Key Points
Loss of Arhgef2 impairs HSPC spindle orientation and results in severely impaired hematopoietic regeneration at the stem cell level.
Arhgef2 is specifically downregulated in SDS patient HSPCs and is essential for human HSC-driven engraftment upon xenotransplantation.
Abstract
How hematopoietic stem cells (HSCs) coordinate their divisional axis and whether this orientation is important for stem cell–driven hematopoiesis is poorly understood. Single-cell RNA sequencing data from patients with Shwachman-Diamond syndrome (SDS), an inherited bone marrow failure syndrome, show that ARHGEF2, a RhoA-specific guanine nucleotide exchange factor and determinant of mitotic spindle orientation, is specifically downregulated in SDS hematopoietic stem and progenitor cells (HSPCs). We demonstrate that transplanted Arhgef2−/− fetal liver and bone marrow cells yield impaired hematopoietic recovery and a production deficit from long-term HSCs, phenotypes that are not the result of differences in numbers of transplanted HSCs, their cell cycle status, level of apoptosis, progenitor output, or homing ability. Notably, these defects are functionally restored in vivo by overexpression of ARHGEF2 or its downstream activated RHOA GTPase. By using live imaging of dividing HSPCs, we show an increased frequency of misoriented divisions in the absence of Arhgef2. ARHGEF2 knockdown in human HSCs also impairs their ability to regenerate hematopoiesis, culminating in significantly smaller xenografts. Together, these data demonstrate a conserved role for Arhgef2 in orienting HSPC division and suggest that HSCs may divide in certain orientations to establish hematopoiesis, the loss of which could contribute to HSC dysfunction in bone marrow failure.
Introduction
Stem and progenitor cells across diverse species and organ systems use symmetric and asymmetric modes of division to achieve balanced expansion and differentiation during development. During embryogenesis, hematopoietic stem cells (HSCs) emerge in response to a signal within the hemogenic endothelium1 and transit through the fetal liver and spleen before reaching and colonizing the bone marrow.2 HSCs rapidly proliferate in this niche before becoming quiescent,3 thus allowing their more proliferative downstream progeny, which follow complex lineage pathways, to productively drive native hematopoiesis.4-7 Numerous cell-autonomous8 and extrinsic regulators2 of HSC activity have been identified to date; however, little is known about how HSCs divide and how their cell fate decisions are coupled to a divisional axis when establishing hematopoiesis in native or transplant settings.
Dividing stem cells in other tissue types are known to couple their cell polarity axis with a properly oriented mitotic spindle to allow for appropriate daughter cell acquisition of stem vs commitment fate determinants.9 Orientation of the mitotic spindle can also direct fate decisions by placing daughter cells in varying proximities to the stem cell niche.9 For example, neural precursors in the developing brain undergo an expansion phase during which fate decisions are largely symmetric as a result of divisions occurring parallel to the ventricular zone apical neuroepithelium.10,11 A switch occurs in the subsequent neurogenic phase when precursor cell divisions become oblique and/or perpendicular, which leads to the production of a differentiated progenitor cell that contributes to proper cortical neurogenesis and layering.12 Within the intestine, dividing crypt base stem cells perpendicularly align their mitotic spindles to the apical lumen generating asymmetric daughter cell fates, whereas dividing cells in positions higher up the crypt assume parallel orientations.13 Similarly, in early epidermal development, perpendicular mitotic divisions are used to properly specify epithelial stratification and differentiation.14
Whether similar principles hold true for the hematopoietic system remains to be determined. In the zebrafish, hematopoietic stem and progenitor cells (HSPCs) are found anchored to a mesenchymal stromal cell and achieve asymmetry of cell fates when they divide in orientations that displace one daughter cell away from this niche.15 Mammalian adult hematopoietic precursors can also divide in specific ways, depending on their surrounding signaling milieu; a prorenewal environment promotes symmetric expansion, and a prodifferentiation environment biases for asymmetric fates.16-18 We and others have shown that HSC maintenance and activity are influenced by several known intrinsic effectors of polarity establishment and asymmetrically associated cell fate determinants.19-22 However, not all such factors seem to have the same mechanism or degree of action in mammalian cell types as previously described in model organisms.23,24 Nonetheless, one study has connected Lis1/Pafah1b1, a dynein-binding microtubulecapturing and spindle-orienting protein, with its ability to modulate murine HSPC inheritance of cell fate determinants in normal hematopoiesis and leukemia.25 However, neither the functional importance of PAFAH1B1 in human HSPCs nor an accounting of whether its altered expression is observed among other hematopoietic disorders were explored in this work.
Here, we identify and describe Arhgef2, an independent determinant of mitotic spindle orientation through its function as a RhoA guanine nucleotide exchange factor (GEF),26 as an important regulator of HSC-driven mammalian hematopoiesis in vivo. Our work provides the first potential disease link between ARHGEF2 dysregulation and Shwachman-Diamond syndrome (SDS) and conceptually underscores the importance of mitotic spindle orientation in HSPC function, the loss of which may contribute to defects in developmental hematopoiesis and clinical bone marrow failure.
Methods
tSNE and clustering analysis
We followed methods previously described by Joyce et al.27 Briefly, we divided transcripts per million (TPM) values by 10 to better reflect the complexity of single-cell libraries estimated to be ∽100 000 transcripts. The data were log2 transformed (log2[TPM/10 + 1]) and the expression of the 79 genes identified by bulk data across the 583 single cells was used for principal component analysis in the Seurat Package in R statistical analysis software. Using a jackstraw approach implemented in the Seurat package with num.replicate = 200 and each time randomly permuting 3 genes, the top 4 principal components were identified as significant (P < 1 × 10-4). To aid visualization, these top 4 principal components were subjected to t-distributed stochastic neighbor embedding (t-SNE) analysis in Seurat with 2000 iterations. For clustering analysis, the t-SNE coordinates were used for partitioning around medoids, a more robust version of k-means clustering implemented in the cluster package in R with default parameters (https://stat.ethz.ch/R-manual/R-devel/library/cluster/html/pam.html). To determine the optimal k, we assessed the average Silhouette value for each clustering result (from k = 2 to k = 10) and selected k = 5, which gave the largest mean Silhouette value.
Differential gene expression and pathway analysis
Colony-forming, proliferation, cell cycle, and apoptosis assays
In total, 1.2 × 104 whole bone marrow (WBM) cells from Arhgef2fl/fl and Arhgef2−/− mice were plated in biological triplicate in MethoCult GF M3434 (STEMCELL Technologies), and colonies were scored at 12 to 14 days. Lin–Sca-1+c-Kit+ (LSK) HSPCs were cultured for up to 7 days in HyClone high-glucose Dulbecco’s modified Eagle medium (DMEM) (GE Healthcare Life Sciences) supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 ng/mL murine stem cell factor (SCF), 100 ng/mL murine thrombopoietin, 10 ng/mL murine interleukin-3 (mIL-3), and 10 ng/mL murine interleukin-6 (mIL-6). K562 cells were cultured in Iscove Modified Dulbecco's Medium (IMDM) (Thermo Fisher Scientific) supplemented with 10% FBS (Wisent Bioproducts) and 100 U/mL Pen/Strep (Thermo Fisher Scientific). Transductions were carried out by adding lentiviruses with 5 µg/mL polybrene (Sigma-Aldrich) for 3 days before starting in vitro assays. For cell cycle analyses, cells were fixed with Cytofix/Cytoperm, permeabilized with Perm/Wash, and stained with Ki-67-phycoerythrin [PE]-Cy7 and 7-aminoactinomycin D (7-AAD) (all from BD Biosciences). Apoptosis was measured through staining with Annexin V-fluorescein isothiocyanate (FITC) (BD Biosciences) in binding buffer (BioLegend) and 7-AAD. In vivo cell cycle experiments were conducted according to Matatall et al.29 with some modifications. Briefly, we injected 5 × 105 Arhgef2−/− Lin– cells with B6.SJL WBM into lethally irradiated recipients and administered 5-bromo-2′-deoxyuridine (BrdU) (1.25 mg BrdU per 6 g of body weight) by intraperitoneal injection 72 hours before time of euthanasia, followed by lineage depletion (STEMCELL Technologies) and flow cytometry cell cycle analysis using LSRFortessa (BD Biosciences).
