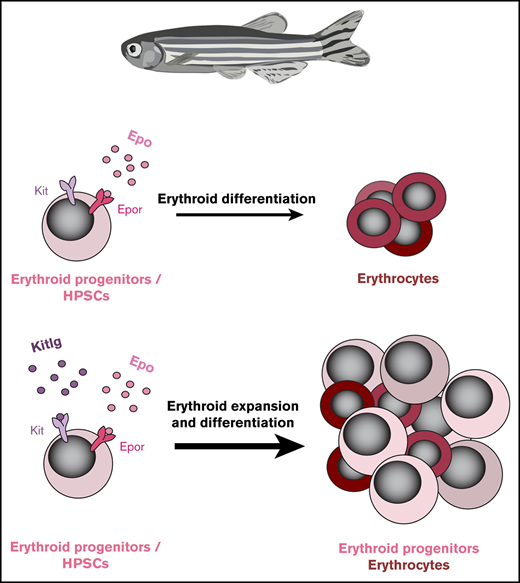
Key Points
Kit signaling contributes to erythroid cell development and is conserved from fish to man.
Ex vivo expansion and self-renewal of zebrafish erythroid progenitors requires addition of recombinant Kitlga.
Abstract
Kit ligand (Kitlg) is pleiotropic cytokine with a prominent role in vertebrate erythropoiesis. Although the role of Kitlg in this process has not been reported in Danio rerio (zebrafish), in the present study we show that its function is evolutionarily conserved. Zebrafish possess 2 copies of Kitlg genes (Kitlga and Kitlgb) as a result of whole-genome duplication. To determine the role of each ligand in zebrafish, we performed a series of ex vivo and in vivo gain- and loss-of-function experiments. First, we tested the biological activity of recombinant Kitlg proteins in suspension culture from zebrafish whole-kidney marrow, and we demonstrate that Kitlga is necessary for expansion of erythroid progenitors ex vivo. To further address the role of kitlga and kitlgb in hematopoietic development in vivo, we performed gain-of-function experiments in zebrafish embryos, showing that both ligands cooperate with erythropoietin (Epo) to promote erythroid cell expansion. Finally, using the kita mutant (kitab5/b5 or sparse), we show that the Kita receptor is crucial for Kitlga/b cooperation with Epo in erythroid cells. In summary, using optimized suspension culture conditions with recombinant cytokines (Epo, Kitlga), we report, for the first time, ex vivo suspension cultures of zebrafish hematopoietic progenitor cells that can serve as an indispensable tool to study normal and aberrant hematopoiesis in zebrafish. Furthermore, we conclude that, although partial functional diversification of Kit ligands has been described in other processes, in erythroid development, both paralogs play a similar role, and their function is evolutionarily conserved.
Introduction
The most prominent cytokines that regulate proliferation and differentiation of erythroid cells in vertebrates are erythropoietin (EPO)1-3 and KIT ligand (KITLG; or stem cell factor).4-9 As was described in other vertebrates, but not in zebrafish, binding of EPO and KITLG to their cognate receptors ensures erythroid lineage commitment by triggering specific signaling events.1,8,10,11
In addition to its role in erythropoiesis, KITLG acts pleiotropically, affecting a wide range of tissues and cells, including hematopoietic stem cells (HSCs) and germ stem cells.12-14 It is an important regulator that plays a role in many processes during ontogenesis and in the adult organism.13,15 In mammals, KIT signaling is associated with erythropoiesis and myelopoiesis,16 as well as with neurogenesis and pigmentation.13,17-19 Interestingly, 2 forms of KITLG occur in vivo: transmembrane (TM), which is important for the regulation of stem cells in their niches, and soluble, which affects more distant tissues.13,20 Binding of KITLG to its receptor KIT, a member of the receptor tyrosine kinase type-III family, leads to its autophosphorylation, further triggering various signaling cascades, including the phosphatidylinositol 3-kinase, MAPK, SRC, and JAK pathways.20,21
In teleost species, which include zebrafish, this scenario is more complex because the whole ligand-receptor signalosome has been duplicated as a result of an extra round of whole-genome duplication. Therefore, future studies are dependent on the understanding of diversification of the functions of both ligands (Kitlga, Kitlgb) and receptors (Kita, Kitb), as well as their binding specificities. Similarly, there are also 2 copies of the epo gene present in the zebrafish genome: epoa and epob. Zebrafish epoa seems to play a role similar to its mammalian ortholog,22,23 but no role for epob has been reported in hematopoiesis (O.S. and P.B., unpublished data, 8 August 2012). For simplicity, we will use the designation Epo/epo for Epoa/epoa in this study.
Current data support the hypothesis that Kit receptor and Kit ligand paralogs have subspecialized during evolution. Kita is expressed in the neural crest, lateral line, and notochord.24 Overexpression of Kitlga results in hyperpigmentation,25 and kita receptor mutants (kitab5/b5 or sparse) have defective pigmentation.24 On the other hand, the second zebrafish Kit paralog, Kitb, is expressed in neural tube and otic vesicles and likely does not play a role in melanogenesis.26,27 Also, a role for Kitlgb has not been reported in melanogenesis.27
Although studied extensively, the role of Kit signaling in zebrafish hematopoiesis has remained unknown for a long time. So far, only 2 studies have suggested potential roles for Kitlg in hematopoiesis in Danio rerio. The first showed a mild increase in HSCs upon overexpression of Kitlgb,28 whereas the second reported a decrease in the number of HSCs upon downregulation of Kitb; however, no such phenotype was observed for Kita or Kitlga. Based on that, the investigators concluded that Kita and Kitlga are not involved in hematopoiesis, but only in melanocyte formation, as suggested by previous studies.27 In contrast to the findings in other vertebrate models, and despite the fact that Kit receptors are expressed in hematopoietic tissues,24,27 hematopoiesis is not affected in adult kita mutants (kitab5/b5 or sparse)24 under steady-state conditions.
