Key Points
Models of KIR alloreactivity and gene content were not associated with pediatric ALL and AML unrelated donor transplant outcomes.
This study does not support the use of KIR alloreactivity or gene content models in the selection of adult unrelated donors.
Abstract
Multiple models of donor killer immunoglobulin receptor (KIR) alloreactivity or KIR genotype have been reported to be protective against leukemia relapse after allogeneic transplantation. However, few studies have addressed this topic in the pediatric population. Here, we assessed the outcomes of allogeneic transplantation in children with acute lymphoblastic leukemia (ALL; n = 372) or acute myeloid leukemia (AML; n = 344) who received unrelated donor (URD) transplantation and were reported to the Center for International Blood and Marrow Transplant Research (CIBMTR) from 2005 to 2016. As expected in this pediatric population, most patients underwent myeloablative conditioning while in remission and with bone marrow as a stem cell source. We tested KIR ligand mismatch, KIR gene content (centromeric [Cen] B), KIR2DS1 mismatching, and Cen B/telomeric A using Cox regression models and found that none were significantly associated with either relapse or disease-free survival when considering the entire cohort of patients (ALL and AML), AML, or ALL separately. Moreover, there was no significant association with outcomes in the in vivo T-cell–depleted (ie, serotherapy) cohort. This study, which is the largest analysis of donor KIR in the pediatric acute leukemia population, does not support the use of KIR in the selection of URDs for children undergoing T-replete transplantation.
Introduction
One key determinant of natural killer (NK)–cell activation (cytotoxicity and cytokine production) is the killer cell immunoglobulin-like receptor (KIR). The KIR family is a multigene family containing up to 15 separate genes with either inhibitory or activating functions (reviewed in Parham1 ). KIR function can be predicted based on the molecular structure of the individual genes. KIR genes that have long intracellular signaling domains associate with Src homology 2 domain-containing protein tyrosine phosphatases 1 and 2 and deliver inhibitory signals, whereas KIR with short signaling domains associate with the activating adapter molecule DAP12 and trigger NK activation. The ligands recognized by KIR vary. In general, inhibitory KIRs recognize the allelic differences of HLA-B (Bw4) or HLA-C (C1 vs C2), whereas the ligands recognized by most activating KIRs are largely uncharacterized.2 Investigators have used several different models to interrogate the KIR genes and how they might be associated with transplant outcomes.
A landmark study by Ruggeri et al showed that in the setting of T-cell–depleted haploidentical transplantation, donors who express KIR for which there were no ligands in the recipient (ie, KIR ligand mismatch) had dramatically lower rates of acute myeloid leukemia (AML) relapse, but no effect was seen on acute lymphoblastic leukemia (ALL) relapse.3 These findings led to a series of large retrospective registry studies involving adult transplant recipients: some of the studies confirmed these findings,4 whereas others did not.5,6 These studies have mainly focused on AML, given the original and subsequent observations and because ALL cells have been shown to be relatively more resistant to NK-cell cytotoxicity.7 Although still unclear, the cumulative data suggest that KIR ligand mismatch may be most active in the setting of haploidentical transplantation with severe T-cell depletion, with no immune suppression, and in patients with AML, but even in this setting, some studies have failed to show an effect on mismatches in the KIR system.8
Cooley et al took a different approach, examining KIR gene content as a predictor for transplant outcomes.9 As in the Parham review,1 there are 15 KIR genes, and individuals differ in the number and distribution of KIR genes that they possess, with 2 large groupings. The first, known as haplotype A, is characterized by a fixed set of KIR genes, all of which are inhibitory, except KIR2DS4. In contrast, individuals with KIR B haplotypes have 1 or more of the following mostly activating genes: KIR2DL5, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5, and KIR3DS1. In their initial analysis of ∼450 AML patients undergoing T-cell–replete HLA-matched or -mismatched unrelated donor (URD) hematopoietic cell transplantation (HCT), donor and recipient KIR genotype (ie, genotype A/A or B/x) was tested for associations with transplant outcomes. In multivariable analysis, KIR B donors (ie, KIR B/x) showed a 30% improved leukemia-free survival.9 In a follow-up study, these same investigators used a larger cohort of patients (n = 1409) and found that KIR B genes in the centromeric (Cen) or telomeric (Tel) region might be associated with their original observations. KIR B (Cen B) donors were associated with less AML relapse.10 Similar findings were published by Stringaris et al and Michaelis et al with significantly fewer patient numbers.11,12 Similar to Cooley, Venstrom et al used registry samples and outcome data to evaluate patients receiving HLA-matched or single allele mismatched URD HCT for AML. Their analysis focused on the presence or absence of the KIR B haplotype-defining gene KIR2DS1 in HLA-C1 donors, which was associated with significantly lower rates of AML relapse.13 Collectively, these data suggest that donor KIR gene content either alone or in the context of HLA may be a better predictor of transplant outcomes following allogeneic HCT (allo-HCT) than ligand mismatching.
