Key Points
Non–polymerase chain reaction-based methods like western blot should be used to diagnose HIV infection in recipients of LVV-mediated GT.
Beti-cel treatment allows maintenance of GT-derived HbAT87Q and reconstitution of hemoglobin after infection-related anemia.
Betibeglogene autotemcel (beti-cel) gene therapy (GT) for patients with transfusion-dependent β-thalassemia uses autologous CD34+ cells transduced with BB305 lentiviral vector (LVV), which encodes a modified β-globin gene. BB305 LVV also contains select HIV sequences for viral packaging, reverse transcription, and integration. This case report describes a patient successfully treated with beti-cel in a phase 1/2 study (HGB-204; #NCT01745120) and subsequently diagnosed with wild-type (WT) HIV infection. From 3.5 to 21 months postinfusion, the patient stopped chronic red blood cell transfusions; total hemoglobin (Hb) and GT-derived HbAT87Q levels were 6.6 to 9.5 and 2.8 to 3.8 g/dL, respectively. At 21 months postinfusion, the patient resumed transfusions for anemia that coincided with an HIV-1 infection diagnosis. Quantitative polymerase chain reaction assays detected no replication-competent lentivirus. Next-generation sequencing confirmed WT HIV sequences. Six months after starting antiretroviral therapy, total Hb and HbAT87Q levels recovered to 8.6 and 3.6 g/dL, respectively, and 3.5 years postinfusion, 13.4 months had elapsed since the patient’s last transfusion. To our knowledge, this is the first report of WT HIV infection in an LVV-based GT recipient and demonstrates persistent long-term hematopoiesis after treatment with beti-cel and the ability to differentiate between WT HIV and BB305-derived sequences.
Introduction
Gene therapy (GT) with lentiviral vectors (LVVs), a type of retroviral vector, has been successful in hemoglobinopathies1,2 and can offer a potentially transformative outcome with low theoretical risk of the generation of replication-competent virus owing to safety measures integrated into the vector design.3,4 GT with betibeglogene autotemcel (beti-cel; LentiGlobin for β-thalassemia) is a one-time treatment for patients with transfusion-dependent β-thalassemia (TDT) that addresses the underlying genetic cause of the disease by adding functional copies of a modified human β-globin (HBB) gene into a patient’s own hematopoietic stem cells (HSCs) through transduction of autologous CD34+ cells with BB305 LVV.1
BB305 LVV is a third-generation, replication-defective, self-inactivating (SIN) human immunodeficiency virus-1 (HIV-1)‒based LVV that encodes a modified human β-globin gene containing a threonine (T) to glutamine (Q) amino acid substitution at position 87 and regulatory elements of the human βA-globin mini-gene that drive expression of vector-derived HbAT87Q in erythroid cells.4,5 The only protein-coding DNA sequences in the BB305 LVV are those coding the β-globin transgene.4 The BB305 LVV transgene replaces all HIV genes but contains a minimal amount of HIV-derived sequences necessary for packaging of the viral genome and transduction of host cells. These sequences include a mutated gag gene segment, the Rev-response element (RRE), and central polypurine tract (cPPT) (Figure 1A).4 Therefore, transduction with BB305 LVV would not be expected to result in HIV infection in the recipient, because wild-type (WT) HIV requires all 9 genes for infection and replication in host cells.4,6 Additional safety modifications include deletion of the viral promoter and enhancer elements in the 3' long terminal repeat (LTR) to generate an SIN vector that reduces both the likelihood of activation of cellular protooncogenes and the generation of replication-competent recombinant lentivirus (RCL).4