The utilization of zebrafish as a model organism requires understanding of the regulatory mechanisms of hematopoietic lineage development and their evolutionary conservation, as well as proper approaches to study blood development and disease. The objective of this study is to better understand the importance of Kit ligands in zebrafish hematopoiesis and, thus, provide important insights for further research of hematopoietic development. Although ex vivo expansion of erythroid progenitors has been reported in different vertebrate organisms,29 a similar approach that would enable analysis of normal and aberrant hematopoiesis in zebrafish is not yet available. Here, we establish suspension culture conditions for the expansion of zebrafish erythroid progenitors and investigate the role of the 2 zebrafish paralogs of Kit ligands (Kitlg’s) in zebrafish hematopoiesis using in vivo and ex vivo experimental approaches.
Methods
Animal stocks and embryos
Fish were mated, raised, and staged according to The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio)30 as well as recommendations for zebrafish husbandry and housing.31 We used a dedicated database solution called Zebrabase, which was developed in-house, to track the animals (age, number, health, genotype).32 Fish were kept in a ZebTEC aquatic system (Tecniplast). Transgenic reporter lines expressing fluorescent genes under the control of tissue-specific promoters (gata1:DsRed33 and lcr:EGFP),34 as well as mutant lines (kitab5/b5 or sparse)24 and wild-type (WT) animals, were used in this study. For ex vivo experiments, 6-month-old fish were used to ensure optimal numbers and states of whole-kidney marrow cells. Animal care and experiments were approved by the Animal Care Committee of the Institute of Molecular Genetics, Czech Academy of Sciences (13/2016 and 96/2018) in compliance with national and institutional guidelines.
Ex vivo cultures
Zebrafish whole-kidney marrow cells were isolated as previously described.35 Cells were fractionated using Biocoll (1.077 g/mL; Merck L6115) density centrifugation and initially seeded at 3 × 106 cells per milliliter and cultivated in zfS13 medium (supplemental Table 3) at 32°C and 5% CO2. Carp serum was obtained as described previously.35 Specific cytokines (Epo, Gcsfa, Kitlga, and Kitlgb) were added at final concentrations of 100 ng/mL, and dexamethasone (Dex) was added at a final concentration of 1 µM. Cells were counted using a CASY Cell Counter and Analyzer (OMNI Life Science). In the following days, cells were maintained at 2 × 106 cells per milliliter, and one third of the medium containing fresh cytokines and Dex was exchanged every other day to ensure optimal growth of the cells. To study the clonogenic potential of the whole-kidney marrow cells after the addition of different cytokines, we performed clonal assays in semisolid media (methylcellulose), which prevents movement of the single cells plated in it. The detailed protocol for these assays was described previously.35
Cytokine cloning and expression
First, the amino acid sequence of zebrafish Kitlga/b was subjected to protein structure prediction and hydrophobicity analysis using Phobius (http://phobius.sbc.su.se/). Two large hydrophobic regions (aa 1-24 and 206-224 for Kitlga and aa 1-31 and 185-209 for Kitlgb) corresponding to the putative signal peptide (SP) and TM domains, respectively, were identified (supplemental Figure 1A). To generate a Kitlga/b version devoid of both domains, sequence-specific primers (supplemental Table 1) were used for polymerase chain reaction (PCR) amplification of each Kitlg complementary DNA (cDNA) fragment corresponding to aa 25 to 182 (Kitlga) and aa 31 to 187 (Kitlgb) from adult zebrafish retina. We used the baculovirus expression system to produce soluble Kitlga/b in large quantities. We cloned the amplified fragment into a modified pAc-GP67-B vector containing 6xHis and generated the recombinant baculovirus by cotransfection of pAc-His-Kitlga/b and BD BaculoGold Bright Baculovirus DNA into sf21 insect cells. Virus-infected cells expressed GFP (supplemental Figure 1B) and secrete recombinant His-Kitlga/b extracellularly. Finally, we purified the secreted proteins on an Ni2+-NTA agarose column (supplemental Figure 1C) and used them for the cell culture experiments. To generate Kitlga/b, the construct for messenger RNA (mRNA) injection experiments, the full-length cDNA fragment was amplified using reverse transcription PCR and sequence-specific primers (supplemental Table 1). EPO protein and mRNA were prepared as described previously.22,35,36
Additional methods
For details about the generation of recombinant cytokines, mRNA microinjection, benzidine staining, image analysis, quantitative PCR (qPCR), fluorescence-activated cell sorting (FACS), RNA sequencing (RNAseq) and transcriptomics, please refer to supplemental Methods.
Results
Kit ligands promote erythroid and myeloid expansion of whole-kidney marrow cells
First, we cloned and expressed recombinant zebrafish Kitlga and Kitlgb. We amplified the mature form of zebrafish kitlga and kitlgb lacking the SP-encoding region, intracellular region, and TM region (supplemental Figure 1A) from adult retina, and we produced recombinant Kitlga and Kitlgb in sf21 insect cells (supplemental Figure 1B-C). His-tagged purified proteins were used in the following experiments.