More recently, Babor et al retrospectively examined the outcomes of high-risk pediatric ALL patients undergoing allo-HCT (both sibling [n = 65] and URD [n = 144]).14 Cen B/x donors were not protective against relapse in this ALL population and they then developed a “composite score” that considered Tel A/A genes in conjunction with Cen B/x genes. Donors were classified into low, moderate, and high risk of relapse (40% vs 21% vs 13%, respectively; P < .01). In a subgroup analysis, these data held up in the URDs. Other than those mentioned in this section, relatively few studies have addressed the topic of donor NK-cell genetics and alloreactivity in pediatric transplant recipients. Recently, Oevermann et al evaluated 85 children with ALL who underwent haploidentical transplantation. Interestingly, 24% of these patients had T-cell ALL and patients received haploidentical grafts that were manipulated using CD34+ selection, CD3/CD19 depletion, or T-cell receptor αβ depletion.15 Thus, the transplants were T-cell depleted (apart from γ/δ T cells in some cases). ALL relapse and disease-free survival (DFS) were significantly better in recipients of KIR B/x donors, leading these investigators to conclude that donor KIR B genotype should be used in haploidentical donor selection.
Whether any of the above-described findings extends to children with ALL or AML who receive well-matched, T-cell–replete, or in vivo T-cell–depleted, URD transplantation is unknown. Here, we report the outcomes of a large cohort of pediatric patients with available KIR typing to test the hypothesis that variations in the donor KIR system are associated with improved transplant outcomes.
Methods
Clinical and demographic variables were evaluated for their distributional differences between ALL and AML using classical χ2 tests (or the Fisher's exact test for a small number of counts). Unadjusted comparison between donor KIR genotypes for overall survival (OS) and DFS outcomes was made using a log-rank test. Kaplan-Meier estimators were used to estimate the probability of OS and DFS. Cumulative incidence rates were estimated for relapse, transplant-related mortality (TRM), and acute and chronic graft-versus-host disease (GVHD; aGVHD and cGVHD). Relapse was summarized using cumulative incidence, with TRM as a competing risk, and TRM was calculated with relapse as a competing risk. Death is a competing risk for aGVHD and cGVHD. Patients were censored at the time of last follow-up. The primary end points include relapse, followed by DFS. Exploratory end points include aGVHD II-IV and OS. KIR typing and interpretation was performed using methods previously described.16 KIR models included missing self,3 Cen B,9 2DS1,13 and CenB/Tel A.14
Cox proportional hazard models were used to adjust for important clinical factors. The proportional hazards assumption was evaluated using a time-dependent covariate method, and factors with nonproportional hazards were adjusted through stratification. Forward stepwise variable selection was used to determine which factors require adjustment in each model based on a significance level of 5%. Testing of the A/B KIR genotypes was performed by forcing each KIR variable separately in these models. A significance level of P = .01 was used to adjust for multiple testing. We also tested interactions between KIR-genotype groups and adjusted covariates at a 0.01 significance level. No significant interactions were detected for any of the end points. All P values are raw and 2-sided. All analyses were done using SAS version 9.4.
Results
Patient demographics
As shown in Table 1, the study population included children with either ALL (n = 372) or AML (n = 342) who underwent their first allo-HCT between 2005 and 2016 and had donor presence/absence KIR gene typing available through the ongoing CIBMTR retrospective genotyping program. The median follow-up was 59 months (6-124 months) and 54 months (3-121 months) for ALL and AML recipients, respectively. Slightly more than one-half of the patients were between 10 and 19 years old (56% and 61%) and the majority were white (64% and 74% for the ALL and AML recipients, respectively). Table 2 shows transplant-associated variables and demonstrates that most patients in both the ALL and AML groups received myeloablative conditioning (95% and 86%) and bone marrow (79% and 76%) as a stem cell source. Regarding GVHD prophylaxis, most ALL and AML patients received a calcineurin inhibitor and methotrexate (79% and 76%) and a minority also received in vivo T-cell depletion with serotherapy using antithymocyte globulin or alemtuzumab (21% and 30%). Other transplant-associated details are shown in Table 2.
