Key Points
Methyl CpG binding magnetic beads fractionate EBV DNA to distinguish DNA extracted from virions and that from latently infected cells.
Methyl CpG binding may be useful in distinguishing circulating tumor DNA from virion DNA in plasma in patients with Hodgkin lymphoma.
Abstract
Epstein-Barr virus (EBV) is associated with a variety of tumors and nonmalignant conditions. Latent EBV genomes in cells, including tumor cells, are often CpG methylated, whereas virion DNA is not CpG methylated. We demonstrate that methyl CpG binding magnetic beads can be used to fractionate among sources of EBV DNA (DNA extracted from laboratory-purified virions vs DNA extracted from latently infected cell lines). We then applied the technique to plasma specimens and showed that this technique can distinguish EBV DNA from patients with EBV-associated tumors (nasopharyngeal carcinoma, Hodgkin lymphoma) and viral DNA from patients without EBV-associated tumors, including immunocompromised patients and patients with EBV(−) Hodgkin lymphoma.
Introduction
Cell-free DNA (cfDNA) in plasma is emerging as an important biomarker in oncology.1,2 Epstein-Barr virus (EBV) is associated with a variety of tumor types, and EBV DNA has potential utility as a tumor biomarker in EBV-associated malignancies. EBV cfDNA has been shown to be useful for staging, monitoring response to therapy, and most recently, for screening for nasopharyngeal carcinoma in high-risk populations.3-6 In previous studies, we have shown that EBV DNA in plasma or serum from patients with Hodgkin lymphoma (HL) correlates closely with the presence of EBV in tumor cells, as assessed by in situ hybridization consistent with the notion that EBV cfDNA is largely tumor-derived in patients with HL.7,8 However, we observed that EBV cfDNA was occasionally present in high copy number in patients with tumors that were EBV(−), as assessed by EBER in situ hybridization or LMP1 immunohistochemistry. Separately, we have investigated EBV cfDNA in plasma from a general hospital population in Baltimore, MD, and found that the presence of EBV DNA is not usually associated with malignancy, as many immunocompromised patients had detectable EBV DNA in plasma.9 With the possible utility of EBV DNA as a tumor marker in mind, we have been interested in developing a tool to better distinguish EBV DNA derived from latently infected cells, including cancer cells, from EBV virion DNA. We note that previous investigators have used DNAse digestion to distinguish virions from naked DNA.10
Latent EBV genomes in cells, including tumor cells, are often CpG methylated.11-15 Several different patterns of viral gene expression in latency are recognized, and these correspond to different patterns of CpG methylation. In contrast, EBV DNA in virions is not CpG methylated.16 In the present investigations, we developed a simple method for separating CpG methylated DNA from unmethylated DNA, and explored its application to clinical specimens with a focus on plasma from patients with HL.
Methods
Cell culture, control DNA samples, and DNA isolation
The Burkitt cell lines Akata (gift from L. Hutt-Fletcher), Raji (gift from S. D. Hayward), and Namalwa (gift from S. D. Hayward), as well as a lymphoblastoid cell line (established in the R.F.A. laboratory) were grown in RPMI 1640 containing 10% fetal calf serum, 2 mM L-glutamine, penicillin-streptomycin (100 U/ml and 100 µg/ml, respectively) at 37°C in 5% CO2. Purified virions were prepared from the supernatant of Akata cultures induced with 50 μg/mL anti-human immunoglobulin G (MP Biomedicals).
Specimens
Plasma specimens from patients with AIDS and Kaposi sarcoma (KS) were studied, as well as plasma from patients with untreated classical HL and untreated undifferentiated nasopharyngeal carcinoma (NPC; World Health Organization type III). Specimens were obtained with informed consent from patients and approval from the relevant institutional review boards.
EBV status of tumor tissue
EBV status in HL was determined by EBER in situ hybridization or LMP1 staining.7,17,18 As NPCs World Health Organization type III are consistently associated with EBV, the EBV association in these tumors was not separately tested.