Diagram of the BB305 LVV with human βA-T87Q substitution and corresponding HIV-derived sequences. (A) The βA-T87Q substitution (ACA [Thr] to CAG [Gln]) in the human β-globin gene is indicated by an asterisk and arrow. Safety modifications included mutation of the gag gene and a 400-bp deletion in the U3 of the right HIV LTR.4 HIV-derived sequences are shown in gray boxes and are aligned with corresponding genes in the HIV genome (top; adapted from Frankel and Young).9 3'enh/pA, 3' enhancer/polyadenylation signal from the β-globin gene; ΔU3, promoter/enhancer-deleted unique 3'; ψ, packaging signal; cPPT, central polypurine tract; E, exon; gag, HIV-1 partial gag sequence; Globin LCR, human globin locus control regions; LTR, long terminal repeat; LVV, lentiviral vector; P, β-globin promoter; Q, gluatmine; R, repeat; RRE, rev-response element; T, threonine; U5, unique 5'. (B) WB assay of BB305 LVV for HIV-1 proteins. Lane 1 of each blot contains 50 to 100 ng of each analyzed protein (from left to right: [A] recombinant (r)-gp120, [B] r-gp41, [C] r-RT, [D] r-p24); lane 2 of each blot contains a molecular weight ladder; lane 3 of each blot contains 4 ng BB305 LVV. Primary mAbs include anti-p24 mAb 63917 (Abcam); anti-gp41 mAb MA1-7590 (Invitrogen); anti-RT mAb 63911 (Abcam); anti-gp120 mAb AHP2204 (Bio-Rad). Secondary mAb is IRDye 800CW (925-32211; Li-Cor Biosciences). The HIV p24 and reverse transcriptase proteins are not encoded in the BB305 LVV but are present as part of the purified vector particle and may be identifiable via WB analysis of drug product. The HIV-1 env protein gp160 cleavage products gp41 and gp120 are not present in the BB305 LVV production system and thus are not detectable.4,9 mAbs, monoclonal antibodies; MW, molecular weight; RT, reverse transcriptase; WB, western blot.
Diagram of the BB305 LVV with human βA-T87Q substitution and corresponding HIV-derived sequences. (A) The βA-T87Q substitution (ACA [Thr] to CAG [Gln]) in the human β-globin gene is indicated by an asterisk and arrow. Safety modifications included mutation of the gag gene and a 400-bp deletion in the U3 of the right HIV LTR.4 HIV-derived sequences are shown in gray boxes and are aligned with corresponding genes in the HIV genome (top; adapted from Frankel and Young).9 3'enh/pA, 3' enhancer/polyadenylation signal from the β-globin gene; ΔU3, promoter/enhancer-deleted unique 3'; ψ, packaging signal; cPPT, central polypurine tract; E, exon; gag, HIV-1 partial gag sequence; Globin LCR, human globin locus control regions; LTR, long terminal repeat; LVV, lentiviral vector; P, β-globin promoter; Q, gluatmine; R, repeat; RRE, rev-response element; T, threonine; U5, unique 5'. (B) WB assay of BB305 LVV for HIV-1 proteins. Lane 1 of each blot contains 50 to 100 ng of each analyzed protein (from left to right: [A] recombinant (r)-gp120, [B] r-gp41, [C] r-RT, [D] r-p24); lane 2 of each blot contains a molecular weight ladder; lane 3 of each blot contains 4 ng BB305 LVV. Primary mAbs include anti-p24 mAb 63917 (Abcam); anti-gp41 mAb MA1-7590 (Invitrogen); anti-RT mAb 63911 (Abcam); anti-gp120 mAb AHP2204 (Bio-Rad). Secondary mAb is IRDye 800CW (925-32211; Li-Cor Biosciences). The HIV p24 and reverse transcriptase proteins are not encoded in the BB305 LVV but are present as part of the purified vector particle and may be identifiable via WB analysis of drug product. The HIV-1 env protein gp160 cleavage products gp41 and gp120 are not present in the BB305 LVV production system and thus are not detectable.4,9 mAbs, monoclonal antibodies; MW, molecular weight; RT, reverse transcriptase; WB, western blot.
Because the BB305 LVV is derived from HIV, a theoretical risk of recombination with WT HIV exists. This risk is considered low because the BB305 LVV contains <25% of the HIV genome and is replication defective. Because the vector contains the minimal sequences required for viral genome packaging and transduction of host cells, HIV-specific capsid protein p24 and reverse transcriptase are identifiable via western blot (WB) of the purified vector (Figure 1B), but no HIV-specific proteins would be expected to be detected in blood samples from GT recipients. Patients are monitored for RCL at various timepoints throughout the clinical trials through the use of a highly sensitive and validated quantitative polymerase chain reaction (PCR) assay for vesicular stomatitis virus envelope G protein (VSV-G) to ensure that recombination and replication of the BB305 LVV have not occurred.