Patient characteristics
| Variable . | ALL . | AML . | P . |
|---|---|---|---|
| No. of recipients | 372 | 344 | |
| No. of centers | 51 | 60 | |
| Recipient age at transplant, y | .12 | ||
| 0-9, n (%) | 165 (44) | 133 (39) | |
| 10-19, n (%) | 207 (56) | 211 (61) | |
| Median (range) | 11 (0-19) | 12 (1-19) | .55 |
| Recipient sex, n (%) | .24 | ||
| Male | 226 (61) | 194 (56) | |
| Recipient race/ethnicity, n (%) | .05 | ||
| White | 223 (64) | 247 (74) | |
| African American | 23 (7) | 16 (5) | |
| Asian | 13 (4) | 12 (4) | |
| Pacific islander | 1 (<1) | 2 (1) | |
| Native American | 1 (<1) | 3 (1) | |
| Hispanic | 87 (25) | 53 (16) | |
| Unknown | 24 (N/A) | 11 (N/A) | |
| Performance score, n (%) | .11 | ||
| 10-80 | 57 (15) | 60 (17) | |
| 90-100 | 304 (82) | 281 (82) | |
| Years of transplant, n (%) | .25 | ||
| 2005-2009 | 155 (42) | 123 (36) | |
| 2010-2014 | 175 (47) | 181 (53) | |
| 2015-2016 | 42 (11) | 40 (12) | |
| Follow-up among survivors, mo | |||
| No. evaluated | 234 | 185 | |
| Median (range) | 59 (6-124) | 54 (3-121) | .20 |
| Variable . | ALL . | AML . | P . |
|---|---|---|---|
| No. of recipients | 372 | 344 | |
| No. of centers | 51 | 60 | |
| Recipient age at transplant, y | .12 | ||
| 0-9, n (%) | 165 (44) | 133 (39) | |
| 10-19, n (%) | 207 (56) | 211 (61) | |
| Median (range) | 11 (0-19) | 12 (1-19) | .55 |
| Recipient sex, n (%) | .24 | ||
| Male | 226 (61) | 194 (56) | |
| Recipient race/ethnicity, n (%) | .05 | ||
| White | 223 (64) | 247 (74) | |
| African American | 23 (7) | 16 (5) | |
| Asian | 13 (4) | 12 (4) | |
| Pacific islander | 1 (<1) | 2 (1) | |
| Native American | 1 (<1) | 3 (1) | |
| Hispanic | 87 (25) | 53 (16) | |
| Unknown | 24 (N/A) | 11 (N/A) | |
| Performance score, n (%) | .11 | ||
| 10-80 | 57 (15) | 60 (17) | |
| 90-100 | 304 (82) | 281 (82) | |
| Years of transplant, n (%) | .25 | ||
| 2005-2009 | 155 (42) | 123 (36) | |
| 2010-2014 | 175 (47) | 181 (53) | |
| 2015-2016 | 42 (11) | 40 (12) | |
| Follow-up among survivors, mo | |||
| No. evaluated | 234 | 185 | |
| Median (range) | 59 (6-124) | 54 (3-121) | .20 |
N/A, not applicable.
Transplant-associated variables
| Variable . | ALL, n (%) . | AML, n (%) . | P . |
|---|---|---|---|
| Stem cell source | .31 | ||
| Marrow | 293 (79) | 260 (76) | |
| PBSC | 79 (21) | 84 (24) | |
| Conditioning regimen | <.001 | ||
| Myeloablative | 355 (95) | 296 (86) | |
| Reduced intensity | 7 (2) | 10 (3) | |
| Nonmyeloablative | 10 (3) | 38 (11) | |
| In vivo T-cell depletion | .004 | ||
| No | 295 (79) | 241 (70) | |
| Yes | 77 (21) | 103 (30) | |
| GVHD prophylaxis | .12 | ||
| Tacrolimus + MMF ± others | 22 (6) | 31 (9) | |
| Tacrolimus + MTX ± others, except MMF | 173 (47) | 137 (40) | |
| Tacrolimus + others, except MTX, MMF | 10 (3) | 7 (2) | |
| Tacrolimus alone | 6 (2) | 1 (<1) | |
| CSA + MMF ± others, except tacrolimus | 25 (7) | 28 (8) | |
| CSA + MTX ± others, except tacrolimus, MMF | 119 (32) | 124 (36) | |
| CSA + others, except tacrolimus, MTX, MMF | 11 (3) | 6 (2) | |
| CSA alone | 6 (2) | 10 (3) |
| Variable . | ALL, n (%) . | AML, n (%) . | P . |
|---|---|---|---|
| Stem cell source | .31 | ||
| Marrow | 293 (79) | 260 (76) | |
| PBSC | 79 (21) | 84 (24) | |
| Conditioning regimen | <.001 | ||
| Myeloablative | 355 (95) | 296 (86) | |
| Reduced intensity | 7 (2) | 10 (3) | |
| Nonmyeloablative | 10 (3) | 38 (11) | |
| In vivo T-cell depletion | .004 | ||
| No | 295 (79) | 241 (70) | |
| Yes | 77 (21) | 103 (30) | |
| GVHD prophylaxis | .12 | ||
| Tacrolimus + MMF ± others | 22 (6) | 31 (9) | |
| Tacrolimus + MTX ± others, except MMF | 173 (47) | 137 (40) | |
| Tacrolimus + others, except MTX, MMF | 10 (3) | 7 (2) | |
| Tacrolimus alone | 6 (2) | 1 (<1) | |
| CSA + MMF ± others, except tacrolimus | 25 (7) | 28 (8) | |
| CSA + MTX ± others, except tacrolimus, MMF | 119 (32) | 124 (36) | |
| CSA + others, except tacrolimus, MTX, MMF | 11 (3) | 6 (2) | |
| CSA alone | 6 (2) | 10 (3) |
CSA, cyclosporine A; MMF, mycophenolate mofetil; MTX, methotrexate; PBSC, peripheral blood stem cell.