Blood specimen collection, DNA extraction, and quantitative real-time polymerase chain reaction
Plasma separation and DNA extraction were performed as described previously.7 EBV copy number was determined by real-time polymerase chain reaction (PCR) with a primer pair and probe corresponding to the BamH-W region of the EBV genome (sense: CCCAACACTCCACCACACC, antisense: TCTTAGGAGCTGTCCGAGGG, and probe: 5′-[6-FAM]CACACACTACACACACCCACCCGTCTC [BHQ-1]-3′) were used.7
Methylated DNA enrichment
Extracted DNA was mixed with 950 ng carrier K562 DNA and added to 10 μL MBD-Bead slurry (MethylMiner DNA Enrichment Kit, Invitrogen, Carlsbad, CA), incubated together on a rotating mixer for 1 hour, and washed. Unbound, low-salt washes and a high-salt eluate (2000 mM NaCl) were collected. EBV was quantitated by PCR in the high-salt eluate and in the washes and flow through. Fractions were ethanol precipitated, resuspended in water, and PCR amplified with primer pair and probe corresponding to the BamH-W region of the EBV genome (amplicon length, 76 bp; repeat located between 14649 and 33137 on the reference sequence NC_007605.1). The EBV methylation index was calculated as the absolute copy number of EBV DNA recovered from the high-salt eluate divided by the sum of the absolute copy number of EBV DNA in all fractions.
Statistics
Statistical analysis and graphing used GraphPad Prism version 8.00 for Windows (GraphPad Software, San Diego, CA).
Results and Discussion
EBV methylation in cell lines
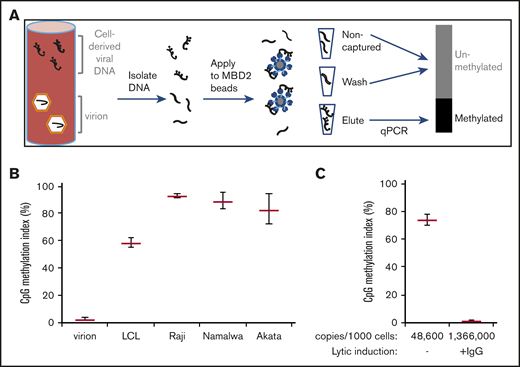
In initial investigations, we used magnetic beads linked to a methyl DNA binding protein to separate CpG methylated DNA from unmethylated DNA (Figure 1A). As expected, virion DNA did not bind to the methyl CpG binding protein beads. In contrast, DNA extracted from a lymphoblastoid cell line was 58% captured, and DNA extracted from 3 EBV(+) Burkitt lymphoma cell lines was more than 70% captured (Figure 1B). When we induced lytic viral replication in the Akata Burkitt cell line, EBV copy number increased approximately 25-fold. The induced lytic viral DNA sequences were not bound by the beads. High levels of lytic viral DNA obscured detection of latent viral DNA in this cell line (Figure 1C).
Methylated EBV DNA from latently infected cells is preferentially enriched by binding to methyl-DNA (MBD2) binding beads. (A) Schematic presentation of the technique. DNA is incubated with methyl-DNA binding beads. DNA from the unbound (noncaptured) fraction, washes (300 and 400 mM NaCl), and eluate (2000 mM NaCl elution) are all quantitatively amplified by quantitative PCR. (B) EBV methylation index of DNA from virions, a lymphoblastoid cell line, and Burkitt cell lines Raji, Namalwa, and Akata. (C) EBV methylation index of Akata cells in the absence of lytic replication, and after incubation with 50 μg/mL anti-human immunoglobulin G (IgG) to induce lytic viral replication.
Methylated EBV DNA from latently infected cells is preferentially enriched by binding to methyl-DNA (MBD2) binding beads. (A) Schematic presentation of the technique. DNA is incubated with methyl-DNA binding beads. DNA from the unbound (noncaptured) fraction, washes (300 and 400 mM NaCl), and eluate (2000 mM NaCl elution) are all quantitatively amplified by quantitative PCR. (B) EBV methylation index of DNA from virions, a lymphoblastoid cell line, and Burkitt cell lines Raji, Namalwa, and Akata. (C) EBV methylation index of Akata cells in the absence of lytic replication, and after incubation with 50 μg/mL anti-human immunoglobulin G (IgG) to induce lytic viral replication.
