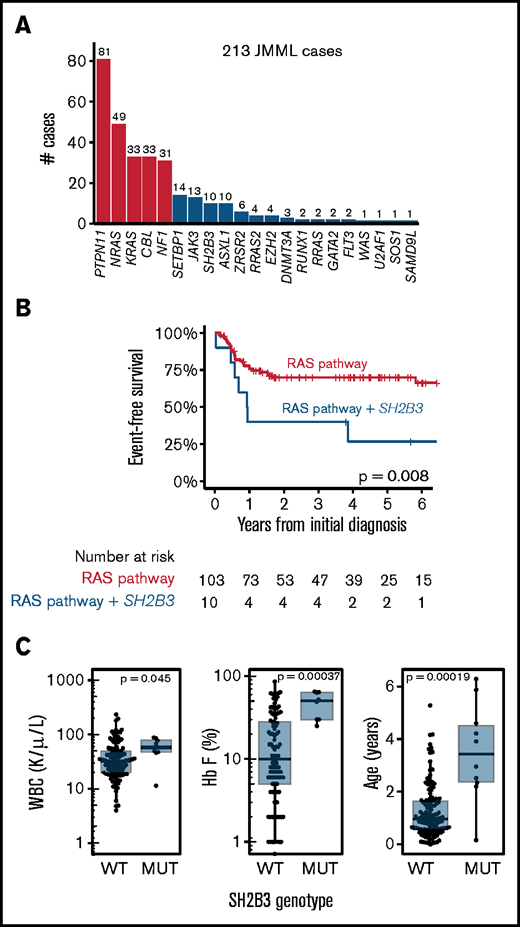
Although the presence of secondary mutations has an adverse prognosis in JMML, the specific impact of SH2B3 has not been investigated. A retrospective analysis of 213 JMML patients with available genotypes, including 112 from previously reported cohorts,3,4,18-20 revealed 9 with SH2B3 mutations at diagnosis and 1 detected only at relapse (Figure 1A; supplemental Figure 1; supplemental Table 1). Despite the small sample size, Kaplan-Meier analysis demonstrated worse event-free survival among JMML patients with SH2B3 mutations compared with those with single Ras-pathway mutations (Figure 1B; supplemental Figure 3). Consistent with this observation, JMML patients with a cooperating SH2B3 mutation also had elevated blood leukocyte counts and hemoglobin F levels, and were diagnosed at an older age (Figure 1C). Four of these patients had been tested for DNA methylation, and all fell into the high-risk classification.20 These clinical data suggest that cooperating mutations in SH2B3 might influence disease severity and prognosis in JMML. However, because multiple adverse risk factors also characterize these cases (older age, hypermethylation, and coexisting mutations in some cases), they do not definitively establish that SH2B3 mutation influences disease severity.
Incidence and impact of SH2B3 mutations in JMML. (A) Frequencies of mutations identified by targeted or genomic sequencing in a cohort of 213 JMML cases. A case is considered positive for any mutation found at diagnosis or relapse. Red bars indicate canonical Ras-pathway genes that are thought to initiate JMML; blue bars represent secondary mutations. (B) Kaplan-Meier analysis of outcomes for JMML, comparing patients with single mutations to those with secondary mutations in SH2B3 (log rank P < .01). (C) WBCs, hemoglobin F levels, and age at diagnosis in groups assigned as in panel B (Wilcoxon P < .05 for WBC and P < .01 for hemoglobin F). MUT, mutated; WT, wild type.
Incidence and impact of SH2B3 mutations in JMML. (A) Frequencies of mutations identified by targeted or genomic sequencing in a cohort of 213 JMML cases. A case is considered positive for any mutation found at diagnosis or relapse. Red bars indicate canonical Ras-pathway genes that are thought to initiate JMML; blue bars represent secondary mutations. (B) Kaplan-Meier analysis of outcomes for JMML, comparing patients with single mutations to those with secondary mutations in SH2B3 (log rank P < .01). (C) WBCs, hemoglobin F levels, and age at diagnosis in groups assigned as in panel B (Wilcoxon P < .05 for WBC and P < .01 for hemoglobin F). MUT, mutated; WT, wild type.
To test this hypothesis prospectively, we investigated the interaction between initiating Ras pathway mutations and cooperating SH2B3 mutations in a relevant in vivo model. We intercrossed Sh2b3-mutant mice21 with a strain carrying a conditional deletion in Nf1 and the Mx1-Cre driver (herein Nf1Δ/Δ mice).22 These mice uniformly develop a JMML-like disease by 6 months of age in the sensitive C57BL/6 × 129SvJ F1 strain background, but this disease is greatly attenuated in inbred C57BL/6 mice.23 Because of the multiple alleles involved, we performed this experiment in the less permissive C57BL6 strain. We reasoned that this subtle but uniform phenotype would provide an informative genetically sensitized background for testing the effects of cooperating mutations. By contrast, JMML models using Kras or Ptpn11 have more severe phenotypes that might obscure cooperative effects, and Nras mice develop diverse hematopoietic neoplasms.24-26 In addition, we noted that the incidence and number of cooperating mutations were greater in JMML with NF1 loss than in cases with other canonical drivers (supplemental Figure 2), suggesting that genetic interactions are clinically relevant in this context. However, we note that the order of mutations in this compound mouse model differs from that in human JMML initiated by NF1 loss, in which SH2B3 mutations are acquired later.