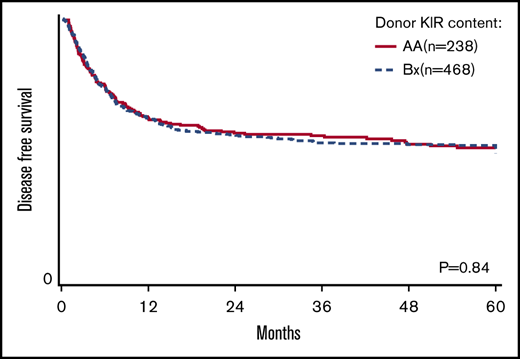
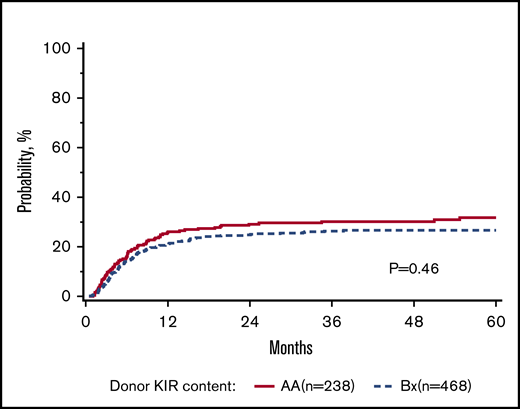
Donor KIR gene content and transplant outcomes
The primary goal of the study was to determine the association of KIR content (AA vs B/x) on transplant outcomes (relapse and DFS) in a purely pediatric cohort of ALL and AML, with TRM, aGVHD, and OS as secondary end points. Considering these 2 groupings of patients, the rate of relapse for these cohorts is shown to demonstrate that the cumulative incidence of relapse is what might be expected in a modern day cohort (Figure 1). In addition to this primary analysis, we also considered other models of donor KIR gene content including: (1) the Cen and Tel region scores (AA vs AB vs BB), (2) donor KIR B ranking score (neutral vs better vs best), (3) a composite scoring system that grouped KIR B gene content based on 0 to 1 vs >2, (4) donor KIR2DS1 and HLA-C1 status, and (5) donor KIR gene content (Tel A and Cen B), which yielded similar findings (not shown). Table 3 shows the number of ALL and AML transplant recipients who had donors who fell into the various groupings, and also demonstrates that the number of donors with the highest density of KIR B genes is relatively limited. The results of the multivariate analyses, considering a variety of relevant clinical outcomes (described in “Methods”) are shown in Table 4. Because we tested multiple end points and KIR genotype variables, we adjusted the level of significance accordingly. The significance level considered a priori was P = .0014 (P = .01/7 tests). Also shown in Table 4, for DFS, there was no significant association of donor AA vs Bx (P = .67), donor centromeric AA vs AB vs BB (P = .36), donor telomeric AA vs AB vs BB (P = .16), donor KIR B content score (P = .28), donor B KIR number (P = .96), alternative B content (P = .98), the KIR composite score (P = .22), or KIR2S1 and HLAC1 (P = .22). When considering other end points including OS, TRM, relapse, and absolute neutrophil count and platelet recovery, similar nonsignificant associations were noted. Not included in Table 4 is the missing ligand analysis, which was also not associated with outcomes (not shown). Similarly, in data not shown, when the analysis was performed based on age (0-10 years old and for those >10-19 years old), the results were not changed. Likewise, when considering only patients in remission, the results were also similar (not shown).
Donor-recipient KIR status
| . | ALL, n (%) . | AML, n (%) . | P . |
|---|---|---|---|
| Donor KIR B content | .50 | ||
| AA | 121 (33) | 121 (35) | |
| B/x | 251 (67) | 223 (65) | |
| Donor B content of KIR | .41 | ||
| 0 | 121 (33) | 121 (35) | |
| 1 | 142 (38) | 109 (32) | |
| 2 | 83 (22) | 87 (25) | |
| 3 | 20 (5) | 23 (7) | |
| 4 | 6 (2) | 4 (1) | |
| Alternative donor B content of KIR | .27 | ||
| Donor B content of KIR of 0 and 1 | 263 (71) | 230 (67) | |
| Donor B content of KIR of 2 or more | 109 (29) | 114 (33) | |
| Cen regions score | .60 | ||
| AA | 191 (51) | 165 (48) | |
| AB | 149 (40) | 144 (42) | |
| BB | 32 (9) | 35 (10) | |
| Tel regions score | .68 | ||
| AA | 218 (59) | 208 (60) | |
| AB | 129 (35) | 118 (34) | |
| BB | 25 (7) | 18 (5) | |
| Donor KIR B content ranking score | .53 | ||
| Best | 32 (9) | 35 (10) | |
| Better | 77 (21) | 79 (23) | |
| Neutral | 263 (71) | 230 (67) | |
| KIR composite score | .06 | ||
| 2 | 97 (26) | 87 (25) | |
| 3 | 205 (55) | 213 (62) | |
| 4 | 70 (19) | 44 (13) |
| . | ALL, n (%) . | AML, n (%) . | P . |