Mouse bone marrow and fetal liver transplantation
Noncompetitive hematopoietic transplantations were carried out in lethally irradiated (1100 Gy; Gammacell 40 Exactor, Best Theratronics) B6.SJL mice. Then, 1 × 106 WBM cells or 3 × 105 E14.5 fetal liver (FL) cells from Arhgef2fl/fl or Arhgef2−/− CD45.2+ donor mice or embryos were injected via tail vein. Competitive transplants were carried out by using the same parameters, but involved injecting either a 1:1 or 2:1 mixture of Arhgef2−/−:C57BL/6 WBM cells, or 1 × 104 Arhgef2−/− lentivirally transduced LSK cells with B6.SJL WBM. Donor engraftment levels were monitored by collecting blood from the tail vein and by flow cytometry analysis using the antibodies described below. Bone marrow transplant periods were ≥16 weeks in duration; secondary transplants were performed with doses of 1.5 × 106 primary WBM cells. Kaplan-Meier survival analysis was calculated on the non-competitive transplantation cohorts. Homing experiments were conducted by injecting 5 × 104 Lin– cells into lethally irradiated recipients and reisolating recipient bone marrow 16 hours later for flow cytometry analysis.
Isolation of primary human HSCs and flow cytometry
Patient samples were obtained with informed consent and approved by the research ethics board at McMaster University. Umbilical cord blood (CB) cells were collected, lineage depleted (STEMCELL Technologies), and analyzed by flow cytometry as previously described.24 Antibodies against mouse antigens, stains, and reagents used in this study included CD45.2 v450, FITC; CD48 FITC, PE; B220 PE; TER-119 PE; CD3 PE; CD4 PE; CD11b PE; c-Kit-PE-Cy7, allophycocyanin (APC); CD8a APC-Cy7; Gr-1 APC-Cy7, and Pacific Blue; all from BD Biosciences. We also used Lineage eFluor 450, APC; Sca-1 APC, and PerCP-Cy5.5 from eBioscience, as well as Lineage Alexa Fluor 700; Sca-1 PE; CD150 PE, BV605; CD41 PE-Dazzle594; and CD11b BV510 from BioLegend. Flow sorting was performed on a MoFlo XDP cell sorter (Beckman Coulter), acquisition was performed on a LSRII or a LSRFortessa flow cytometer (both from BD Biosciences), and data were analyzed using FlowJo v10.0.7 and v10.4 software (Tree Star Inc).
Lentivirus constructs, production, and western blot validation
Third-generation short hairpin RNA (shRNA) sequences against human ARHGEF2 and SBDS were selected on the basis of their high-sensor assay rankings (supplemental Table 1), and they were cloned into lentiviral vectors pZIP-SFFV-ZsGreen-Puro (TransOMIC Technologies) or pZIP-SFFV-tNGFR-Puro re-adapted to contain a microRNA-E delivery scaffold.30 Human ARHGEF2 (NM_001162384.1) and constitutively active RHOA Q63L (a gift from Gary Bokoch, Addgene #12961) were subcloned into the pUMG-LV5 lentiviral expression vector (a gift from Maria Mesuraca). Lentivirus particles were produced by co-transfection of the vector with viral packaging plasmids pMD2.G and psPAX2 (a gift from Didier Trono, Addgene #12259 and #12260) into 293FT cells (Thermo Fisher Scientific). Standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blotting procedures were performed to validate knockdown or overexpression by using antibodies against human ARHGEF2 (ab201687; Abcam), SBDS (ab128946; Abcam), and ACTB (sc-81178; Santa Cruz Biotechnology).
RNA extraction and qRT-PCR
Total RNA was isolated with TRIzol LS reagent (Invitrogen) according to the manufacturer’s instructions, and complimentary DNA was synthesized using qScript cDNA Synthesis Kit (Quanta Biosciences). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed in triplicate with PerfeCTa qPCR SuperMix Low Rox (Quanta Biosciences) with gene-specific probes and primers. Validation of knockdown was performed on hematopoietic cells and leukemia cell lines with the following primers: ARHGEF2 (left 5'-TACCTGCGGCGAATTAAGAT-3', right 5'-AAACAGCCCGACCTTCTCTC-3'; Roche Universal ProbeLibrary #22); SBDS (left 5'-TGGCCAACAGTTAGAAATCGT-3', right 5'-TTCCAAAGAACCTTTGCCTTTA-3'; Roche Universal ProbeLibrary #38); and ACTB (Assay ID: Hs01060665_g1, Thermo Fisher Scientific). Analysis of genes in the TNF-RIP pathway was performed on mouse Lin– cells with gene-specific TaqMan assays (Applied Biosystems) that were normalized to Actb (UPL Set Reference Gene Assays; Roche) for expression comparisons: Hsp90aa1 (Mm00658568_gH), Fth1 (Mm00850707_g1), Glud1 (Mm00492353_m1), Tnf (Mm00443260_g1), Tnfsf1b (Mm00441889_m1), Ripk1 (Mm00436354_m1), Ripk3 (Mm01319233_g1).
Cord blood infection and xenotransplantation experiments
A total of 5 × 104 CD34+ human cord blood HSPCs were cultured in StemSpan serum-free expansion medium II (SFEM II; STEMCELL Technologies) supplemented with 100 ng/mL human SCF (hSCF), 100 ng/mL human Fms related receptor tyrosine kinase 3 (hFLT3), 20 ng/mL human thrombopoietin (hTPO), and 20 ng/mL human IL-6 (hIL-6) for 16 to 20 hours. Lentivirus encoding shRNAs against luciferase, ARHGEF2, or SBDS were added, and transduced cultures were kept for 3 days before gene transfer values were measured by flow cytometry. One fifth of day 0 equivalent cultures were transplanted intrafemorally into each (for a total of 5) sublethally irradiated (315 cGy) NSG recipient mouse. Bone marrow aspirates were taken from the opposite femur between 8 and 10 weeks posttransplant; bilateral iliac crests, femurs, and tibias were processed to evaluate the bone marrow ≥16 weeks posttransplant by flow cytometry.
