Key Points
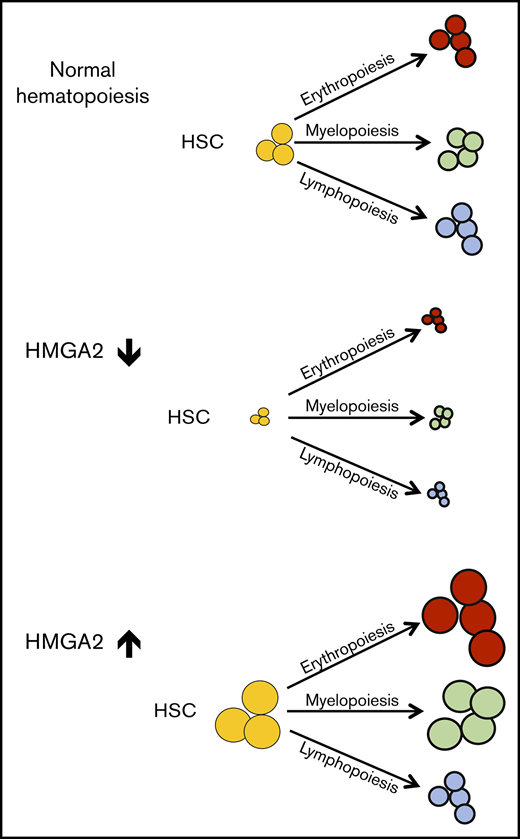
HMGA2 is required to sustain human HSCs both in culture and during regeneration in vivo.
Overexpression of HMGA2 triggers increased human myeloerythroid reconstitution in xenograft transplantation assays.
Abstract
Identification of determinants of fate choices in hematopoietic stem cells (HSCs) is essential to improve the clinical use of HSCs and to enhance our understanding of the biology of normal and malignant hematopoiesis. Here, we show that high-mobility group AT hook 2 (HMGA2), a nonhistone chromosomal-binding protein, is highly and preferentially expressed in HSCs and in the most immature progenitor cell subset of fetal, neonatal, and adult human hematopoiesis. Knockdown of HMGA2 by short hairpin RNA impaired the long-term hematopoietic reconstitution of cord blood (CB)–derived CB CD34+ cells. Conversely, overexpression of HMGA2 in CB CD34+ cells led to overall enhanced reconstitution in serial transplantation assays accompanied by a skewing toward the myeloerythroid lineages. RNA-sequencing analysis showed that enforced HMGA2 expression in CD34+ cells induced gene-expression signatures associated with differentiation toward megakaryocyte-erythroid and myeloid lineages, as well as signatures associated with growth and survival, which at the protein level were coupled with strong activation of AKT. Taken together, our findings demonstrate a key role of HMGA2 in regulation of both proliferation and differentiation of human HSPCs.
Introduction
The hierarchical organization of the blood system consists of stem and progenitor cells, which generate differentiated blood cells throughout the life of an organism during homoeostasis and stress. Regulation of gene-expression programs through DNA-binding factors such as transcription factors, coregulators, and epigenetic regulators, has been shown to be critical in determining self-renewal and differentiation of hematopoietic stem and progenitor cells (HSPCs).1 High-mobility group AT hook 2 (HMGA2), a member of the family of high-mobility group AT hook proteins, is a nonhistone chromatin-associated factor. Although HMGA proteins are devoid of intrinsic transcriptional activities, these factors can influence gene expression either by changing the chromatin conformation or by recruiting other factors to the transcriptional complexes near gene promoters or in enhancer regions.2 In recent years, several studies in mice have demonstrated a role of Hmga2 in regulation of somatic stem and progenitor cells from various tissues. For example, during myogenesis, expression of Hmga2 messenger RNA (mRNA) is activated in proliferating satellite cells and negatively correlates with myoblast differentiation.3 Another study identified Hmga2 as a factor required for self-renewal of the neural stem cell in young but not in old mice, highlighting a developmental stage–specific role in regulating stem cell functions.4 In the mouse hematopoietic system, overexpression of Hmga2 has been shown to lead to a clonal expansion of hematopoietic stem cells (HSCs) in the bone marrow (BM) and subsequent development of myeloproliferative-like disease.5 Copley et al further showed that Hmga2 confers the enhanced self-renewal seen in HSCs during fetal liver hematopoiesis.6 Similarly, it has been shown that fetal-specific erythroid-dominant hematopoiesis is dependent on Hmga2.7 Moreover, Hmga2 was identified as a direct target of Runx1 that led to myeloid cell expansion in the context of Runx1 deficiency.8 Together, these studies point to a direct functional role of Hmga2 in regulation of stem and progenitor cells in mice and provide an inviting prospect to further elucidate its role in human hematopoiesis. Yet, to date, no functional studies describing the role of HMGA2 in human HSPCs have been described.