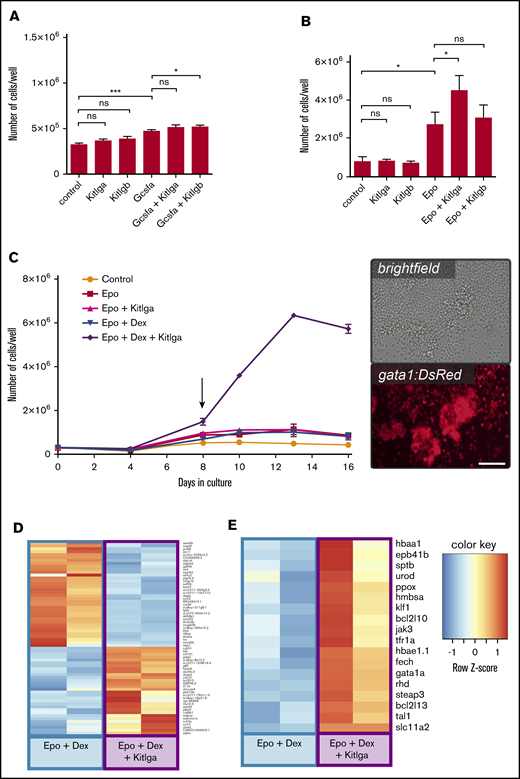
To test the biological activity of Kitlg proteins, we designed an experiment using whole-kidney marrow cells isolated from adult 6-month-old zebrafish. To reveal differences that would suggest biological activity and potential cooperation of Kitlg proteins with other recombinant cytokines, we treated the cells with various factors, or their combination, and counted them at specific time points. After 3 days in culture, potential enhancement of the myeloid lineage was assessed. We added both Kit ligands, either separately or combined with Gcsfa, which has been shown to support myeloid cell fate.37 Interestingly, we observed an increase in the number of cells in all of the conditions tested compared with untreated control (Figure 1A). In agreement with previously published studies, Gcsfa significantly promoted the growth of whole-kidney marrow cells, and the effect was further potentiated by the addition of either Kit ligand. This additional effect tended to be similar for both Kit ligand paralogs; however, the cooperation proved to be statistically significant only for Kitlgb. The Kit ligands alone exhibited reproducible mild activity that was not statistically significant. To verify that the addition of Gcsfa together with Kitlga or Kitlgb promoted myelopoiesis, we characterized cultured cells morphologically (supplemental Figure 2A, upper panels). At day 3, the culture primarily consisted of monocytes and macrophages. Although the addition of Kitlga or Kitlgb to Gcsfa increases the cumulative number of cells (as shown in Figure 1A), the overall composition of the cell culture and the proportions of different cell types remained similar, with or without the Kit ligands.
The effect of zebrafish Kitlga and Kitlgb on self-renewal and proliferation of hematopoietic cells ex vivo. Quantification of the number of whole-kidney marrow cells after treatment with phosphate-buffered saline (control) or specific cytokine combinations for 3 days (A) and 7 days (B). (C) Growth curve for ex vivo culture of whole-kidney marrow cells with different cytokine combinations. Biological duplicates for every condition and time point are shown. Representative photomicrographs (right panel) of whole-kidney marrow cells isolated from transgenic gata1:DsRed fish treated with Epo, Dex, and Kitlga and cultivated for 8 days (see arrow). Images were acquired using an Olympus IX70 inverted microscope equipped with an Olympus DP72 camera. Scale bar, 100 μm. Expression heat map of Epo + Dex–treated and Epo + Dex + Kitlga–treated cells showing the top predicted differentially expressed genes (D) and representative markers of erythroid differentiation (E). *P ≤ .05, ***P ≤ .001, standard 1-way analysis of variance. n.s., not significant (P > .05).
The effect of zebrafish Kitlga and Kitlgb on self-renewal and proliferation of hematopoietic cells ex vivo. Quantification of the number of whole-kidney marrow cells after treatment with phosphate-buffered saline (control) or specific cytokine combinations for 3 days (A) and 7 days (B). (C) Growth curve for ex vivo culture of whole-kidney marrow cells with different cytokine combinations. Biological duplicates for every condition and time point are shown. Representative photomicrographs (right panel) of whole-kidney marrow cells isolated from transgenic gata1:DsRed fish treated with Epo, Dex, and Kitlga and cultivated for 8 days (see arrow). Images were acquired using an Olympus IX70 inverted microscope equipped with an Olympus DP72 camera. Scale bar, 100 μm. Expression heat map of Epo + Dex–treated and Epo + Dex + Kitlga–treated cells showing the top predicted differentially expressed genes (D) and representative markers of erythroid differentiation (E). *P ≤ .05, ***P ≤ .001, standard 1-way analysis of variance. n.s., not significant (P > .05).
Next, we tested the activity of Kit ligands during erythroid cell expansion. Cells were treated with Kitlga and Kitlgb, either separately or combined with Epo. The increase in cell number was quantified after 7 days in culture. In agreement with previously published studies,22,36 Epo boosted cell growth significantly and, again, there was a mild increase in the number of the cells when treated with either Kit ligand alone. Strikingly, when Epo was combined with Kitlga, we noticed a significant increase in the number of cells at 7 days in culture compared with Epo alone (Figure 1B). Although the additive effect of Kitlgb was not significant, the trend toward an increase in the number of cells in the presence of Epo was still observed. To examine the composition of the cell culture, we further performed morphological characterization of cultured cells. At day 7, the culture was primarily composed of erythroid cells upon Epo stimulation, as expected (supplemental Figure 2A, lower panels), and the addition of Kitlga led to an increase in the number of erythroid progenitors, which explains the elevated number of cells in this culture.