Live cell imaging of mitotic spindle orientation
For fluorescent labeling, H2B-EGFP and mCherry-α-tubulin were cloned from pLKO-H2B-EGFP (a gift from Daniel Schramek) and mCh-α-tubulin (a gift from Gia Voeltz, Addgene #49149) vectors into an MSCV retroviral backbone. These constructs were co-transfected with pGP1 and pHCMV-G packaging plasmids into 293GPG packaging cells. Viral supernatant was added to the GP+E-86 packaging line, and EGFP+mCherry+ producers were sorted for coculture with mouse Arhgef2−/− LSKs over 3 days in equally mixed HyClone DMEM (GE Healthcare Life Sciences) and StemSpan SFEM (STEMCELL Technologies) media with 10% FBS (Gibco) and the aforementioned murine cytokines. Sorted CD45.2+EGFP+mCherry+ cells were plated onto retronectin-coated chambered glass bottom microslides (80447 and 80827; Ibidi). Live cell fluorescence images were acquired by using a scanning laser confocal microscope with a 60× oil immersion objective (Nikon Eclipse Ti2). Then, 15-μm stacks were obtained over 21 steps at 0.7 μm per step. Images were processed using ImageJ231 on the Fiji platform32 where they were cropped to focus on dividing cells and rotated such that the plane of division was parallel to the x-axis of the image. Once aligned, the angle of division was visualized by using an orthogonal view of the xz plane. The angle of division was measured between the x-axis and the uppermost centrosome, and the lowermost centrosome was used as the vertex of the angle.
Results
ARHGEF2 is significantly downregulated in HSCs in patients with SDS
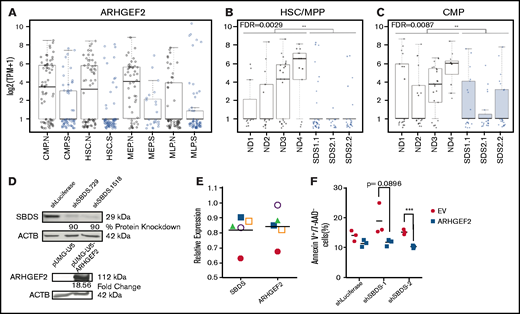
Defects at the stem and progenitor level underlie aspects of the severe paucity of hematopoiesis characteristic of inherited bone marrow failure disorders including SDS; however, the nature of underlying pathogenic mechanisms in this disease are still being uncovered.27,35-38 Primitive cell deficiencies driven by dysregulated spindle orientation have a pathogenic role in human diseases in other tissue systems. For example, defects that disrupt spindle orientation in neural progenitors are known to lead to microcephaly during brain development.39 To explore whether analogous defects might contribute to clinical hematopoietic failure, we queried a recently published data set of single-cell RNA sequencing on CD34+ bone marrow HSPCs from patients diagnosed with SDS.27 Among 11 094 detected genes, we identified ARHGEF2 (fold-change [FC] = 0.49; false discovery rate [FDR] = 0.0029; Figure 1A-C), ITGB1 (FC = 0.43; FDR = 0.0013), and MAD2L1 (FC = 0.24; FDR = 0.0031) as the only annotated spindle-orienting proteins significantly and differentially reduced in expression in HSCs/multipotent progenitors (MPPs) from patients with SDS compared with healthy individuals. Notably, these genes represented 3 from a restricted group of 229 genes found to be significantly downregulated within both SDS HSC/MPPs and common myeloid progenitors (CMPs). Among these candidates, ARHGEF2 exhibited the highest amplitude fold-change. By examining its expression across cell clusters, it can also be appreciated that ARHGEF2-expressing HSC/MPPs and CMPs are underrepresented in patients with SDS compared to healthy individuals. Arhgef2 is well known to associate with the mitotic spindle and facilitate its orientation through RhoA activation.26,33,34 As previous work has demonstrated that Arhgef2 downregulation results in the maintenance of cycling neural precursors and impaired mammalian neurogenesis,34 we selected Arhgef2 for further study in the hematopoietic system. Due to the rarity of primitive cells in primary SDS samples, we first used an orthogonal qPCR approach to determine whether altered ARHGEF2 expression could also be identified in primary Lin–CD34+ CB HSPCs at an early time point after the shRNA-mediated knockdown of SBDS, a known pathogenic driver event in SDS. Interestingly, we observed a trend in ARHGEF2 mRNA levels that mirrored the reduction of SBDS mRNA in these knockdown studies (r = 0.7; P = .09) (Figure 1D-E). On the basis of apoptotic defects previously observed in K562 cells used to model SDS upon SBDS repression,40 we also found enhanced cell survival as measured by the proportion of early apoptotic cells (annexin V+7-AAD–) with SBDS knockdown and ARHGEF2 overexpression on day 3 in contrast to shLuciferase controls, suggesting that ARHGEF2 may be dependent on and/or act as an effector of disrupted SBDS activity (Figure 1D,F). With these findings, we hypothesized that Arhgef2 may similarly regulate HSC function and spindle orientation in hematopoiesis.
ARHGEF2 is significantly downregulated among differentially expressed genes in SDS HSCs. (A) ARHGEF2 transcript expression within single hematopoietic stem cell (HSC), common myeloid progenitor (CMP), megakaryocyte-erythroid progenitor (MEP), and multilymphoid progenitor (MLP) cells from CD34+ patient samples from normal (N, black) or SDS (S, blue) backgrounds. (B-C) ARHGEF2 transcript expression from individual patient samples within (B) HSC/MPP (FDR = 0.0029) and (C) CMP (FDR = 0.0087) populations. ND and SDS represent normal donor and SDS samples, respectively. Each data point represents a single cell. (D) Western blot validation of shRNAs targeting SBDS (top); ARHGEF2 overexpression (bottom). (E) qPCR assessment of SBDS and ARHGEF2 expression in human CD34+ HSPCs after SBDS knockdown (n = 5 CB samples); each sample is represented by a unique symbol. (F) Flow cytometric evaluation of early apoptosis levels upon concomitant SBDS knockdown and ARHGEF2 overexpression in K562 cells (n = 3 biological replicates evaluated at day 3 of in vitro cultures). Error bars represent standard error of the mean (SEM). EV represents empty vector; **P < .01; ***P < .001.
ARHGEF2 is significantly downregulated among differentially expressed genes in SDS HSCs. (A) ARHGEF2 transcript expression within single hematopoietic stem cell (HSC), common myeloid progenitor (CMP), megakaryocyte-erythroid progenitor (MEP), and multilymphoid progenitor (MLP) cells from CD34+ patient samples from normal (N, black) or SDS (S, blue) backgrounds. (B-C) ARHGEF2 transcript expression from individual patient samples within (B) HSC/MPP (FDR = 0.0029) and (C) CMP (FDR = 0.0087) populations. ND and SDS represent normal donor and SDS samples, respectively. Each data point represents a single cell. (D) Western blot validation of shRNAs targeting SBDS (top); ARHGEF2 overexpression (bottom). (E) qPCR assessment of SBDS and ARHGEF2 expression in human CD34+ HSPCs after SBDS knockdown (n = 5 CB samples); each sample is represented by a unique symbol. (F) Flow cytometric evaluation of early apoptosis levels upon concomitant SBDS knockdown and ARHGEF2 overexpression in K562 cells (n = 3 biological replicates evaluated at day 3 of in vitro cultures). Error bars represent standard error of the mean (SEM). EV represents empty vector; **P < .01; ***P < .001.