Serial blood leukocyte counts revealed a strong interaction between loss-of-function mutations in Nf1 and Sh2b3 (Figure 2A-B). Importantly, heterozygous Sh2b3 inactivation caused an increase in the white blood cell counts (WBCs) of Nf1Δ/Δ mice, demonstrating a haploinsufficient phenotype for Sh2b3 in the presence of Nf1 inactivation (Figure 2B). These mice demonstrated expansion of all peripheral blood lineages (Figure 2C), suggesting a broad disruption of homeostasis. This included monocytosis, a hallmark of JMML (supplemental Figure 4).There was a trend toward a myeloid bias in Nf1-mutant mice that was preserved with additional mutation in Sh2b3. Nf1Δ/Δ Sh2b3−/− mice also demonstrated massive splenomegaly (Figure 2D) and early mortality (Figure 2E). Histopathologic examination demonstrated a qualitatively similar MPN–like disease process in Nf1Δ/Δ and Nf1Δ/Δ Sh2b3−/− mice (supplemental Figure 5).
Interaction of Nf1 and Sh2b3 in mice.Mx1-Cre, Nf1flox/flox, and Sh2b3−/− mice were intercrossed and Cre expression was induced with polyinosinic-polycytidylic acid at birth. Mice were euthanized when moribund or at end of trial. (A) Combined homozygous mutations in Nf1 and Sh2b3 cause higher WBCs than either mutation does alone. Mean plus or minus standard error of the mean is shown for each time point, with a line for the trend predicted by a mixed-effects linear model.17 Significance by lmertest29 yields P < 10−4 for Nf1Δ/Δ Sh2b3−/− vs Nf1Δ/Δ and for Nf1Δ/Δ Sh2b3−/− vs Sh2b3−/−. (B) A similar interaction was observed when Sh2b3 mutation was heterozygous, though with a lesser degree of leukocytosis. Data are presented as in panel A. P < 10−4 for Nf1Δ/Δ Sh2b3+/− vs Nf1Δ/Δ and for Nf1Δ/Δ Sh2b3+/− vs Sh2b3+/−. (C) Flow cytometry of peripheral blood taken at time of euthanization shows that leukocytosis involves both lymphoid (CD19+ or CD4/8+) and myeloid (Mac1+) lineages, with a trend toward myeloid expansion when Nf1 and Sh2b3 are both mutant. Means and standard errors are shown. In multivariate regression, Sh2b3 mutation increased B and myeloid cells (analysis of variance [ANOVA] P < 10−5), Nf1 mutation increased T (P < .05) and myeloid (P < 10−3) cells, and their interaction further increased B and myeloid cells (P < .05). (D) Splenomegaly is more severe in mice with mutations in both Nf1 and Sh2b3 than in mice with mutations in either gene alone. Individual mice are shown as dots, and the line for each genotype shows the linear regression trend for age. In multivariate linear regression, spleen size increased with time (P < .001 by ANOVA), with the number of mutant Sh2b3 alleles (P < 10−11), and with Nf1 mutation (P < 10−13). Spleen size increased faster in Nf1Δ/Δ mice (P < .01); other interaction terms were not significant. (E) Kaplan-Meier survival analysis shows that Sh2b3 mutation causes a dose-dependent acceleration of mortality in Nf1-mutant mice (log rank P < .01 for 0 vs 1 and for 1 vs 2 Sh2b3 alleles). (F) Extramedullary expansion of HSCs (Lin−c-kit+Sca1−CD150+CD48−) in the spleen is markedly increased when both Nf1 and Sh2b3 are mutated (P < .03 for Nf1Δ/Δ Sh2b3−/− vs Nf1Δ/Δ and for Nf1Δ/Δ Sh2b3−/− vs Sh2b3−/− by Student t test; assessed at time of euthanization).