With the aim of maximally expanding erythroid progenitors, we decided to improve the suspension culture conditions for whole-kidney marrow cells. Although there were no existing reports of suspension cultures of zebrafish or hematopoietic progenitor cells, we theorized that similar conditions as used for humans and mice might be effective.10,38 Because different steroids were reported to be important players in the maintenance of self-renewal and the proliferation of erythroid progenitors in chickens, mice, and humans,38-41 we included Dex and tested its different concentrations and combinations together with Epo and Kitlga, the more potent Kitlg paralog.
We found that, in line with previous findings in other vertebrates, Kitlga, Epo, and Dex act synergistically in ex vivo cultures, enabling erythroid expansion (Figure 1C, left panel). Compared with the cells treated with Epo and Dex, cells treated with Epo, Dex, and Kitlga formed islets of round highly gata1:DsRed+ cells, corresponding to erythroid progenitors (Figure 1C, right panel). Additionally, we observed that Epo can cooperate with Kitlga to promote the growth of large erythroid colonies in colony-forming assays in methylcellulose (supplemental Figure 2B). When Kitlga was combined with Epo in kidney marrow culture from gata1:DsRed-transgenic fish, we observed significantly more highly gata1:DsRed+ colonies that were also significantly larger (supplemental Figure 2C-D).
Kitlga treatment, when combined with Epo and Dex, leads to upregulation of erythroid-, translation-, and cell cycle–related gene expression
To gain a deeper insight into the mechanism of Kitlga action when combined with Epo and Dex in kidney marrow suspension cultures, we performed an RNAseq experiment comparing samples from Epo-, Dex-, and Kitlga-treated cells and Epo and Dex–treated cells. Samples were collected after 8 days in culture. This was the first time point at which we could observe a significant increase in the proliferation of erythroid cells in the presence of Epo, Dex, and Kitlga, as shown in the growth curve (Figure 1C, left panel).
RNAseq analysis revealed that both culture conditions (Epo+Dex, Epo+Dex+Kitlga) resulted in high expression of erythroid marker genes (data not shown). However, addition of Kitlga to the combination of Epo and Dex caused an additional increase in the expression of erythroid genes. Among the differentially upregulated genes (see heat map in Figure 1D; supplemental Table 4), we identified several erythroid-specific (g6fl, lias) and cell cycle–specific (cdc27) genes, confirming additional Kitlga-mediated erythroid cell expansion over Epo and Dex treatment alone. On the contrary, downregulated genes were primarily linked to the myeloid cell lineage (pigrl2.3, blnk, nfkbiz, alox5a, inpp5d), providing further evidence that Epo, Dex, and Kitlga–induced expansion led to a more enriched erythroid cell population compared with Epo and Dex culture alone. Next, we examined the expression of both kit receptors and revealed that the expression of kita is predominant in adult erythroid progenitor cells (average raw counts 285 [kita] vs 3 [kitb]).
Finally, we were specifically interested in changes in the expression of erythroid-specific genes. The analysis of RNAseq data revealed that, compared with cells treated with Epo and Dex, cells treated with Epo, Dex, and Kitlga exhibit higher expression of many of the important erythroid genes (ie, globins [hbae1.1, hbaa1]), as well as genes involved in heme and iron metabolism (eg, urod, slc11a2, steap3), erythroid cytoskeleton (eg, rhd, sptb, eph41b), and signaling (gata1a, jak3) (Figure 1E; supplemental Table 5). Based on Gene Ontology (GO) prediction, we identified an enrichment in specific categories of biological processes: translation (GO:0006412, GO:0002181, GO:0000028, GO:0000027), erythropoiesis (GO:0042541, GO:0030218, GO:0048821), and cell cycle regulation (GO:0051726). For a full list of enriched GO terms in all 3 standard GO term categories, refer to supplemental Table 6.
Both Kit ligands cooperate with Epo to promote erythroid expansion in zebrafish embryos
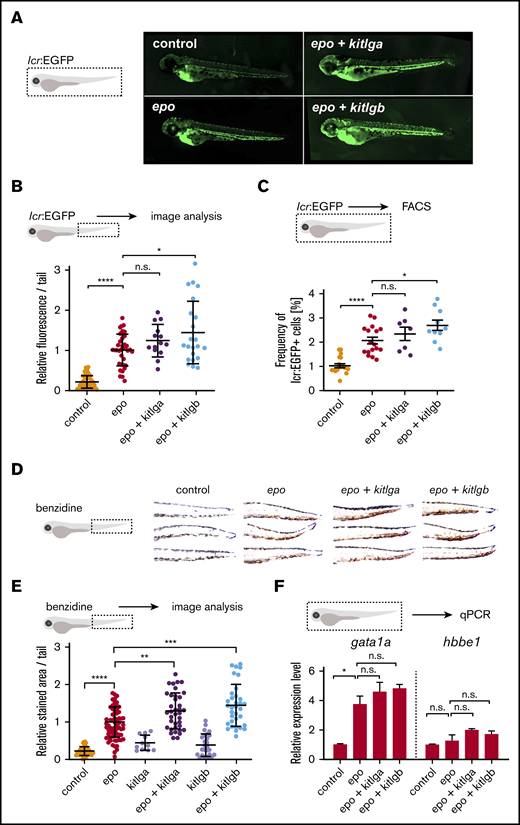
To test the function of both Kit ligands in vivo, we microinjected cytokine mRNA(s) into 1-cell embryos. When combined with epo, kitlga and kitlgb showed an enhancement in lcr:EGFP expression (Figure 2A) and hemoglobinization (Figure 2D) at 72 hours postfertilization (hpf) compared with epo alone. Quantification of lcr:EGFP+ cells in the tail region (Figure 2B), FACS analysis of total lcr:EGFP+ cells (Figure 2C; supplemental Figure 3), and benzidine staining in the tails (Figure 2E) of 72-hpf embryos confirmed the observed trend. Of note, although a similar trend toward an increase in the number of erythroid cells, in combination with epo, was observed for both Kit ligands under all 3 experimental readouts, the effect was more robust and statistically significant only for kitlgb in all types of experiments. Next, qPCR analysis from whole injected embryos also showed an increase in the expression of erythroid markers at 72 hpf (Figure 2F); however, when epo was combined with either of the ligands, these changes were rather subtle. Finally, in agreement with previous studies, we observed an increased number of melanocytes after the overexpression of Kitlga, but not Kitlgb.