Arhgef2−/− mice undergo compromised embryonic development and exhibit mild hematopoietic alterations at native steady-state
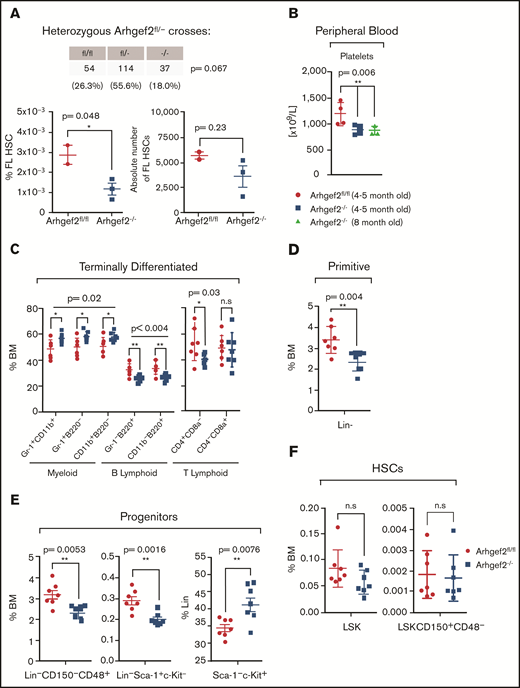
To directly investigate the effects of loss of function of Arhgef2 in mammalian hematopoiesis, we capitalized on a validated Arhgef2−/− mouse model41 to first characterize the nature of steady-state hematopoiesis in its absence. Arhgef2−/− mice were viable, but litter sizes were noticeably reduced, and serial heterozygous crosses yielded fewer Arhgef2−/− mice than expected (Figure 2A, top). In developing embryos, we observed an overall decrease in the percentage and absolute number of Arhgef2−/− fetal liver HSCs relative to controls (Figure 2A, bottom). Among viable adult Arhgef2−/− mice, native peripheral blood analysis revealed ∽25% fewer circulating platelets (Figure 2B) but no other significant differences in the number of leukocytes. These parameters were unchanged in both younger (4-month-old) and maturing adult (8-month-old) mice. Immunophenotyping of Arhgef2−/− adult bone marrow revealed a higher myeloid:lymphoid ratio that was maintained at the terminal end of the hierarchy (Figure 2C). There were significantly fewer Lin– cells (Figure 2D), characterized by fewer restricted (Lin–CD150–CD48+) and lymphoid (Lin–Sca-1+c-Kit– [LS]) progenitors (Figure 2E) and a corresponding relative increase in myeloid (Lin–Sca-1–c-Kit+ [LK]) progenitors within the Lin– compartment (Figure 2E). However, neither HSCs (Lin–Sca-1+c-Kit+ [LSK]) nor a population more enriched for long-term HSCs (LSK CD150+CD48–) were significantly different between knockout and control adult bone marrow samples (Figure 2F). Overall, these data indicate that although Arhgef2−/− embryonic development and phenotypic fetal HSC output are compromised and signs of thrombocytopenia are present in viable Arhgef2−/− mice, adult steady-state hematopoiesis is stable and only mildly altered where the blood system is sufficiently established.
Arhgef2−/− mice exhibit mildly altered hematopoietic parameters. (A) Non-Mendelian ratios observed from heterozygous Arhgef2fl/– crosses across 205 born pups (top); relative percentage and absolute number of fetal liver (FL) HSCs (Lin–CD150+CD48–CD11b+) are decreased in Arhgef2−/− embryos (bottom). (B) Decreased circulating platelets in Arhgef2−/− mice (n = 4 mice per age group). (C) Higher myeloid:B lymphoid ratios and (D) fewer Lin– cell populations. (E) Less restricted (left), lymphoid progenitors in Arhgef2−/− bone marrow (BM) (middle), and relative increase in myeloid progenitors within Lin– compartment of Arhgef2−/− bone marrow (bottom). (F) LSK HSCs (left) and LSK+SLAM long-term HSCs (LT-HSCs) (right) are not statistically different in Arhgef2−/− mice. (C-F) Total of n = 7 mice per group. Error bars represent SEM. *P < .05; **P < .01. n.s., not significant.
Arhgef2−/− mice exhibit mildly altered hematopoietic parameters. (A) Non-Mendelian ratios observed from heterozygous Arhgef2fl/– crosses across 205 born pups (top); relative percentage and absolute number of fetal liver (FL) HSCs (Lin–CD150+CD48–CD11b+) are decreased in Arhgef2−/− embryos (bottom). (B) Decreased circulating platelets in Arhgef2−/− mice (n = 4 mice per age group). (C) Higher myeloid:B lymphoid ratios and (D) fewer Lin– cell populations. (E) Less restricted (left), lymphoid progenitors in Arhgef2−/− bone marrow (BM) (middle), and relative increase in myeloid progenitors within Lin– compartment of Arhgef2−/− bone marrow (bottom). (F) LSK HSCs (left) and LSK+SLAM long-term HSCs (LT-HSCs) (right) are not statistically different in Arhgef2−/− mice. (C-F) Total of n = 7 mice per group. Error bars represent SEM. *P < .05; **P < .01. n.s., not significant.
Arhgef2−/− bone marrow HSPCs do not show significant alterations in their total colony output, proliferation, cell cycle, or apoptosis status
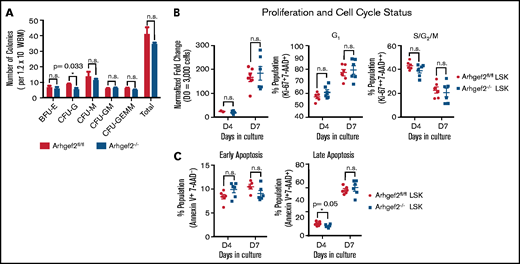
To measure progenitor outputs, we performed colony-forming unit (CFU) assays using WBM. We noticed a slight decrease in the proportion of CFU-granulocyte (CFU-G) colonies, but the remainder of all myeloid progenitors, including CFU-granulocyte, erythrocyte, megakaryocyte, macrophage (CFU-GEMM) colonies, were present in similar numbers in Arhgef2−/− bone marrow compared with Arhgef2fl/fl controls (Figure 3A). Measured at 2 distinct time points in culture, Arhgef2−/− LSK HSCs did not differ in their proliferation rates (Figure 3B, left), cell cycle status (Figure 3B, middle and right), or levels of early and late apoptosis (Figure 3C). These results argue against severe defects in either spindle stability (eg, lack of and/or multipolar spindles) and DNA damage (eg, aneuploidy, chromatin bridges) in these cells and show that apart from defects in granulocyte colony numbers, Arhgef2−/− HSPCs are functionally comparable to control HSPCs in their overall myeloid progenitor outputs, division kinetics, apoptosis, and growth.
Arhgef2−/− bone marrow HSPCs are not altered in their total colony output, proliferation, cell cycle, or apoptosis status. (A) Myeloid CFUs from 1.2 × 104 WBM cells plated in biological triplicate enumerated at 7 to 10 days. (B) LSK HSPCs cultured in vitro enumerated for proliferation (left), proportional G1 (middle), and S/G2/M (right) cell cycle status. (C) Early apoptosis (left) and late apoptosis at days 4 and 7 in culture (right). (B-C) Total of n = 6 biological replicates. Error bars represent SEM. *P < .05. BFU-E, burst forming unit-erythroid; CFU-GM, CFU-granulocyte–macrophage; CFU-M, CFU-megakaryocyte.