|---|---|---|---|
| Donor KIR B content | .50 | ||
| AA | 121 (33) | 121 (35) | |
| B/x | 251 (67) | 223 (65) | |
| Donor B content of KIR | .41 | ||
| 0 | 121 (33) | 121 (35) | |
| 1 | 142 (38) | 109 (32) | |
| 2 | 83 (22) | 87 (25) | |
| 3 | 20 (5) | 23 (7) | |
| 4 | 6 (2) | 4 (1) | |
| Alternative donor B content of KIR | .27 | ||
| Donor B content of KIR of 0 and 1 | 263 (71) | 230 (67) | |
| Donor B content of KIR of 2 or more | 109 (29) | 114 (33) | |
| Cen regions score | .60 | ||
| AA | 191 (51) | 165 (48) | |
| AB | 149 (40) | 144 (42) | |
| BB | 32 (9) | 35 (10) | |
| Tel regions score | .68 | ||
| AA | 218 (59) | 208 (60) | |
| AB | 129 (35) | 118 (34) | |
| BB | 25 (7) | 18 (5) | |
| Donor KIR B content ranking score | .53 | ||
| Best | 32 (9) | 35 (10) | |
| Better | 77 (21) | 79 (23) | |
| Neutral | 263 (71) | 230 (67) | |
| KIR composite score | .06 | ||
| 2 | 97 (26) | 87 (25) | |
| 3 | 205 (55) | 213 (62) | |
| 4 | 70 (19) | 44 (13) |
Multivariate analysis of donor KIR models and transplant outcomes
| End points . | Donor KIR haplotype, AA vs Bx . | Donor KIR Cen regions score, AA vs AB vs BB . | Donor KIR Tel regions score, AA vs AB vs BB . | Donor KIR B content score, neutral vs better vs best . | Donor KIR B content score, 1 vs 2 vs 3 vs 4 . | Alternative donor KIR B content score, 0-1 vs 2 ≤ . | KIR B composite score, 4 vs 3 vs 2 . | KIR2DS1 score, no vs yes . |
|---|---|---|---|---|---|---|---|---|
| Survival | P = .78 | P = .38 | P = .54 | P = .42 | P = .89 | P = .67 | P = .28 | P = .33 |
| AA, n = 240 | AA, n = 354 | AA, n = 424 | Neutral, n = 491 | 0, n = 240 | 0-1, n = 491 | 2, n = 184 | No, n = 438 | |
| Bx, n = 473 | AB, n = 292 | AB, n = 246 | Better, n = 155 | 1, n = 251 | 2+, n = 222 | 3, n = 415 | Yes, n = 275 | |
| BB, n = 67 | BB, n = 43 | Best, n = 67 | 2, n = 169 | 4, n = 114 | ||||
| 3+, n = 53 | ||||||||
| DFS | P = .67 | P = .36 | P = .16 | P = .28 | P = .96 | P = .98 | P = .22 | P = .71 |
| AA, n = 238 | AA, n = 350 | AA, n = 421 | Neutral, n = 488 | 0, n = 238 | 0-1, n = 488 | 2, n = 183 | No, n = 432 | |
| Bx, n = 468 | AB, n = 289 | AB, n = 244 | Better, n = 151 | 1, n = 250 | 2+, n = 218 | 3, n = 411 | Yes, n = 274 | |
| BB, n = 67 | BB, n = 41 | Best, n = 67 | 2, n = 165 | 4, n = 112 | ||||
| 3+, n = 53 | ||||||||
| TRM | P = .46 | P = .09 | P = .35 | P = .08 | P = .61 | P = .61 | P = .19 | P = .55 |
| AA, n = 238 | AA, n = 350 | AA, n = 421 | Neutral, n = 488 | 0, n = 238 | 0-1, n = 488 | 2, n = 183 | No, n = 432 | |
| Bx, n = 468 | AB, n = 289 | AB, n = 244 | Better, n = 151 | 1, n = 250 | 2+, n = 218 | 3, n = 411 | Yes, n = 274 | |
| BB, n = 67 | BB, n = 41 | Best, n = 67 | 2, n = 165 | 4, n = 112 | ||||
| 3+, n = 53 | ||||||||
| Relapse | P = .46 | P = .82 | P = .13 | P = .76 | P = .86 | P = .46 | P = .71 | P = .73 |
| AA, n = 238 | AA, n = 350 | AA, n = 421 | Neutral, n = 488 | 0, n = 238 | 0-1, n = 488 | 2, n = 183 | No, n = 432 | |
| Bx, n = 468 | AB, n = 289 | AB, n = 244 | Better, n = 151 | 1, n = 250 | 2+, n = 218 | 3, n = 411 | Yes, n = 274 | |
| BB, n = 67 | BB, n = 41 | Best, n = 67 | 2, n = 165 | 4, n = 112 | ||||
| 3+, n = 53 | ||||||||
| aGVHD II-IV | P = .79 | P = .54 | P = .73 | P = .33 | P = .21 | P = .46 | P = .90 | P = .09 |
| AA, n = 191 | AA, n = 271 | AA, n = 326 | Neutral, n = 369 | 0, n = 191 | 0-1, n = 369 | 2, n = 135 | No, n = 329 | |
| Bx, n = 344 | AB, n = 212 | AB, n = 182 | Better, n = 114 | 1, n = 178 | 2+, n = 166 | 3, n = 320 | Yes, n = 206 | |