In vivo effects of kitlga and kitlgb at 72 hpf. (A) Representative photomicrographs of lcr:EGFP-transgenic reporter larvae at 72 hpf after injection of corresponding mRNAs. Control represents uninjected larvae. Images were acquired using a Zeiss Axio Zoom.V16 with Zeiss Axiocam 506 mono microscope camera and ZEN (blue edition) software. (B) Quantification of lcr:EGFP+ cells in embryonic tails from fish in panel A. Values were plotted relative to the mean of epo (relative expression level 1). (C) FACS analysis of lcr:EGFP+ cells in whole embryos. (D) Representative photomicrographs of benzidine-stained embryonic tails at 72 hpf after injection of corresponding mRNAs. Control represents uninjected embryos. Note the increase in pigmentation in the dorsal part of kitlga-injected embryos. Z-stacks were acquired using a Zeiss Axio Zoom.V16 with aa Zeiss Axiocam 105 color camera (total magnification ×125). Images were processed using the Extended Depth of Focus module in ZEN (blue edition) software. (E) Quantification of the benzidine-stained embryonic tails in (D). Values were plotted relative to the mean of epo (relative expression level 1). (F) qPCR expression analysis of gata1a and hbbe1 from whole injected embryos at 72 hpf. Data were normalized using mob4 as the housekeeping gene and plotted relative to the mean of uninjected control (relative expression level 1). Bars represent a mean of 3 triplicates with standard deviation. *P ≤ .05, **P ≤ .01, ***P ≤ .001, ****P ≤ .0001, standard 1-way analysis of variance. n.s., not significant (P > .05).
In vivo effects of kitlga and kitlgb at 72 hpf. (A) Representative photomicrographs of lcr:EGFP-transgenic reporter larvae at 72 hpf after injection of corresponding mRNAs. Control represents uninjected larvae. Images were acquired using a Zeiss Axio Zoom.V16 with Zeiss Axiocam 506 mono microscope camera and ZEN (blue edition) software. (B) Quantification of lcr:EGFP+ cells in embryonic tails from fish in panel A. Values were plotted relative to the mean of epo (relative expression level 1). (C) FACS analysis of lcr:EGFP+ cells in whole embryos. (D) Representative photomicrographs of benzidine-stained embryonic tails at 72 hpf after injection of corresponding mRNAs. Control represents uninjected embryos. Note the increase in pigmentation in the dorsal part of kitlga-injected embryos. Z-stacks were acquired using a Zeiss Axio Zoom.V16 with aa Zeiss Axiocam 105 color camera (total magnification ×125). Images were processed using the Extended Depth of Focus module in ZEN (blue edition) software. (E) Quantification of the benzidine-stained embryonic tails in (D). Values were plotted relative to the mean of epo (relative expression level 1). (F) qPCR expression analysis of gata1a and hbbe1 from whole injected embryos at 72 hpf. Data were normalized using mob4 as the housekeeping gene and plotted relative to the mean of uninjected control (relative expression level 1). Bars represent a mean of 3 triplicates with standard deviation. *P ≤ .05, **P ≤ .01, ***P ≤ .001, ****P ≤ .0001, standard 1-way analysis of variance. n.s., not significant (P > .05).