This case report describes the identification of WT HIV and differentiation from GT-derived sequences in a patient who participated in HGB-204 (Northstar), a phase 1/2 trial evaluating the efficacy and safety of beti-cel for 24 months after infusion in 18 patients with TDT (#NCT01745120).1 In this trial, 11 patients were able to stop receiving transfusions, and at last visit (12 to 36 months postinfusion), the range of total hemoglobin (Hb) concentrations was 8.2 to 13.7 g/dL.1 This patient is currently enrolled in LTF-303, the follow-up study evaluating the long-term safety and efficacy of beti-cel GT for up to 13 years (#NCT02633943).
Case description
Patient history and treatment in HGB-204
The patient (nonsplenectomized) was diagnosed with β-thalassemia as an infant and was enrolled in the HGB-204 study as a young adult. During the 2-year period before study enrollment, the patient was receiving an annualized 14 transfusion episodes per year, with an annualized transfusion volume of 140 mL/kg per year. At screening, the patient tested negative for HIV-1 and HIV-2, as required for enrollment into all beti-cel clinical trials.
The patient received granulocyte colony-stimulating factor and plerixafor to mobilize stem cells from the bone marrow followed by apheresis to collect CD34+ cells containing HSCs for manufacture of the beti-cel drug product. The patient received myeloablative conditioning with single-agent busulfan before beti-cel was infused IV at a dose of 13 × 106 CD34+ cells per kg. The patient’s drug product had a vector copy number (VCN) of 0.4 copies per diploid genome, with 22% BB305 LVV-positive cells.
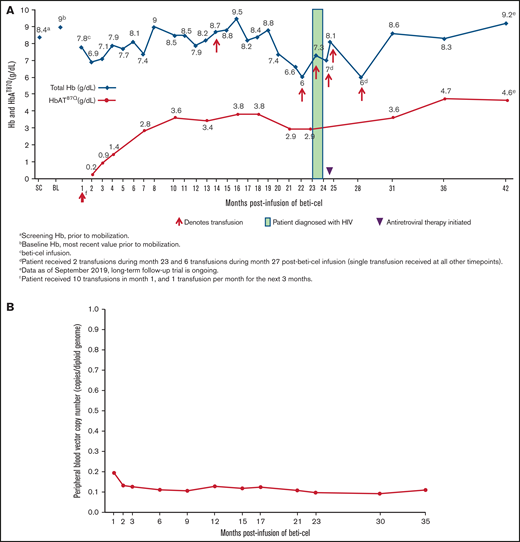
The patient received packed red blood cell transfusions for 3.5 months after beti-cel infusion but then was able to stop transfusions for 17 months with the exception of a single transfusion during month 13 postinfusion because of food poisoning‒related Vibrio parahaemolyticus septicemia, which resolved several days after treatment. During this 17-month period in which the patient stopped receiving chronic transfusions, total Hb and HbAT87Q levels were 6.6 to 9.5 g/dL and 2.8 to 3.8 g/dL, respectively. The patient again began requiring monthly transfusions, starting at month 21 following a previous 3-month decline in total Hb and HbAT87Q levels that reached nadirs of 6.0 and 2.9 g/dL, respectively (Figure 2A). The VCN in the peripheral blood remained stable (range: 0.11 to 0.13 copies per diploid genome; Figure 2B).
Total Hb and HbAT87Q levels and transfusion requirements post–beti-cel infusion. (A) Patient schedule of total Hb (blue diamonds) and HbAT87Q (red circles) measurements relative to transfusions received. Total Hb measurement of <9.0 g/dL was indicated as a common reason for transfusion. (B) Measurements of patient’s peripheral blood VCN levels by month postinfusion. Determination of peripheral blood VCN was performed as a pharmacodynamic measure of treatment efficacy in study HGB-204. In the study population, VCN in peripheral blood mononuclear cells correlated with the production of vector-encoded HbAT87Q-globin levels.1 BL, baseline; Hb, hemoglobin; SC, screening; VCN, vector copy number.
Total Hb and HbAT87Q levels and transfusion requirements post–beti-cel infusion. (A) Patient schedule of total Hb (blue diamonds) and HbAT87Q (red circles) measurements relative to transfusions received. Total Hb measurement of <9.0 g/dL was indicated as a common reason for transfusion. (B) Measurements of patient’s peripheral blood VCN levels by month postinfusion. Determination of peripheral blood VCN was performed as a pharmacodynamic measure of treatment efficacy in study HGB-204. In the study population, VCN in peripheral blood mononuclear cells correlated with the production of vector-encoded HbAT87Q-globin levels.1 BL, baseline; Hb, hemoglobin; SC, screening; VCN, vector copy number.