| BB, n = 52 | BB, n = 27 | Best, n = 52 | 2, n = 129 | 4, n = 80 | ||||
| 3+, n = 37 | ||||||||
| aGVHD III-IV | P = .02 | P = .02 | P = .57 | P = .04 | P = .04 | P = .11 | P = .67 | P = .42 |
| AA, n = 212 | AA, n = 311 | AA, n = 372 | Neutral, n = 427 | 0, n = 212 | 0-1, n = 427 | 2, n = 160 | No, n = 372 | |
| Bx, n = 409 | AB, n = 251 | AB, n = 215 | Better, n = 135 | 1, n = 215 | 2+, n = 194 | 3, n = 362 | Yes, n = 242 | |
| BB, n = 59 | BB, n = 34 | Best, n = 59 | 2, n = 151 | 4, n = 99 | ||||
| 3+, n = 43 | ||||||||
| cGVHD | P = .45 | P = .17 | P = .36 | P = .18 | P = .22 | P = .36 | P = .51 | P = .93 |
| AA, n = 237 | AA, n = 350 | AA, n = 419 | Neutral, n = 485 | 0, n = 237 | 0-1, n = 485 | 2, n = 182 | No, n = 432 | |
| Bx, n = 467 | AB, n = 288 | AB, n = 242 | Better, n = 153 | 1, n = 248 | 2+, n = 219 | 3, n = 409 | Yes, n = 272 | |
| BB, n = 66 | BB, n = 43 | Best, n = 66 | 2, n = 167 | 4, n = 113 | ||||
| 3+, n = 52 | ||||||||
| Neutrophil engraftment | P = .76 | P = .56 | P = .28 | P = .68 | P = .51 | P = .71 | P = .44 | P = .38 |
| AA, n = 242 | AA, n = 356 | AA, n = 426 | Neutral, n = 493 | 0, n = 242 | 0-1, n = 493 | 2, n = 184 | No, n = 439 | |
| Bx, n = 474 | AB, n = 293 | AB, n = 247 | Better, n = 156 | 1, n = 251 | 2+, n = 223 | 3, n = 418 | Yes, n = 277 | |
| BB, n = 67 | BB, n = 43 | Best, n = 67 | 2, n = 170 | 4, n = 114 | ||||
| 3+, n = 53 | ||||||||
| Platelet engraftment | P = .09 | P = .57 | P = .28 | P = .48 | P = .36 | P = .23 | P = .51 | P = .42 |
| AA, n = 240 | AA, n = 354 | AA, n = 423 | Neutral, n = 490 | 0, n = 240 | 0-1, n = 490 | 2, n = 183 | No, n = 437 | |
| Bx, n = 472 | AB, n = 291 | AB, n = 246 | Better, n = 155 | 1, n = 250 | 2+, n = 222 | 3, n = 415 | Yes, n = 275 | |
| BB, n = 67 | BB, n = 43 | Best, n = 67 | 2, n = 169 | 4, n = 114 | ||||
| 3+, n = 53 |
| End points . | Donor KIR haplotype, AA vs Bx . | Donor KIR Cen regions score, AA vs AB vs BB . | Donor KIR Tel regions score, AA vs AB vs BB . | Donor KIR B content score, neutral vs better vs best . | Donor KIR B content score, 1 vs 2 vs 3 vs 4 . | Alternative donor KIR B content score, 0-1 vs 2 ≤ . | KIR B composite score, 4 vs 3 vs 2 . | KIR2DS1 score, no vs yes . |
|---|---|---|---|---|---|---|---|---|
| Survival | P = .78 | P = .38 | P = .54 | P = .42 | P = .89 | P = .67 | P = .28 | P = .33 |
| AA, n = 240 | AA, n = 354 | AA, n = 424 | Neutral, n = 491 | 0, n = 240 | 0-1, n = 491 | 2, n = 184 | No, n = 438 | |
| Bx, n = 473 | AB, n = 292 | AB, n = 246 | Better, n = 155 | 1, n = 251 | 2+, n = 222 | 3, n = 415 | Yes, n = 275 | |
| BB, n = 67 | BB, n = 43 | Best, n = 67 | 2, n = 169 | 4, n = 114 | ||||
| 3+, n = 53 | ||||||||
| DFS | P = .67 | P = .36 | P = .16 | P = .28 | P = .96 | P = .98 | P = .22 | P = .71 |
| AA, n = 238 | AA, n = 350 | AA, n = 421 | Neutral, n = 488 | 0, n = 238 | 0-1, n = 488 | 2, n = 183 | No, n = 432 | |
| Bx, n = 468 | AB, n = 289 | AB, n = 244 | Better, n = 151 | 1, n = 250 | 2+, n = 218 | 3, n = 411 | Yes, n = 274 | |
| BB, n = 67 | BB, n = 41 | Best, n = 67 | 2, n = 165 | 4, n = 112 | ||||
| 3+, n = 53 | ||||||||
| TRM | P = .46 | P = .09 | P = .35 | P = .08 | P = .61 | P = .61 | P = .19 | P = .55 |
| AA, n = 238 | AA, n = 350 | AA, n = 421 | Neutral, n = 488 | 0, n = 238 | 0-1, n = 488 | 2, n = 183 | No, n = 432 | |
| Bx, n = 468 | AB, n = 289 | AB, n = 244 | Better, n = 151 | 1, n = 250 | 2+, n = 218 | 3, n = 411 | Yes, n = 274 | |
| BB, n = 67 | BB, n = 41 | Best, n = 67 | 2, n = 165 | 4, n = 112 | ||||