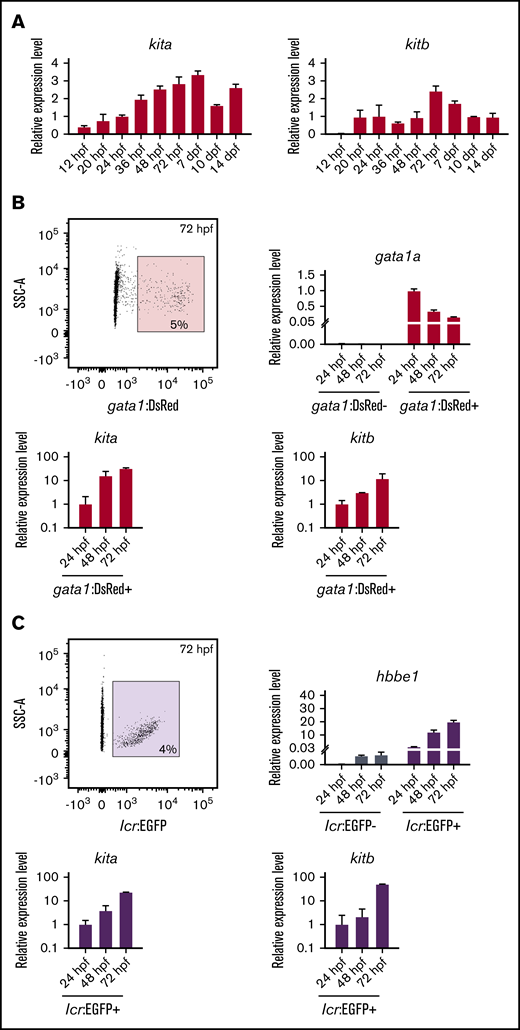
The expression of both kit receptors gradually increases during embryonic development
To better understand the mechanism of cooperation between Epo and Kit ligands, we first analyzed the expression of both kit receptors in tissues and during development using qPCR. Among all of the tissues analyzed, the highest kita expression was observed in kidney (supplemental Figure 4A), whereas the highest kitb expression was observed in the retina (supplemental Figure 4B). During zebrafish embryonic development, the highest relative expression was observed at 7 days postfertilization for kita and at 72 hpf for kitb (Figure 3A). Because we saw significant expansion of erythroid gata1:DsRed+ cells in ex vivo cultures, we aimed to analyze the dynamics of receptor expression in erythroid cells during development. Therefore, we collected samples from 24-hpf, 48-hpf, and 72-hpf gata1:DsRed and lcr:EGFP embryos and FACS-sorted gata1:DsRed+ cells (Figure 3B) or lcr:EGFP+ cells (Figure 3C). In both cases, we found a gradual increase in the expression of both receptors in erythroid cells during the course of early development.
Expression of kit receptors in the zebrafish embryonic development. (A) qPCR expression analysis of kita (left panel) and kitb (right panel) at different developmental stages. Data were normalized using ef1a as the housekeeping gene and relative to the mean of 24-hpf sample (relative expression level 1). (B) Sorting strategy used for FACS isolation of gata1:DsRed+ cells (upper left panel) from transgenic zebrafish at 24 hpf. qPCR expression analysis of gata1 (upper right panel), kita (lower left panel), and kitb (lower right panel) in gata1:DsRed− and gata1:DsRed+ cells. Data were normalized using ef1a as the housekeeping gene and relative to the mean of gata1:DsRed+ 24-hpf sample (relative expression level 1). (C) Sorting strategy used for FACS isolation of lcr:EGFP+ cells (upper left panel) from transgenic zebrafish at 24 hpf. qPCR expression analysis of gata1 (upper right panel), kita (lower left panel), and kitb (lower right panel) in lcr:EGFP− and lcr:EGFP+ cells. Data were normalized using ef1a as the housekeeping gene and relative to the mean of lcr:EGFP+ 24-hpf sample (relative expression level 1).
Expression of kit receptors in the zebrafish embryonic development. (A) qPCR expression analysis of kita (left panel) and kitb (right panel) at different developmental stages. Data were normalized using ef1a as the housekeeping gene and relative to the mean of 24-hpf sample (relative expression level 1). (B) Sorting strategy used for FACS isolation of gata1:DsRed+ cells (upper left panel) from transgenic zebrafish at 24 hpf. qPCR expression analysis of gata1 (upper right panel), kita (lower left panel), and kitb (lower right panel) in gata1:DsRed− and gata1:DsRed+ cells. Data were normalized using ef1a as the housekeeping gene and relative to the mean of gata1:DsRed+ 24-hpf sample (relative expression level 1). (C) Sorting strategy used for FACS isolation of lcr:EGFP+ cells (upper left panel) from transgenic zebrafish at 24 hpf. qPCR expression analysis of gata1 (upper right panel), kita (lower left panel), and kitb (lower right panel) in lcr:EGFP− and lcr:EGFP+ cells. Data were normalized using ef1a as the housekeeping gene and relative to the mean of lcr:EGFP+ 24-hpf sample (relative expression level 1).
Kita mediates Kitlg-dependent erythroid expansion in vivo
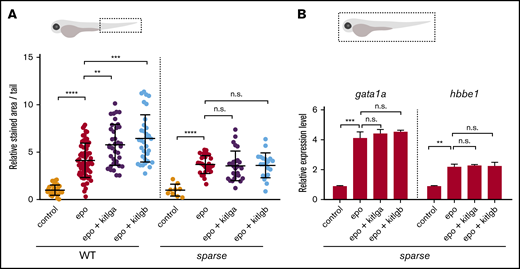
To understand the molecular basis of the effect that we observed on erythroid cells, we used the sparse-mutant (kitab5/b5) zebrafish lacking the functional Kita receptor. We injected mRNA for epo, kitlga, and kitlgb. We observed that, in sparse mutants, the number of erythrocytes is slightly decreased in epo-injected embryos, and the effect of cooperation between either of the kit ligands and epo is completely lost (Figure 4A). These findings are also supported by qPCR data showing that the expression of β-globin (hbbe1) and gata1a is not increased after the injection of epo with either of the kit ligands, as is the case in WT embryos (Figure 4B).