Methods
HIV testing and diagnosis
Approximately 10 months post–beti-cel infusion, the patient reported to the clinical team with a positive result on a self-initiated screening HIV PCR assay conducted outside of the clinical trial based on reported activities that carry a high risk of exposure to HIV. The patient had a viral load of 404 copies per mL detected by HIV PCR assay (AmpliPrep/TaqMan V.2.0; Roche Diagnostics, Basel, Switzerland); a repeat test performed several days later detected a viral load of 145 copies per mL. The clinical trial team retested the patient’s HIV viral load using the Abbott RealTime HIV-1 assay (Abbott, Chicago, IL), which showed an undetectable viral load (limit of detection: 40 copies per mL). The discrepancy between the 2 assay results might have occurred because the former assay detects the HIV-1 gag gene and LTR region, but the latter assay detects the integrase region. Anti-HIV tests were repeatedly negative around this time point. Although the exact sequences detected by these different viral load tests are unknown, the BB305 LVV proviral genome contains a portion of the gag gene as well as a portion of the integrase gene that is part of the cPPT element (Figure 1A). Because of the SIN feature of the vector design, these HIV-based sequences should not be present as RNA in cells carrying an integrated vector genome. However, it is possible that rare vector insertions lie adjacent to an active endogenous promoter that transcribes into the vector, producing sequences that may cross-react with an HIV viral load assay. A follow-up WB performed 12.5 months postinfusion was negative for anti–HIV-1 antibodies. At the discretion of the study sponsor and investigator, the patient subsequently received annual screenings for HIV infection.
The next WB performed 22 months postinfusion was positive for antibodies against several HIV-1 viral proteins (gp160, gp120, gp41, p40, p24, and p51/55). This was preceded by the previously described decline in the patient’s Hb and occurrence of neutropenia, fever, and lymphadenitis. Detection of anti-HIV antibodies by WB is consistent with a diagnosis of HIV-1 infection and not detection of antibodies directed at BB305 LVV‒derived proteins.7 WB analysis of a sample of the BB305 LVV used to transduce stem cells shows the presence of p24 and reverse transcriptase, because the vector contains the minimum HIV gene sequences necessary for viral genome packaging, reverse transcription, and integration into host cells,4,8,9 but it does not contain HIV-derived sequences coding for gp160, gp120, or gp41 (Figure 1B). Therefore, detection of these components in the patient’s samples via WB is consistent with infection with WT HIV. Laboratory tests performed 1 week later confirmed the WB results, showing an HIV-1 viral load of 266 586 copies per mL by PCR (Abbott RealTime HIV-1) and a CD4+ cell count of 111 cells per mm3 (12%). Testing by quantitative PCR for VSV-G performed at this time did not detect any RCL.
Differentiation between WT HIV and BB305 LVV
Risk of BB305 LVV-mediated mobilization or recombination with WT HIV is considered low owing to the SIN vector design and limited amount of HIV-derived sequence. Nonetheless, a highly sensitive PCR assay designed to detect only GT-derived VSV-G and not WT HIV has never detected RCL in any patient in this clinical study, including in this patient in multiple screenings as well as in a screening that roughly coincided with the detection of anti‒HIV-1 antibodies by WB, confirming that the patient was infected with WT HIV and not through recombination and replication of GT-derived HIV sequences.
Next-generation sequencing (NGS) of plasma samples from the patient was performed by Monogram Biosciences (a LabCorp specialty testing group; South San Francisco, CA). The gag, protease, integrase, reverse transcriptase, and the entire env (gp160) regions of HIV-1 from the patient sample were sequenced and compared with sequences relative to a subtype-specific reference sample. Although the BB305 LVV contains short segments of HIV pol and env genes, there are large stretches (>2000 bp) of HIV sequence that are not in the BB305 LVV. NGS of the patient sample identified HIV-1 subtype AE (circulating recombinant form-01), with a high percentage of reads mapped to the reference sequences but with substantial differences between the patient sample and BB305 LVV, which contains HIV-1‒derived sequences from clade B. The sequenced regions mostly cover segments of the HIV genome that are absent from the BB305 LVV but do include sequences corresponding to the packaging signal (Ψ), cPPT, and RRE that are in the BB305 LVV. Notably, sequencing of the Ψ, cPPT, and RRE regions of the patient sample were not identical to BB305 LVV. In addition, other sequences unique to BB305 LVV and its manufacture, including the β-globin gene or VSV-G, were not present. Thus, it was concluded that the HIV identified was WT and not related to beti-cel GT.