| 3+, n = 53 | ||||||||
| Relapse | P = .46 | P = .82 | P = .13 | P = .76 | P = .86 | P = .46 | P = .71 | P = .73 |
| AA, n = 238 | AA, n = 350 | AA, n = 421 | Neutral, n = 488 | 0, n = 238 | 0-1, n = 488 | 2, n = 183 | No, n = 432 | |
| Bx, n = 468 | AB, n = 289 | AB, n = 244 | Better, n = 151 | 1, n = 250 | 2+, n = 218 | 3, n = 411 | Yes, n = 274 | |
| BB, n = 67 | BB, n = 41 | Best, n = 67 | 2, n = 165 | 4, n = 112 | ||||
| 3+, n = 53 | ||||||||
| aGVHD II-IV | P = .79 | P = .54 | P = .73 | P = .33 | P = .21 | P = .46 | P = .90 | P = .09 |
| AA, n = 191 | AA, n = 271 | AA, n = 326 | Neutral, n = 369 | 0, n = 191 | 0-1, n = 369 | 2, n = 135 | No, n = 329 | |
| Bx, n = 344 | AB, n = 212 | AB, n = 182 | Better, n = 114 | 1, n = 178 | 2+, n = 166 | 3, n = 320 | Yes, n = 206 | |
| BB, n = 52 | BB, n = 27 | Best, n = 52 | 2, n = 129 | 4, n = 80 | ||||
| 3+, n = 37 | ||||||||
| aGVHD III-IV | P = .02 | P = .02 | P = .57 | P = .04 | P = .04 | P = .11 | P = .67 | P = .42 |
| AA, n = 212 | AA, n = 311 | AA, n = 372 | Neutral, n = 427 | 0, n = 212 | 0-1, n = 427 | 2, n = 160 | No, n = 372 | |
| Bx, n = 409 | AB, n = 251 | AB, n = 215 | Better, n = 135 | 1, n = 215 | 2+, n = 194 | 3, n = 362 | Yes, n = 242 | |
| BB, n = 59 | BB, n = 34 | Best, n = 59 | 2, n = 151 | 4, n = 99 | ||||
| 3+, n = 43 | ||||||||
| cGVHD | P = .45 | P = .17 | P = .36 | P = .18 | P = .22 | P = .36 | P = .51 | P = .93 |
| AA, n = 237 | AA, n = 350 | AA, n = 419 | Neutral, n = 485 | 0, n = 237 | 0-1, n = 485 | 2, n = 182 | No, n = 432 | |
| Bx, n = 467 | AB, n = 288 | AB, n = 242 | Better, n = 153 | 1, n = 248 | 2+, n = 219 | 3, n = 409 | Yes, n = 272 | |
| BB, n = 66 | BB, n = 43 | Best, n = 66 | 2, n = 167 | 4, n = 113 | ||||
| 3+, n = 52 | ||||||||
| Neutrophil engraftment | P = .76 | P = .56 | P = .28 | P = .68 | P = .51 | P = .71 | P = .44 | P = .38 |
| AA, n = 242 | AA, n = 356 | AA, n = 426 | Neutral, n = 493 | 0, n = 242 | 0-1, n = 493 | 2, n = 184 | No, n = 439 | |
| Bx, n = 474 | AB, n = 293 | AB, n = 247 | Better, n = 156 | 1, n = 251 | 2+, n = 223 | 3, n = 418 | Yes, n = 277 | |
| BB, n = 67 | BB, n = 43 | Best, n = 67 | 2, n = 170 | 4, n = 114 | ||||
| 3+, n = 53 | ||||||||
| Platelet engraftment | P = .09 | P = .57 | P = .28 | P = .48 | P = .36 | P = .23 | P = .51 | P = .42 |
| AA, n = 240 | AA, n = 354 | AA, n = 423 | Neutral, n = 490 | 0, n = 240 | 0-1, n = 490 | 2, n = 183 | No, n = 437 | |
| Bx, n = 472 | AB, n = 291 | AB, n = 246 | Better, n = 155 | 1, n = 250 | 2+, n = 222 | 3, n = 415 | Yes, n = 275 | |
| BB, n = 67 | BB, n = 43 | Best, n = 67 | 2, n = 169 | 4, n = 114 | ||||
| 3+, n = 53 |
Subset analyses: disease type and in vivo T-cell depletion
Given that most prior studies have found KIR associations with myeloid leukemia, we performed a subgroup analysis looking just at this disease type. As above in the KIR gene content analysis, there was no significant association between DFS and donor AA vs Bx (P = .66), donor centromeric AA vs AB vs BB (P = .13), donor telomeric AA vs AB vs BB (P = .17), donor KIR B content score (P = .32), donor B KIR number (P = .54), alternative B content (P = .17), or the KIR composite score (P = .02) (Table 4). Additionally, there was no significant impact on AML relapse for the various KIR groupings. Similar results were seen in the ALL subgroup (not shown). Lastly, we restricted the analysis to those patients who had in vivo T-cell depletion and similarly negative findings were noted (not shown).