The effect of kitlga and kitlgb in sparse mutants. (A) Quantification of benzidine staining in the tail region (dashed box) of control (uninjected) embryos or WT or kita-mutant embryos (sparse) injected with epo mRNA, alone or in combination with kitlga or kitlgb mRNA. Values are plotted relative to the mean of epo (=1). (B) Expression of gata1a and hbbe1 from whole (dashed box) injected kita-mutant embryos (sparse) at 72 hpf, as measured by qPCR. Data were normalized using mob4 as the housekeeping gene and plotted relative to the mean of control sample (relative expression level 1). Bars represent the mean of 3 triplicates with standard deviation. **P ≤ .01, ***P ≤ .001, ****P ≤ .0001, standard 1-way analysis of variance. n.s., not significant (P > .05).
The effect of kitlga and kitlgb in sparse mutants. (A) Quantification of benzidine staining in the tail region (dashed box) of control (uninjected) embryos or WT or kita-mutant embryos (sparse) injected with epo mRNA, alone or in combination with kitlga or kitlgb mRNA. Values are plotted relative to the mean of epo (=1). (B) Expression of gata1a and hbbe1 from whole (dashed box) injected kita-mutant embryos (sparse) at 72 hpf, as measured by qPCR. Data were normalized using mob4 as the housekeeping gene and plotted relative to the mean of control sample (relative expression level 1). Bars represent the mean of 3 triplicates with standard deviation. **P ≤ .01, ***P ≤ .001, ****P ≤ .0001, standard 1-way analysis of variance. n.s., not significant (P > .05).
To summarize, we have cloned and expressed both Kit ligand zebrafish paralogs. Using the purified recombinant proteins, we tested various cytokine combinations in suspension cultures of whole kidney marrow cells and defined an optimal factor combination, composed of Epo, Kitlga, and Dex, which enables efficient expansion of erythroid progenitors. Also, we tested the gain-of-function effect of kitlga and kitlgb in embryonic hematopoiesis and observed an increase in the number of erythroid cells in epo mRNA–, kitlgb mRNA–, and epo and kitlgb mRNA–injected fish compared with epo alone. Finally, by using the kita (sparse) mutants, we showed that Kita mediates Kitlg-dependent erythroid expansion in vivo.
Discussion
In this study, we explored the role of zebrafish Kit ligands in hematopoiesis. In zebrafish, multiple paralogs of many important genes, including hematopoietic cytokines, resulted from an extra round of whole-genome duplication 250 to 350 million years ago.42,43 After such an event, the process of pseudogenization often leads to a loss of function of 1 of the paralogs.44-46 Alternatively, paralogs of an ancestral gene can remain functionally redundant, split the original function between the 2 (subfunctionalization), or acquire completely new functions (neofunctionalization).47 Here, we studied the hematopoietic function of 2 retained zebrafish paralogs of a crucial hematopoietic cytokine: the Kit ligand (KITL or stem cell factor), designated as Kitlga and Kitlgb.
So far, KIT signaling has not been associated with erythropoiesis in zebrafish.24,25 This is in contrast with previous studies in humans,8 mice,4 and chickens,9,16,48 where it has been shown that KITL is critical for proper erythroid development.5,15,49 By focusing on Kit function during erythropoiesis in zebrafish, we were able to prove for the first time, using ex vivo and in vivo gain- and loss-of-function experiments, that Kit signaling does indeed play a role in zebrafish erythroid differentiation. Based on RNAseq and GO analysis, this likely happens in a similar manner as in other vertebrate species via induction of cell proliferation coupled with differentiation,40,48 promoting cell cycle progression and enhancing protein translation in differentiating cells.
To examine the role of Kit ligands ex vivo, we used clonal assays and liquid culture experiments.23,36,37,50 Because the reported cross-reactivity of mammalian hematopoietic cytokines in zebrafish is very limited,35 it was essential to use bona fide zebrafish Kit ligands as recombinant proteins. As described for other cytokines,7,9,35,36 we cloned the extracellular portion of the corresponding cDNAs, lacking the N-terminal SP, TM domain, and cytosolic part of both cytokines. This approach allowed us to generate the soluble form of Kit ligands to stimulate kidney marrow progenitor cells ex vivo.8,13
As a result, treatment of hematopoietic progenitor cells isolated from kidney marrow with Kitlga, in combination with the other master regulator of erythroid lineage commitment, Epo,22,23 enabled erythroid expansion of cells in suspension cultures and clonal assays, as in other vertebrates.1,8,10,11 Such an effect was also observed for Kitlgb; however, its cooperation with Epo was less pronounced and was not statistically significant. We hypothesize that the soluble form of Kitlgb that lacks the TM and cytosolic domains is less effective in stimulating the differentiation of hematopoietic progenitors toward the erythroid lineage and that the main role of Kitlgb is stimulation of neighboring cells in stem cell niches via contact-dependent signaling.
Next, we also tested the activity of Dex in addition to Epo and Kitlga, the more effective soluble Kitlg paralog. Dex is a glucocorticoid receptor agonist that has been reported to support self-renewal and differentiation of erythroid progenitors10,38-40 ; indeed, addition of Dex to Kitlga and Epo led to synergistic erythroid expansion in suspension cultures.