Interaction of Nf1 and Sh2b3 in mice.Mx1-Cre, Nf1flox/flox, and Sh2b3−/− mice were intercrossed and Cre expression was induced with polyinosinic-polycytidylic acid at birth. Mice were euthanized when moribund or at end of trial. (A) Combined homozygous mutations in Nf1 and Sh2b3 cause higher WBCs than either mutation does alone. Mean plus or minus standard error of the mean is shown for each time point, with a line for the trend predicted by a mixed-effects linear model.17 Significance by lmertest29 yields P < 10−4 for Nf1Δ/Δ Sh2b3−/− vs Nf1Δ/Δ and for Nf1Δ/Δ Sh2b3−/− vs Sh2b3−/−. (B) A similar interaction was observed when Sh2b3 mutation was heterozygous, though with a lesser degree of leukocytosis. Data are presented as in panel A. P < 10−4 for Nf1Δ/Δ Sh2b3+/− vs Nf1Δ/Δ and for Nf1Δ/Δ Sh2b3+/− vs Sh2b3+/−. (C) Flow cytometry of peripheral blood taken at time of euthanization shows that leukocytosis involves both lymphoid (CD19+ or CD4/8+) and myeloid (Mac1+) lineages, with a trend toward myeloid expansion when Nf1 and Sh2b3 are both mutant. Means and standard errors are shown. In multivariate regression, Sh2b3 mutation increased B and myeloid cells (analysis of variance [ANOVA] P < 10−5), Nf1 mutation increased T (P < .05) and myeloid (P < 10−3) cells, and their interaction further increased B and myeloid cells (P < .05). (D) Splenomegaly is more severe in mice with mutations in both Nf1 and Sh2b3 than in mice with mutations in either gene alone. Individual mice are shown as dots, and the line for each genotype shows the linear regression trend for age. In multivariate linear regression, spleen size increased with time (P < .001 by ANOVA), with the number of mutant Sh2b3 alleles (P < 10−11), and with Nf1 mutation (P < 10−13). Spleen size increased faster in Nf1Δ/Δ mice (P < .01); other interaction terms were not significant. (E) Kaplan-Meier survival analysis shows that Sh2b3 mutation causes a dose-dependent acceleration of mortality in Nf1-mutant mice (log rank P < .01 for 0 vs 1 and for 1 vs 2 Sh2b3 alleles). (F) Extramedullary expansion of HSCs (Lin−c-kit+Sca1−CD150+CD48−) in the spleen is markedly increased when both Nf1 and Sh2b3 are mutated (P < .03 for Nf1Δ/Δ Sh2b3−/− vs Nf1Δ/Δ and for Nf1Δ/Δ Sh2b3−/− vs Sh2b3−/− by Student t test; assessed at time of euthanization).
We performed flow cytometry of bone marrow and spleen to quantify mature and immature hematopoietic populations. This analysis was notable primarily for the presence of increased HSCs (Lin−c-kit+Sca1+CD150+CD48−) in spleens of mutant mice. Spleen HSC content was increased modestly in Nf1 mice, and to a greater degree in Sh2b3+/− or Sh2b3−/− mice. The greatest HSC content was found in mice with homozygous loss of both Nf1 and Sh2b3 (Figure 2F).
JMML has 2 defining biologic features: clonal hematopoiesis within the HSC compartment and excessive myeloid proliferation. Extensive mouse modeling has demonstrated that Ras-pathway driver mutations can partly account for each of these abnormalities.27 By regulating JAK2, SH2B3 appears perfectly positioned to exacerbate both of these key pathologic abnormalities in JMML. In HSCs, JAK2 transduces critical signals for maintenance and self-renewal from the cytokine TPO and its receptor, MPL. In myeloid progenitors, JAK2 mediates mitogenic and survival signals from the cytokine GM-CSF. Finally, in addition to exacerbating MPNs, JAK2 signaling may promote resistance to therapy. For example, TPO protects HSC from radiation by promoting DNA repair.28
MEK inhibition reduces myeloproliferation in mouse models of JMML driven by endogenous KrasG12D expression or Nf1 inactivation, and these data have informed an active phase 1/2 trial of trametinib, a highly selective MEK inhibitor, in relapsed/refractory patients (NCT03190915). However, ongoing efforts to improve outcomes for JMML will need to address the adverse impact of cooperating mutations in SH2B3 and other genes. The model presented here is a genetically accurate platform for testing rational drug combinations for targeting both initiating and cooperating mutations in JMML.
Advances in understanding the biology of JMML have begun to suggest the possibility of personalized approaches to therapy. This will require investigating how specific mutations influence disease. This study demonstrates for the first time that SH2B3 mutations specifically exacerbate disease severity and confer a poor prognosis. Furthermore, this is the first experimental demonstration of a haploinsufficient phenotype for Sh2b3, which is unusual for a tumor-suppressor gene.
Acknowledgments
The authors thank Kevin Shannon for critical review of the manuscript.