Arhgef2−/− bone marrow HSPCs are not altered in their total colony output, proliferation, cell cycle, or apoptosis status. (A) Myeloid CFUs from 1.2 × 104 WBM cells plated in biological triplicate enumerated at 7 to 10 days. (B) LSK HSPCs cultured in vitro enumerated for proliferation (left), proportional G1 (middle), and S/G2/M (right) cell cycle status. (C) Early apoptosis (left) and late apoptosis at days 4 and 7 in culture (right). (B-C) Total of n = 6 biological replicates. Error bars represent SEM. *P < .05. BFU-E, burst forming unit-erythroid; CFU-GM, CFU-granulocyte–macrophage; CFU-M, CFU-megakaryocyte.
Arhgef2−/− fetal liver and bone marrow insufficiently reconstitute the blood system and more heavily rely on HSCs that have functional deficits
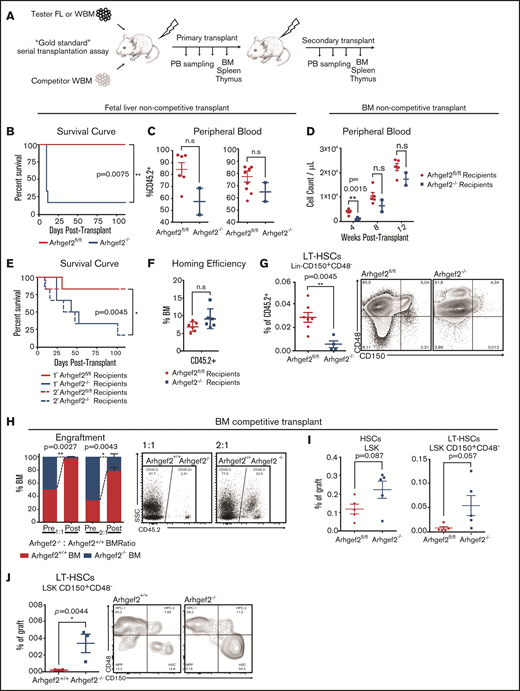
We next sought to verify that the decreased number of phenotypic fetal liver HSCs reflected decreased function by transplanting matched doses of E14.5 fetal liver cells isolated from Arhgef2fl/fl and Arhgef2−/− embryos into lethally irradiated congenic recipients (Figure 4A). Within 2 weeks, the vast majority of recipients of Arhgef2−/− cells became moribund, whereas all mice that received Arhgef2fl/fl cells survived until the experimental 16-week posttransplant end point (Figure 4B). In 2 of the 6 Arhgef2−/− recipients that survived until 10 days posttransplant, we noted a relative decrease in the percentage of peripheral CD45.2+ and Gr-1+ granulocytic engraftment, with the lowest engrafted of these becoming moribund shortly after this sampling (Figure 4C). Together, these data indicate a significant impairment in repopulating HSCs within the fetal liver of Arhgef2−/− mice.
Arhgef2−/− fetal liver and bone marrow insufficiently reconstitute the blood system and show productive deficits at the HSC level. (A) Experimental schematic of non-competitive and competitive transplantations. (B) Kaplan-Meier survival curves demonstrating higher mortality among recipients of Arhgef2−/− E14.5 fetal liver cells (n = 6 recipients with n = 2 biological Arhgef2fl/fl and n = 3 Arhgef2−/− fetal liver donors). (C) Decreased levels of peripheral blood engraftment and granulocytic populations as measured in the 2 remaining mice that received Arhgef2−/− E14.5 fetal liver cells at day 10 posttransplantation. (D) Insufficient and/or delayed hematopoietic recovery in the peripheral blood of primary transplanted mouse recipients of Arhgef2−/− bone marrow. (E) Kaplan-Meier survival curves demonstrating higher mortality among recipients of non-competitively transplanted Arhgef2−/− bone marrow. (F) Comparable homing efficiencies from 5 × 104 Lin–Arhgef2−/− bone marrow cells. (G) Lower levels of Arhgef2−/− Lin–CD150+CD48– HSCs in secondary non-competitively transplanted bone marrow grafts. (H) Poorer engraftment levels from competing 1:1 and 2:1 Arhgef2−/−:wild-type doses of bone marrow among primary recipients. (I) Increased percentage of HSCs in the primary grafts of competitively transplanted Arhgef2−/− bone marrow. (J) Increased proportion of LSK CD150+CD48– LT-HSCs within secondary grafts among competitively transplanted recipients. Primary non-competitive transplants and homing experiments initiated with 6 recipients per condition using 3 biological replicates of bone marrow samples. Secondary noncompetitive transplants completed with 7 Arhgef2fl/fl and 4 Arhgef2−/− recipients (data from 2 representative experiments). Primary competitive transplants initiated with 3 recipients per dose and secondary competitive analyses conducted with 3 recipients from 2 primary mice. Error bars represent SEM.*P < .05; **P < .01. PB, peripheral blood; SSC, side scatter.
Arhgef2−/− fetal liver and bone marrow insufficiently reconstitute the blood system and show productive deficits at the HSC level. (A) Experimental schematic of non-competitive and competitive transplantations. (B) Kaplan-Meier survival curves demonstrating higher mortality among recipients of Arhgef2−/− E14.5 fetal liver cells (n = 6 recipients with n = 2 biological Arhgef2fl/fl and n = 3 Arhgef2−/− fetal liver donors). (C) Decreased levels of peripheral blood engraftment and granulocytic populations as measured in the 2 remaining mice that received Arhgef2−/− E14.5 fetal liver cells at day 10 posttransplantation. (D) Insufficient and/or delayed hematopoietic recovery in the peripheral blood of primary transplanted mouse recipients of Arhgef2−/− bone marrow. (E) Kaplan-Meier survival curves demonstrating higher mortality among recipients of non-competitively transplanted Arhgef2−/− bone marrow. (F) Comparable homing efficiencies from 5 × 104 Lin–Arhgef2−/− bone marrow cells. (G) Lower levels of Arhgef2−/− Lin–CD150+CD48– HSCs in secondary non-competitively transplanted bone marrow grafts. (H) Poorer engraftment levels from competing 1:1 and 2:1 Arhgef2−/−:wild-type doses of bone marrow among primary recipients. (I) Increased percentage of HSCs in the primary grafts of competitively transplanted Arhgef2−/− bone marrow. (J) Increased proportion of LSK CD150+CD48– LT-HSCs within secondary grafts among competitively transplanted recipients. Primary non-competitive transplants and homing experiments initiated with 6 recipients per condition using 3 biological replicates of bone marrow samples. Secondary noncompetitive transplants completed with 7 Arhgef2fl/fl and 4 Arhgef2−/− recipients (data from 2 representative experiments). Primary competitive transplants initiated with 3 recipients per dose and secondary competitive analyses conducted with 3 recipients from 2 primary mice. Error bars represent SEM.*P < .05; **P < .01. PB, peripheral blood; SSC, side scatter.