EBV methylation in plasma DNA
We assayed plasma specimens from patients with untreated NPC. A large fraction of EBV DNA isolated from the plasma was captured by the beads, as illustrated in Figure 2, confirming our hypothesis that CpG methylated EBV DNA would be detected in plasma from patients with nasopharyngeal cancer. Next we were interested in studying plasma from an immunocompromised population (with cancer), but without EBV-associated disease. KS is an AIDS-defining malignancy associated with KSHV/HHV8, but not associated with EBV. Thus, EBV DNA is not usually found at high copy number in the plasma of patients with KS, but on occasion, it is detected. We studied plasma specimens from 5 such patients, and found that the EBV methylation index was less than 5% in all cases. This was consistent with our hypothesis that plasma EBV DNA in these patients was likely virion DNA. Finally, we evaluated plasma from patients with EBV(−) and EBV(+) HL. For the EBV(−) cases, we selected rare cases in which patients with EBV(−) tumors had high EBV viral copy number in plasma. EBV DNA isolated from the plasma of patients with EBV(−) HL tumors, similar to EBV DNA isolated from the plasma of patients with KS, showed EBV methylation indices of less than 5%, whereas EBV DNA isolated from the plasma of patients with EBV(+) HL showed EBV methylation indices of more than 50%. Thus, in a variety of clinical settings, retention by methyl DNA binding beads allowed separation of EBV(+) and EBV(−) disease among patients with high EBV copy number in plasma. Although unmethylated EBV DNA is not necessarily derived from intact virions, methylated DNA is not from virions.
Analysis of EBV copy number and methylation in plasma DNA from clinical specimens. EBV copy number and EBV methylation index from plasma of patients with nasopharyngeal carcinoma (A-B), Kaposi sarcoma (C-D), and EBV(−) and EBV(+) HL (E-F). Scatter plots with mean and standard deviation presenting EBV DNA copy number on the left and the percentage of EBV DNA binding to beads (methylation index) on the right. The results of a 1-tailed Student t test are indicated. ****P ≤ .0001.
Analysis of EBV copy number and methylation in plasma DNA from clinical specimens. EBV copy number and EBV methylation index from plasma of patients with nasopharyngeal carcinoma (A-B), Kaposi sarcoma (C-D), and EBV(−) and EBV(+) HL (E-F). Scatter plots with mean and standard deviation presenting EBV DNA copy number on the left and the percentage of EBV DNA binding to beads (methylation index) on the right. The results of a 1-tailed Student t test are indicated. ****P ≤ .0001.
In the future, assay for the presence of EBV CpG DNA methylation may be useful in the clinical laboratory to distinguish patients with active lytic viral infection leading to the presence of virion DNA in cfDNA vs those with EBV-associated malignancies. This may be especially useful in the posttransplant setting. In the investigations presented here, we have used methyl DNA binding protein beads to select CpG methylated EBV DNA. We note that immunoprecipitation with antibodies that recognize methylcytosine, and a variety of other techniques, to separate CpG methylated DNA would likely give similar results. Detailed characterization of EBV DNA methylation patterns in plasma using bisulfite sequencing has recently been published for NPC, and is consistent with these findings.19 Although that technique provides more granularity with regard to methylation patterns, and may provide additional useful information, our technique is relatively simple and inexpensive.
Data sharing e-mails should be sent to the corresponding author, Richard F. Ambinder (rambind1@jhmi.edu).
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Cancer Institute (P50CA96888, U01CA121947, P30CA006973, and P01CA113239) (R.F.A.), a grant from the Singapore National Research Foundation and the Ministry of Education under the Research Center of Excellence Program (W.-s.H. and R.F.A.), and grants from the Israel Cancer Association (20150095 and 20161143) funded by Walter Bela foundation, and Research Career Development Award from the Israel Cancer Research Fund (01282) (M.S.). The authors are grateful for the support of the Elias, Genevieve and Georgianna Atol Charitable Trust to the Daniella Lee Casper Laboratory in Viral Oncology.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship
Contribution: M.S., J.A.K., N.A.H., G.J., A.A., J.S.W.L., Y.E., E.J.D., J.S., W.L.W., W.-s.H., and E.B. performed experiments; M.S., G.J., J.S., W.L.W., and R.R.X. analyzed results and made figures; and M.S., W.-s.H., and R.F.A. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard F. Ambinder, Department of Oncology, Johns Hopkins School of Medicine, 389 CRB1, 1650 Orleans St, Baltimore, MD 21287; e-mail: rambind1@jhmi.edu.