We performed an RNAseq experiment to understand the mechanism triggering this substantial cell expansion. We decided to compare the effects of Epo and Dex vs Epo, Dex, and Kitlga on adult kidney marrow cells. Transcriptional profiling showed expression of erythroid-specific genes in both conditions, which was expected because of Epo-mediated signaling. However, we observed relatively subtle changes in expression between the Epo and Dex and the Epo, Dex, and Kitlga cultures. The trends in expression changes showed a distinct erythroid fingerprint in cells treated with Epo, Dex, and Kitlga compared with a more mixed/myeloid one in cells treated with Epo and Dex alone, as suggested by the increased expression of myeloid-specific genes. In addition, we found that kita is the prominent receptor expressed in adult erythroid progenitor cells, indicating that the effect of erythroid expansion is mediated by this receptor. This corresponds to the fact that isolation of kidney marrow cells yields, in addition to progenitor cells, a fraction of myeloid cells with the ability to survive in the culture. However, this effect becomes negligible during erythroid expansion of cells treated with Epo, Dex, and Kitlga, when myeloid cells are overgrown by expanding erythroid progenitors. Importantly, GO analysis revealed an enrichment, especially in the translation and expression of erythroid-specific genes, in Epo, Dex, and Kitlga cultures. Therefore, we hypothesize that Kitlga might enhance erythroid cell development and cell proliferation potentially through upregulation of genes related to protein translation and cell cycle progression.
To confirm these findings, we decided to test whether Kit signaling is also involved in zebrafish erythropoiesis in vivo using the full-length forms of both kit ligands. We used soluble forms for ex vivo experiments, whereas we used the full-length coding sequence, including the TM and cytosolic domains, as used previously,27 for in vivo experiments. It has been shown that the functions of soluble and TM forms under physiological conditions are different.13 The soluble form of the protein is distributed throughout the entire organism via circulation and affects distant tissues. It has been shown that this form is required and sufficient for proper erythroid signaling in cultured cells.16 On the other hand, according to previous studies, the major site of action of the TM form of Kit ligand might be in stem cell niches.13 We hypothesize that full-length Kitlgb can contribute, in addition to hematopoietic stem and progenitor cells,27 to erythroid expansion in vivo. As a result, we found that full-length kitlga was able to expand erythroid cells at 72 hpf when injected together with epo mRNA. Interestingly, similarly to kitlga, kitlgb also was able to mediate such erythroid expansion when injected in vivo in full-length form; however, this effect was markedly decreased in ex vivo experiments using the short soluble form. This supports our hypothesis that there is a difference in potency between the soluble and full-length forms of Kitlgb. To prove this, we generated shortened kitlga and kitlgb mRNA, corresponding to the soluble region of Kitlg proteins; indeed, there was a decrease in the ability of soluble kitlgb to stimulate erythropoiesis (data not shown), whereas soluble kitlga remained equally effective.
Finally, we investigated whether the Kita receptor is dispensable in Kitlg-mediated erythroid cell expansion in vivo. As a result, we did not observe cooperation between epo and either of the kit ligands in erythroid differentiation in kita (sparse)–mutant fish. These data suggest that Kita is responsible for Kit signal transduction in erythroid cells. Although Kitb might also be involved in this process, it could not compensate for missing Kita, and Kita seems to be required for Kit signaling in zebrafish erythroid cells. In addition to this, we observed decreased numbers of erythroid cells in control and in epo-injected sparse mutants compared with WT embryos. This indicates that endogenous Kit signaling also plays a role under physiological conditions during normal erythropoiesis.
To summarize, in this study we have addressed the role of Kitlg in hematopoiesis in zebrafish using in vivo and ex vivo approaches. Importantly, we established optimized liquid culture conditions for erythroid expansion of kidney marrow progenitor cells that require the addition of Kitlga. Together with colony-forming assays, suspension cultures might serve as indispensable tools in disease modeling, allowing analysis of normal and aberrant hematopoiesis in zebrafish blood mutants (eg, vlade, moonshine, mindbomb).51 This will enable further biochemical, genomic, and proteomic analyses of ex vivo–cultured and expanded cells.
Interestingly, zebrafish Kitlga and Kitlgb seem to possess only partial redundancy in their function: Kitlga retains a role in melanogenesis and pigmentation, which is lost in the case of Kitlgb, but both ligands potentially contribute to the overall process of erythroid commitment and development in vivo, which is not required in steady-state erythroid development, but it might become important in stress conditions. The exact mechanism of functional diversification of Kit ligands has yet to be investigated; however, altogether, we demonstrate that the role of Kit signaling is evolutionarily conserved with respect to erythroid development, although previously unnoticed.24 Therefore, it strengthens the use of zebrafish as a model to study normal and aberrant human hematopoiesis.
RNAseq data have been deposited in the ArrayExpress Microarray Database under accession number E-MTAB-8800. Plasmids for cytokine expression are available from Addgene under accession numbers 140292 (pAc-His-zfKitlga) and 140293 (pAc-His-zfKitlgb).
Data sharing requests should be sent to Petr Bartunek (bartunek@img.cas.cz).
Acknowledgments
The authors thank Nikol Pavlu, Tereza Hojerova, and Tereza Hingarova for animal care; Tereza Mikulasova and Martina Hason for creating vector graphics and providing cDNA samples; Trevor Epp for editing the manuscript; and Leonard Zon for providing gata1:DsRed and lcr:EGFP reporter fish lines.
This work was supported by the Czech Science Foundation (16-21024S), the Ministry of Health (NV19-07-00412), and the Czech Academy of Sciences (68378050-KAV-NPUI) (P.B.). O.S. was partially funded by the American Heart Association (19POST34380328).
Authorship
Contribution: J.O., O.M., and O.S. performed experiments; and J.O., O.S., P.S., D.T., M.K., and P.B. designed the experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Petr Bartunek, Institute of Molecular Genetics of the Czech Academy of Sciences, Videnska 1083, 142 20 Prague 4, Czech Republic; e-mail: bartunek@img.cas.cz.
References
Author notes
The full-text version of this article contains a data supplement.