Treatment of HIV
Approximately 1 month after diagnosis, antiretroviral treatment with tenofovir disoproxil fumarate/efavirenz/emtricitabine 300/600/200 mg once daily and dapsone 100 mg/d was initiated. Three months later, the patient’s viral load had decreased to 70 copies per mL, and the CD4+ cell count was 163 cells per mm3 (10%), and the antiretroviral treatment regimen was continued.
One year later, the patient’s CD4+ cell count had increased to 378 cells per mm3 (20%), the viral load had decreased to <40 copies per mL, and the patient appeared well on physical examination. As of the last examination (20 months after initiating antiretroviral therapy), the patient was continuing antiretroviral therapy with tenofovir disoproxil fumarate/emtricitabine with rilpivirine (because of side effects experienced with efavirenz) and had a viral load <40 copies per mL.
Recovery from anemia
Anemia is a common complication of HIV infection and is more prevalent in symptomatic (50%) than asymptomatic patients (18%).10 This patient’s anemia necessitated the reinitiation of transfusions after nearly 1.5 years post‒beti-cel infusion, during which time the patient did not require chronic transfusions and had stable total Hb and HbAT87Q levels. Within 3 months of the lowest postdiagnosis Hb measurement, the patient’s total Hb and HbAT87Q levels rebounded, reaching 8.6 and 3.6 g/dL, respectively, at 6 months after initiation of antiretroviral therapy, while the peripheral blood VCN levels remained stable. At last follow-up 3.5 years after beti-cel infusion, 13.4 months had elapsed since the patient’s last transfusion.
Results and discussion
Although hundreds of patients have safely received LVV-based GT,11 theoretical considerations may linger regarding viral sequence recombination and replication upon secondary infection. Here, we report a case in which a patient who was initially transfusion-free with stable levels of GT-derived HbAT87Q and VCN after receiving beti-cel GT had subsequent WT HIV infection coincident with declining total Hb and HbAT87Q levels and an increasing requirement for transfusions. Diagnosis of HIV was confirmed by WB, and WT HIV sequences were differentiated from GT-derived HIV sequences by NGS and absence of RCL. The patient responded to antiretroviral therapy as evidenced by a reduction in viral load and a concomitant increase in total Hb and HbAT87Q levels. Stable peripheral blood VCN and recovery of pre-HIV levels of total Hb and HbAT87Q suggest that successful BB305 LVV transduction of long-lived HSCs can provide sustained erythroid cell production and lead to durable HbAT87Q levels.
Patients treated with beti-cel in whom HIV infection is suspected should undergo a non‒PCR-based assay to test for HIV, such as a WB to detect anti-HIV antibodies. It is possible that HIV testing with a PCR-based assay could return a false-positive result if the PCR primers used to detect WT HIV and GT-derived HIV sequences detect target sequences shared among the vector and the WT sequence.12 Sequencing may be used to monitor for possible recombination in the event of WT viral infection.4,13 Assays to identify WT virus and differentiate it from the vector should be made accessible to physicians caring for patients treated with beti-cel GT, particularly in geographic regions where TDT is prevalent and HIV infection and transmission rates are elevated. To our knowledge, this is the first published account of WT HIV infection in a recipient of LVV-based GT. As GT becomes more prevalent, physicians should understand how routine monitoring and preventive health care are affected in patients treated with GT. However, safety measures have been integrated into the design of the BB305 LVV to avoid risk of emergence of RCL or recombination with WT HIV.
Acknowledgments
The authors acknowledge the following individuals for their contributions to this manuscript: the patient, Alexandria Petrusich (operational support), Alison Tibbets (pharmacovigilance support), Ying Chen (biostatistics), Benchawan Ratchasueb (CRA), Yegor Smurnyy, Jessica McKenzie, and Brandan Lang.