Discussion
This report examined the role of donor KIR on pediatric transplant outcomes for patients with acute leukemia reported to the CIBMTR from 2005 to 2016 with KIR typing that was performed centrally and retrospectively. We examined multiple different published models of KIR gene content and KIR alloreactivity and found that none were significantly associated with OS, DFS, relapse, TRM, or aGVHD. Although similar negative associations have been seen in other populations,8 there are relatively few studies focused on whether donor KIR is associated with transplant outcomes in pediatric recipients.
Examining the association of donor KIR and transplant outcomes in a purely pediatric cohort is relevant because there are significant differences between children and adults that might positively or negatively impact NK-cell alloreactivity. For instance, it is increasingly clear that the drivers of pediatric and adult AML are genetically distinct; children typically have NRAS-, KIT-, KRAS-, and WT1-driven disease, whereas adults commonly have mutations in DNMT3A, NPM1, IDH1/2, RUNX1, and TP53.17 Whether these differences in driver mutations affect NK-cell recognition and cytotoxicity is unknown, but in ALL, it has been speculated that pediatric patients may be more sensitive to NK-cell alloreactivity due to the higher expression of adhesion receptors and ligands for NK-activating receptor on pediatric vs adult ALL blasts.18,19 The presence of a functional thymus in children has been associated with more rapid reconstitution of T cells, which could compete for cytokines and attenuate NK function. Still other differences are that pediatric patients almost always receive myeloablation, whereas reduced-intensity conditioning is commonly used in the adult population. Myeloablative-preparative regimens cause more profound cytopenias, potentially resulting in higher exposure to homeostatic cytokines, such as interleukin 15, known to affect NK-cell function. In contrast, the strongest association with NK-cell alloreactivity has been seen in recipients of reduced-intensity conditioning,12,20 where relapse rates are generally higher, making it easier to identify a statistical association.
Despite being the largest study of donor KIR in the pediatric leukemia population, there are weaknesses with this study, including the retrospective and multicenter registry nature of the data. Due to the relatively low relapse rates in children, it is possible that this study was underpowered. Likewise, we tested multiple models of KIR alloreactivity and KIR gene content. Accordingly, we adjusted our interpretation of significance appropriately. However, some of the models were interdependent (eg, the presence/absence of donor KIR was quantified by different scoring/grouping); thus, whether the adjustment in significance was truly required could be debated. Despite this conservative approach, most statistical tests were clearly not significant, regardless of the level of adjustment based on multiple testing.
Given the prior associations of donor KIR with AML and in the setting of T-cell–depleted haploidentical transplants, we performed subgroup analyses, but were unable to identify significant associations with relapse or DFS. Given these findings, our results do not support the selection of URDs for myeloablative transplantation based on KIR in pediatric patients, as it may lead to a deprioritization of other donor criteria known to be associated with improved outcomes in children (ie, age, sex, parity, cytomegalovirus serostatus).21 Lastly, it is important to stress that this analysis does not address the association of donor KIR with other stem cell sources (umbilical cord blood), nor will it necessarily apply to newer approaches of transplantation and/or cell manipulation, especially those that do not use immune suppression (ie, haploidentical transplantation), or approaches that incorporate the adoptive transfer of donor-derived expanded NK cells.
Data-sharing requests may be e-mailed to the corresponding author, Michael R. Verneris, at michael.verneris@ucdenver.edu.
Authorship
Contribution: M.R.V. designed the study, analyzed data, and wrote the manuscript; J.S.M., K.C.H., S.P., H.R., D.A.L., and S.J.L. assisted in study design and reviewed the data and the manuscript; T.W. and J.A.S. assisted in study design, performed statistical analysis, and reviewed the manuscript; and S.R.S. assisted in study design, facilitated data acquisition, and reviewed the data and the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael R. Verneris, University of Colorado Anschutz Medical Campus, Research Complex 1 North Tower, 12800 East 19th Ave, Mail Stop 8302, Room P18-4108, Denver, CO 80045; e-mail: michael.verneris@cuanschutz.edu.