To functionally test the hematopoietic reconstitution capacity of Arhgef2−/− adult bone marrow, we performed non-competitive and competitive serial transplantation assays in vivo (Figure 4A). In non-competitive transplantations, the majority of mice receiving Arhgef2−/− bone marrow showed evidence of anemia and became moribund, whereas recipients of Arhgef2fl/fl bone marrow did not display signs of hematopoietic insufficiency or delayed recovery (Figure 4D-E). Similar phenotypes and increased mortality were also evident in the recipients of Arhgef2−/− bone marrow in secondary transplant settings (Figure 4E). Importantly, this posttransplant failure phenotype was not a result of compromised homing abilities because as early as 16 hours posttransplantation, Arhgef2−/− Lin– cells homed to recipient bone marrow with an efficiency comparable to Arhgef2fl/fl controls (Figure 4F). However, within the grafts of the recipients of Arhgef2−/− bone marrow that remained at the end of secondary transplants, Lin–CD150+CD48– HSCs were significantly exhausted in comparison with those found in control grafts (Figure 4G).
Competitive primary transplants of Arhgef2−/− bone marrow against wild-type bone marrow also demonstrated significantly impaired reconstitution by cells depleted for Arhgef2 at both equivalent (1:1) doses and even when biased (2:1) to give an advantage to Arhgef2−/− bone marrow (Figure 4H). However, in contrast to the non-competitive transplant results, here we observed an accumulation of HSCs in the Arhgef2−/− grafts (Figure 4I), an outcome that was propagated in secondary recipients where long-term HSCs were significantly overrepresented in the secondary Arhgef2−/− compared with wild-type counterpart grafts (Figure 4J), indicating that in the presence of competitors that can rescue hematopoietic regeneration, Arhgef2-/- HSCs are predisposed toward enhanced stem cell production at the expense of more committed cells. Considered alongside these results, it would be expected that as we observed, Arhgef2-/- primitive cells exhaust in the noncompetitive setting where reconstitution is completely dependent on stem cells with an impaired ability to generate committed progeny. Together, our transplantation strategies demonstrate that transplant-driven hematopoiesis in the absence of Arhgef2 heavily relies on primitive HSCs and that functional differentiation deficits existing at this level leads to compromised hematopoiesis and increased mortality risk.
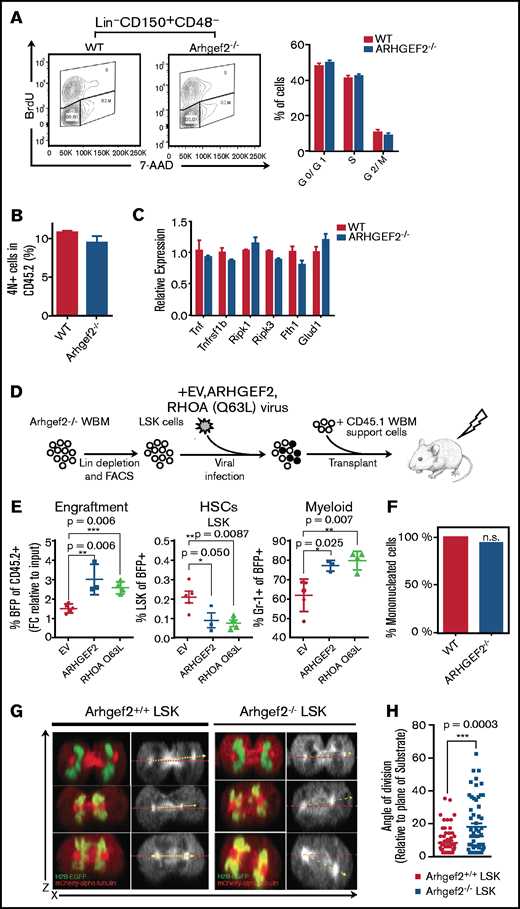
Constitutively active RhoA can revert Arhgef2−/− HSC functional defects
We next wanted to explore the mechanisms through which Arhgef2 regulates primitive hematopoietic cells that we hypothesized could be through its dominant substrate RhoA.42 Zhou et al.43 previously studied the role of RhoA in primitive murine HSPCs, in which its pan-hematopoietic conditional deletion in transplanted mice resulted in a preservation of HSCs but an impairment of progenitors as a result of alterations in their cell cycle kinetics and cytokinesis, and their death through necroptosis. Interestingly, upon administering BrdU to primary mice competitively transplanted with Arhgef2−/− or control HSCs, we did not observe any differences in the in vivo percentage of Lin–CD150+CD48– cells at the various stages of the cell cycle (Figure 5A), nor did we see an increase in primitive cells with 4N or above DNA content that otherwise would have indicated defects in cytokinesis (Figure 5B). We next examined Lin– cells from the graft the transcripts for 6 well-known indicators of necroptosis that were elevated in RhoA-deleted HPCs and discerned no increase in the expression of any of them, suggesting that necroptosis is not a feature of the Arhgef2−/− phenotype (Figure 5C).
Arhgef2 regulates HSC function through RhoA activation and its loss disrupts HSPC mitotic spindle orientation. (A) Arhgef2−/− or wild-type (WT) Lin– cells supplemented with CD45.1 bone marrow support cells were transplanted into recipient mice for 13 weeks (n = 3 recipients per group). BrdU was administered 72 hours before the end point, and cell cycle distribution of wild-type or Arhgef2−/− Lin–CD150+CD48– cells were examined by flow cytometry. (B) Quantification of 4N/4N+ cell frequency by 7-AAD staining. (C) Expression of TNF-RIP–related genes evaluated by qPCR in wild-type or Arhgef2−/− Lin– graft cells. (D) Experimental schematic of rescue competitive transplants. (E) Proportion of blue fluorescent protein positive (BFP+) transduced Arhgef2−/− cells after 12 weeks (left), HSC content (middle), and myeloid content as measured by Gr-1 (right) of the grafts. Empty vector (EV), n = 5 recipients; ARHGEF2, n = 3 recipients; RhoA Q63L, n = 4 recipients. (F) Multinuclearity (4N and above) assessed by live cell fluorescence microscopy of dividing wild-type (51 events) or Arhgef2−/− (50 events) HSPCs. Differences between groups for multinuclearity enumeration were not significant by Fisher’s exact test (2-tailed). (G) Representative z-stack stitched images of LSK HSPCs retrovirally transduced with both H2B-EGFP and mCherry-α-tubulin imaged under live cell fluorescence microscopy to capture telophase events; observed division axes (yellow) as measured in reference to the horizontal axis (red). (H) Quantification of cytokinesis events indicate that Arhgef2−/− LSK HSPCs exhibit a significantly increased frequency of random divisional orientations whereas wild-type HSPCs preferentially divide parallel to an underlying retronectin substrate (55 cells for wild-type background and 56 cells for Arhgef2−/− background). Error bars represent SEM. *P < .05; **P < .01; ***P < .001. FACS, fluorescence-activated cell sorter.