This work was supported by the Neurofibromatisis Therapeutic Consortium of the Children’s Tumor Foundation (C.E.M. and B.S.B.); Leukemia & Lymphoma Society grant R6511-19 (M.L.L., E.S., and B.S.B.); National Institutes of Health, National Cancer Institute grants U54 CA196519 (M.L.L. and E.S.) and P30 CA082103 (Cancer Center support grant to the Helen Diller Family Comprehensive Cancer Center at the University of California, San Francisco); National Institutes of Health, National Heart, Lung, and Blood Institute grant K08 HL135434 (E.S.); Cookies for Kids’ Cancer (M.L.L.); the Pediatric Cancer Research Foundation (E.S. and B.S.B.); the V Foundation (E.S.), and the Frank A. Campini Foundation (M.L.L. and E.S.).
Authorship
Contribution: C.E.M., E.S., S.C.K., M.L.L., and B.S.B. designed research, performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Benjamin S. Braun, Benioff Children’s Hospital, University of California, San Francisco, Helen Diller Family Cancer Research Building, Mail Code 3112, 1450 3rd St, San Francisco, CA 94158; e-mail: ben.braun@ucsf.edu.
References
Author notes
For data sharing, contact the corresponding author at ben.braun@ucsf.edu. Data set overlaps with prior publications are noted in the following references cited in the manuscript: Stieglitz et al3,4,19,20 and Dvorak et al.18
The full-text version of this article contains a data supplement.

![Interaction of Nf1 and Sh2b3 in mice.Mx1-Cre, Nf1flox/flox, and Sh2b3−/− mice were intercrossed and Cre expression was induced with polyinosinic-polycytidylic acid at birth. Mice were euthanized when moribund or at end of trial. (A) Combined homozygous mutations in Nf1 and Sh2b3 cause higher WBCs than either mutation does alone. Mean plus or minus standard error of the mean is shown for each time point, with a line for the trend predicted by a mixed-effects linear model.17 Significance by lmertest29 yields P < 10−4 for Nf1Δ/Δ Sh2b3−/− vs Nf1Δ/Δ and for Nf1Δ/Δ Sh2b3−/− vs Sh2b3−/−. (B) A similar interaction was observed when Sh2b3 mutation was heterozygous, though with a lesser degree of leukocytosis. Data are presented as in panel A. P < 10−4 for Nf1Δ/Δ Sh2b3+/− vs Nf1Δ/Δ and for Nf1Δ/Δ Sh2b3+/− vs Sh2b3+/−. (C) Flow cytometry of peripheral blood taken at time of euthanization shows that leukocytosis involves both lymphoid (CD19+ or CD4/8+) and myeloid (Mac1+) lineages, with a trend toward myeloid expansion when Nf1 and Sh2b3 are both mutant. Means and standard errors are shown. In multivariate regression, Sh2b3 mutation increased B and myeloid cells (analysis of variance [ANOVA] P < 10−5), Nf1 mutation increased T (P < .05) and myeloid (P < 10−3) cells, and their interaction further increased B and myeloid cells (P < .05). (D) Splenomegaly is more severe in mice with mutations in both Nf1 and Sh2b3 than in mice with mutations in either gene alone. Individual mice are shown as dots, and the line for each genotype shows the linear regression trend for age. In multivariate linear regression, spleen size increased with time (P < .001 by ANOVA), with the number of mutant Sh2b3 alleles (P < 10−11), and with Nf1 mutation (P < 10−13). Spleen size increased faster in Nf1Δ/Δ mice (P < .01); other interaction terms were not significant. (E) Kaplan-Meier survival analysis shows that Sh2b3 mutation causes a dose-dependent acceleration of mortality in Nf1-mutant mice (log rank P < .01 for 0 vs 1 and for 1 vs 2 Sh2b3 alleles). (F) Extramedullary expansion of HSCs (Lin−c-kit+Sca1−CD150+CD48−) in the spleen is markedly increased when both Nf1 and Sh2b3 are mutated (P < .03 for Nf1Δ/Δ Sh2b3−/− vs Nf1Δ/Δ and for Nf1Δ/Δ Sh2b3−/− vs Sh2b3−/− by Student t test; assessed at time of euthanization).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/5/18/10.1182_bloodadvances.2020003754/3/m_advancesadv2020003754f2.png?Expires=1769155054&Signature=ufuDAhg4rDbwyXWneSefglj3Z~hOa7Q1VmYU6RdwPGTZA6V-Zg6tKNDAG5JWJmy9twZLLhEx6WZ5swEHswayVgvz35ukTm2lPve8An-KJixcRS3oSnvGUA1e-QEyOyadR49bwDHXZlPuAJEPRp9jx9SJMfi2JBegQ5nFr0LR0tEebsr56Drk0Rop78jCRMaE6usXzSGKMe09VfsMy49zTmZcLPyYGCJd99IWN-o8AT-eC1SYyCppiwf5vGzhyjGzWCSlT1fF8uDqD3VOu41GgxoqjHPyD28fQmnBM2lN19MJo2988ujk4q3sxLHiXq0-rzMq8z76D3s8UqfCeVlhLg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)