Funding support for this article was provided by bluebird bio, Inc., Cambridge, MA. Medical writing and editorial assistance were provided by Jennifer Czarneski and Jennifer Darby of Peloton Advantage, LLC (Parsippany, NJ), an OPEN Health company, and funded by bluebird bio, Inc., Cambridge, MA.
Authorship
Contribution: S.H., U.A., D.S., A.P., and K.J. were involved with treatment of the patient; G.P., B.D., M.B., G.V., and M.A. analyzed the results; and all authors collaborated on writing the paper.
Conflict-of-interest disclosure: G.P., B.D., and M.B. are employees of bluebird bio, Inc., and own shares in the company. G.V. is a former employee of bluebird bio, Inc. M.A. was an employee of bluebird bio, Inc., during the study in which the patient was enrolled. The remaining authors declare no competing financial interests.
Correspondence: Suradej Hongeng, Faculty of Medicine, Rama thibodi Hospital, Mahidol University, Rama VI Rd, Bangkok, 10400 Thailand; e-mail: suradej.hon@mahidol.ac.th
References
Author notes
Appropriately deidentified patient-level datasets and supporting documents may be shared following attainment of applicable marketing approvals and consistent with criteria established by bluebird bio and/or industry best practices to maintain the privacy of study participants. For more information, please contact datasharing@bluebirdbio.com.
S.H. and U.A. contributed equally to this study.

![Diagram of the BB305 LVV with human βA-T87Q substitution and corresponding HIV-derived sequences. (A) The βA-T87Q substitution (ACA [Thr] to CAG [Gln]) in the human β-globin gene is indicated by an asterisk and arrow. Safety modifications included mutation of the gag gene and a 400-bp deletion in the U3 of the right HIV LTR.4 HIV-derived sequences are shown in gray boxes and are aligned with corresponding genes in the HIV genome (top; adapted from Frankel and Young).9 3'enh/pA, 3' enhancer/polyadenylation signal from the β-globin gene; ΔU3, promoter/enhancer-deleted unique 3'; ψ, packaging signal; cPPT, central polypurine tract; E, exon; gag, HIV-1 partial gag sequence; Globin LCR, human globin locus control regions; LTR, long terminal repeat; LVV, lentiviral vector; P, β-globin promoter; Q, gluatmine; R, repeat; RRE, rev-response element; T, threonine; U5, unique 5'. (B) WB assay of BB305 LVV for HIV-1 proteins. Lane 1 of each blot contains 50 to 100 ng of each analyzed protein (from left to right: [A] recombinant (r)-gp120, [B] r-gp41, [C] r-RT, [D] r-p24); lane 2 of each blot contains a molecular weight ladder; lane 3 of each blot contains 4 ng BB305 LVV. Primary mAbs include anti-p24 mAb 63917 (Abcam); anti-gp41 mAb MA1-7590 (Invitrogen); anti-RT mAb 63911 (Abcam); anti-gp120 mAb AHP2204 (Bio-Rad). Secondary mAb is IRDye 800CW (925-32211; Li-Cor Biosciences). The HIV p24 and reverse transcriptase proteins are not encoded in the BB305 LVV but are present as part of the purified vector particle and may be identifiable via WB analysis of drug product. The HIV-1 env protein gp160 cleavage products gp41 and gp120 are not present in the BB305 LVV production system and thus are not detectable.4,9 mAbs, monoclonal antibodies; MW, molecular weight; RT, reverse transcriptase; WB, western blot.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/5/13/10.1182_bloodadvances.2020003680/3/m_advancesadv2020003680f1.png?Expires=1769852966&Signature=tT~ZBltsGTKT5T5FjX04DbvRLTlpGuEi4YDUXZo9K1GLWyCrlni6vA4kkghu6yqBS-vVxxuviCuUwFYXMCSy-GmzmP7Nz9K5uCU~FMS31JVmvkj13Dj3bytKk7~uX7FPFKJ1AF26wdZ9-NNSjBqhuTCVU9nwodYovTxJ~3R-gX0DAf6fGo1XszKyfvGekU10~bQge14NP7-ub7-XJSfJi4Pxfecq2dEuxdb5rqLckELrR~82pbu69xelp36JjnI80bhUMtIbm9Gimn7y6Y57Qo1hfDpE9qv6eZjK~8jr6C6LgIrXR3ojQBJ3oWTWwtz-dr0RR0SyOv0nkj9j0DUlvQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)