Arhgef2 regulates HSC function through RhoA activation and its loss disrupts HSPC mitotic spindle orientation. (A) Arhgef2−/− or wild-type (WT) Lin– cells supplemented with CD45.1 bone marrow support cells were transplanted into recipient mice for 13 weeks (n = 3 recipients per group). BrdU was administered 72 hours before the end point, and cell cycle distribution of wild-type or Arhgef2−/− Lin–CD150+CD48– cells were examined by flow cytometry. (B) Quantification of 4N/4N+ cell frequency by 7-AAD staining. (C) Expression of TNF-RIP–related genes evaluated by qPCR in wild-type or Arhgef2−/− Lin– graft cells. (D) Experimental schematic of rescue competitive transplants. (E) Proportion of blue fluorescent protein positive (BFP+) transduced Arhgef2−/− cells after 12 weeks (left), HSC content (middle), and myeloid content as measured by Gr-1 (right) of the grafts. Empty vector (EV), n = 5 recipients; ARHGEF2, n = 3 recipients; RhoA Q63L, n = 4 recipients. (F) Multinuclearity (4N and above) assessed by live cell fluorescence microscopy of dividing wild-type (51 events) or Arhgef2−/− (50 events) HSPCs. Differences between groups for multinuclearity enumeration were not significant by Fisher’s exact test (2-tailed). (G) Representative z-stack stitched images of LSK HSPCs retrovirally transduced with both H2B-EGFP and mCherry-α-tubulin imaged under live cell fluorescence microscopy to capture telophase events; observed division axes (yellow) as measured in reference to the horizontal axis (red). (H) Quantification of cytokinesis events indicate that Arhgef2−/− LSK HSPCs exhibit a significantly increased frequency of random divisional orientations whereas wild-type HSPCs preferentially divide parallel to an underlying retronectin substrate (55 cells for wild-type background and 56 cells for Arhgef2−/− background). Error bars represent SEM. *P < .05; **P < .01; ***P < .001. FACS, fluorescence-activated cell sorter.
These findings were not entirely unexpected given that there are many other RhoA GEFs,44 so a total depletion of the cellular pool of RhoA would be anticipated to yield more pronounced or altered phenotypes when compared with the localized and possibly lower magnitude reduction of RhoA activation achieved by depletion of Arhgef2. We next directly tested the RhoA dependence of the Arhgef2−/− phenotype through rescue experiments. We lentivirally introduced ARHGEF2 or a constitutively activated form of RHOA Q63L into Arhgef2−/− LSK cells and competitively transplanted them into congenic recipients (Figure 5D). At 12.5 weeks posttransplantation, the provision of ARHGEF2 or RHOA Q63L significantly reverted the impaired competitiveness of Arhgef2−/− HSCs, increasing the proportion of transduced cells in grafts 1.7-fold and 2-fold, respectively, compared with empty vector controls (Figure 5E). Both ARHGEF2 and RHOA Q63L also rescued the aberrant accumulation of primitive LSK cells observed in the Arhgef2−/− competitive transplant setting and produced robust myeloid grafts (Figure 5E). These results indicate that the phenotypes we have observed are a direct result of Arhgef2 deletion and, most importantly, provide clear evidence that Arhgef2 maintains HSC function and downstream cell outputs through its ability to activate RhoA.
Arhgef2−/− HSPCs exhibit a significantly increased frequency of misoriented divisions
Because Arhgef2 has been characterized to function by orienting the mitotic spindle, we used a previously published live cell imaging method to measure the angle of division of mouse HSPCs.25 LSK cells were labeled with H2B-EGFP and mCherry-α-tubulin and plated on retronectin-covered chambered slides, which allowed us to acquire confocal z-stacks of dividing cells and generate orthogonal projections of cell division events at telophase. In agreement with the aforementioned lack of cell cycle and cell survival changes, individual Arhgef2−/− HSPC divisions displayed no evidence of mitotic catastrophe events or multipolar spindles and no significant increase in the frequency of multinucleated daughters when observing single-cell divisions (Figure 5F). We next verified that wild-type LSK HSPCs preferentially divided parallel (between 0° and 10°) to this underlying substrate (Figure 5G). However, although Arhgef2−/− LSK HSPCs also yielded parallel division events, we observed a significantly increased frequency of nonparallel angles that reached as high as 60° (Figure 5H). These results indicate that even though altered divisional preferences do not largely compromise HSPC survival and division kinetics in vitro, the posttransplant failure phenotypes measured in vivo could be explained at least in part by dysregulated fate decisions as a result of misoriented divisions. Our results are thus consistent with the concept that Arhgef2 is essential for regulating HSC divisional orientation and proper lineage differentiation within their niche during the establishment of hematopoiesis.
ARHGEF2 knockdown in human HSPCs compromises hematopoietic xenografts
We next confirmed that ARHGEF2 localizes at the microtubule apparatus in human hematopoietic cells during division by performing immunofluorescence staining on several myeloid cell lines (Figure 6A). To determine whether ARHGEF2 function is conserved in human hematopoiesis, we interrogated the effects of its downregulation by introducing shRNAs against either ARHGEF2 or a luciferase control into CB CD34+ HSPCs (Figure 6B-C). Similar to results derived from Arhgef2-/- mice, cells with reduced ARHGEF2 proliferated comparably or showed slightly dampened outputs relative to controls (Figure 6D). Myeloid CFU assays showed no significant differences in multipotent progenitor colonies and significantly decreased monocytic progenitors, whereas the total colony number remained equivocal across settings (Figure 6E). These data suggest that ARHGEF2 knockdown in human hematopoietic progenitors imparts only mild defects at the lineage-restricted level. Finally, using 2 separate and efficient shRNAs against ARHGEF2, in vivo analyses of intrafemorally xenotransplanted NSG recipient mice at 16 weeks posttransplant showed significantly diminished hematopoietic grafts with a paucity of CD15+ myeloid output observed in the residual xenografts of mice receiving ARHGEF2 knockdown cells compared with controls (Figure 6F-G). Considered with our murine data, this clear in vivo phenotype in the human context demonstrates the cross-species importance of ARHGEF2 to the regenerative and productive capacity of HSCs and may implicate ARHGEF2-regulated spindle orientation as a process that contributes to effective human hematopoiesis (Figure 6H).
Loss of ARHGEF2 function in CD34+ HSCs results in significantly diminished xenografts. (A) Immunofluorescent staining of ARHGEF2 and TUBA at the mitotic spindle in THP-1 and LAMA-84 cell lines. (B) Schematic of shRNA knockdown of ARHGEF2 in CD34+ HSCs in vivo and in vitro. (C) Protein level knockdown validation of shRNAs against ARHGEF2. (D) Proliferation of CD34+ HSCs in vitro over 7 days. (E) Colony output of ARHGEF2 knocked-down HSPCs. (F-G) Decreased engraftment (F) and output of CD15+ myeloid cells (G) derived from CD34+ HSCs with comparable gene transfer levels receiving shRNAs targeting ARHGEF2. There were 5 recipients each derived from 1 CB sample. (H) Hypothetical model summarizing the role of Arhgef2 (red dots) in regulating the orientation of the HSC mitotic spindle within the niche when establishing hematopoiesis, the loss of which may contribute to bone marrow failure at the stem cell level. Error bars represent SEM. *P < .05; **P < .01.
Loss of ARHGEF2 function in CD34+ HSCs results in significantly diminished xenografts. (A) Immunofluorescent staining of ARHGEF2 and TUBA at the mitotic spindle in THP-1 and LAMA-84 cell lines. (B) Schematic of shRNA knockdown of ARHGEF2 in CD34+ HSCs in vivo and in vitro. (C) Protein level knockdown validation of shRNAs against ARHGEF2. (D) Proliferation of CD34+ HSCs in vitro over 7 days. (E) Colony output of ARHGEF2 knocked-down HSPCs. (F-G) Decreased engraftment (F) and output of CD15+ myeloid cells (G) derived from CD34+ HSCs with comparable gene transfer levels receiving shRNAs targeting ARHGEF2. There were 5 recipients each derived from 1 CB sample. (H) Hypothetical model summarizing the role of Arhgef2 (red dots) in regulating the orientation of the HSC mitotic spindle within the niche when establishing hematopoiesis, the loss of which may contribute to bone marrow failure at the stem cell level. Error bars represent SEM. *P < .05; **P < .01.
Discussion
The fundamental role of ARHGEF2 in associating with microtubules and modulating RhoA activity at mitotic spindles is well established.26,33,34,41,42,45 Moreover, the spindle-regulating abilities of ARHGEF2 depend on its GEF activity, and constitutive RHOA restores spindle defects upon ARHGEF2 depletion.26 Our results showing functional rescue of spindle orientation-defective Arhgef2−/− HSCs with active RHOA therefore provide important support for the Arhgef2-RhoA axis being influential in HSC decision-making and implicate the spindle-regulating activity of ARHGEF2 as a possible contributor to proper HSC function. Our work also complements the previous investigation of the global dependence on RhoA in the blood system in that it clarifies the contribution of the Arhgef2-regulated pool of RhoA specifically to primitive blood cell function. In particular, although deletion of RhoA impairs progenitors, the lack of direct effects on progenitors we see upon Arhgef2 loss suggests that other RhoA GEFs can maintain a sufficient level of RhoA activation in these cells to permit their normal function. Zhou et al.43 found no effects on Lin–CD150+ HSCs when RhoA was deleted in established hematopoietic grafts, but our methodologically unique approach of interrogating the effects of Arhgef2 loss during the process of hematopoietic regeneration suggests that regulation of RhoA activity at the mitotic spindle by Arhgef2 represents an important axis for productive HSC divisions during the critical window over which hematopoiesis is established. In further support of this point, we note that ARHGEF2, but not RHOA, was found to be significantly downregulated in primitive SDS cells within the single-cell RNA sequencing data set.27
Our finding of decreased function in fetal liver HSCs in Arhgef2−/− mice indicates that fetal hematopoiesis defects may, in addition to other nonhematopoietic defects not evaluated, contribute to the reduced fraction of Arhgef2−/− embryos that reach postnatal viability. Indeed, proper establishment of the hematopoietic system during development requires a minimum number of productive HSC divisions in the fetal liver.46 Thus, only some Arhgef2−/− embryos may generate enough effective HSC divisions to allow for sufficient production of functional progenitors to populate the hematopoietic system. Our results using adult HSCs interrogated in 2 distinct transplant models provide further important insight into the mechanism of Arhgef2 loss in HSC decision-making in different bone marrow states. In recipients of competitively transplanted Arhgef2−/− bone marrow cells, wild-type HSCs regenerated hematopoiesis more effectively, placing less of the burden of reconstitution on Arhgef2−/− HSCs and allowing readout of their preferred tendency to divide in a manner that promotes primitive cell production. This relative elevation of HSC frequency compared to control competitors may be due to an uncoupling of the cell polarity axes from their orientation of division, leading to a relative retention of stemness determinants over asymmetric partitioning of differentiation determinants in daughter cells. Alternatively, their compromised ability to adopt particular divisional orientations may result in Arhgef2−/− daughter cells being localized in more niche-proximal locations where they would receive enhanced HSC maintenance cues. In contrast, the long-term regeneration of hematopoiesis in non-competitive settings is entirely dependent on Arhgef2−/− HSCs because the paucity of progenitors generated as a result of the spindle orientation defects leads to a heavier reliance on Arhgef2−/− HSC divisions. In this latter context, which mimics the dependencies on HSCs in developmental hematopoiesis, the stem cell loss we observed may be the result of an exhaustion of Arhgef2−/− HSCs.
Our observation of similarly defective hematopoietic reconstitution in vivo upon transplant of ARHGEF2-depleted human HSCs suggests a conserved function of Arhgef2 across species. Downregulation of ARHGEF2 in SDS HSCs and acutely upon SBDS repression in CD34+ cells raises the possibility that repression of ARHGEF2 in patients with SDS may contribute to defective HSC-driven hematopoiesis. It is also interesting to note other phenotypes we have observed in our Arhgef2−/− mouse model, including native thrombocytopenia and reduced posttransplant output of differentiated megakaryocytes, which may indicate an additional role for Arhgef2 in regulating megakaryocyte maturation47,48; defects in neutrophil chemotaxis49; and though not formally characterized yet, bone malformations and clear neurologic abnormalities (data not shown). This latter finding parallels reports of cognitive impairments and intellectual disability in patients with ARHGEF2 loss-of-function mutations.50,51 Notably, all of these features can be found in patients diagnosed with SDS.52,53 Although these findings are beyond the scope of this study, they encourage future efforts to understand whether and how the loss of ARHGEF2 function contributes to the etiology and pathogenesis of SDS.
In summary, our work highlights possible implications for how mitotic spindle orientation itself may more broadly affect stem cell division during development and disease. As mentioned, centrosomal protein loss-of-function events that influence spindle orientation disrupt the balance of symmetric and asymmetric divisions, which lead to microcephaly during brain development.39 With the addition of our findings, conceptual parallels may also exist between spindle-regulating genes and bone marrow failure syndromes within and beyond SDS. Indeed, the loss of Cdk5rap2, a centrosomal spindle-orienting protein, results in a macrocytic, hypoproliferative anemia and leukopenia (Hertwig’s anemia) in a mouse model.54 With the identification of several other spindle-regulating genes implicated in microcephaly,55 it may be interesting to determine whether any of these genes also have roles within bone marrow failure or in disorders that result in tissue insufficiency elsewhere. Because SDS progresses with high frequency to myeloid malignancies in which expansion of transformed HSCs are known to be early pathogenic events, spindle orientation dysregulation may also be interesting to explore in the future as a possible contributor to the larger group of disorders that include clonal hematopoiesis, myelodysplastic syndrome, and/or leukemia.
Acknowledgments
The authors thank Johann Hitzler and Sheila Singh for feedback.
This study was supported by a Canadian Institutes of Health Research (CIHR) PhD Studentship (D.C.H.C.), Ontario Graduate Scholarship (D.C.H.C.), a CIHR Studentship (J.X.), an Ontario Institute for Cancer Research Investigator Award (K.H.), and a CIHR Foundation Grant (R.K.R.). Funding support for this article was provided by the Ontario Institute for Cancer Research (IA-033).
Authorship
Contribution: D.C.H.C. designed and performed the experiments, analyzed and interpreted the data, and wrote the manuscript; A.V. and L.P.M.H.d.R. assisted with animal experiments; J.X. and N.W. designed and performed experiments and analyzed and interpreted the data; V.G. assisted with imaging work and analysis; S.M. performed experiments; C.E.J. and C.D.N. designed and completed analyses on sequencing data from patient samples; J.L.R., M.J.S., and R.K.R. generated the knockout mouse model; R.K.R. advised on experimental design; K.H. supervised the project, designed experiments, reviewed the data, and wrote the manuscript; and all authors reviewed and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kristin Hope, Princess Margaret Cancer Centre, University Health Network, 101 College St, Toronto, ON, Canada M5G 1L7; e-mail: kristin.hope@uhnresearch.ca.
References
Author notes
Data may be requested by sending an e-mail to Kristin Hope at kristin.hope@uhnresearch.ca.
The full-text version of this article contains a data supplement.