Here, using both gain-of function and loss-of-function approaches, we demonstrate a key role of HMGA2 in regulating renewal and differentiation of human HSPCs in vitro and in vivo.
Materials and methods
Human cord blood CD34 cell isolation
Umbilical cord blood (CB) samples were obtained from full-term newborns (Skåne University Hospital and Helsingborg Hospitals) and normal BM samples were obtained from healthy volunteers (aged 20-30 years), with informed consent according to guidelines approved by the regional ethical committee. Ficoll-Paque–purified week 16 human fetal liver mononuclear cells were obtained from Novogenix Laboratories. CB and BM cells were collected in Dulbecco's modified Eagle medium supplemented with 2% fetal calf serum (FCS; Invitrogen), penicillin/streptomycin (Invitrogen), and heparin (20 U/mL; Leo Pharmaceutics). Mononuclear cells were separated on a Ficoll-density gradient (AXIS-Shield PoC AS), pooled, and enriched for CD34+ cells with anti-CD34 magnetic beads (Miltenyi Biotec). Cells were subsequently frozen in Dulbecco's modified Eagle medium supplemented with 20% FCS and 10% dimethyl sulfoxide (Sigma-Aldrich). For sorting of HSCs, human CB CD34+ cells were thawed and resuspended in phosphate-buffered saline with 2% FCS and stained for cell surface markers. Cells were sorted using Becton Dickinson (BD) FACSAria. The following anti-human antibodies were used for sorting: CD34–fluorescein isothiocyanate (BD), CD38-phycoerythrin (BD), CD90-allophycocyanin (BioLegend), CD45RA-V450 (BD).
Cell culture, lentivirus transduction, and transplantation
CB CD34+ cells were cultured in serum-free expansion medium (StemCell Technologies) supplemented with stem cell factor, thrombopoietin, and FLT3 ligand (PeproTech), each at 100 ng/mL. Lentiviral transduction: short hairpin RNAs (shRNAs) targeting HMGA2 (supplemental Table 2) were cloned in a pLKO1-GFP vector from The RNAi Consortium (TRC). For overexpression, HMGA complementary DNA, or a spacer for control, were cloned under the human EF1a promoter in the lentiviral vector pHAGE-EF1a-PGK-ZsGreen-W. Wells in 96-well non–tissue-culture–treated plates were coated with 150 μL of retronectin (Takara Bioscience) at a concentration of 40 μg/mL overnight prior to transduction. Wells were then washed and blocked with 150 μL of a 2% (wt/vol) solution of bovine serum albumin in phosphate-buffered saline for 30 minutes at room temperature. Eighty thousand to 100 000 CD34+ CB cells were plated in each well with the lentivirus and incubated at 37°C overnight. Frequencies of colony-forming cells were determined by plating CD34+ cells at indicated time points in methylcellulose (Methocult H4230; StemCell Technologies) supplemented with stem cell factor (25 ng/mL), granulocyte macrophage colony-stimulating factor (50 ng/mL), interleukin 3 (25 ng/mL) (PeproTech), and erythropoietin (5 U/mL; Janssen). Plates were incubated at 37°C and mature hematopoietic colonies were scored after 14 days. For transplantation, within 48 hours of transduction, CD34+ cells were IV injected into the tail vein of sublethally irradiated (3 Gy) NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice. Human chimerism and lympho/myeloid distribution was assessed by anti-human CD45 antibody (HI30), anti-CD33 (P67.6), anti-CD15 (HI98), anti-CD19 (HIB19), and anti-CD3 (UCHT1) antibodies. HSC-enriched populations were analyzed using anti-CD34 and anti-CD38. Anti-human glycophorin A (GlyA) (CD235a; HIR2) was used to stain human erythroid cells. All animal experiments were reviewed and approved by the Lund/Malmö local ethical committee. For cell-cycle analysis, cells were stained and analyzed using the Click-iT EdU Alexa Fluor 488 Imaging kit (Thermo Fisher) according to the manufacturer’s protocol. For apoptosis assay, cells were stained with the Annexin V Apoptosis Detection kit, according to the manufacturer’s protocol (BD Biosciences).
Quantitative reverse transcription–PCR
RNA from sorted cells was extracted using a Qiagen RNA isolation kit. Complementary DNA synthesis was performed using the SuperScript III (Life Technologies). Quantitative reverse transcription–polymerase chain reaction (PCR) was performed in TaqMan gene-expression mix using a 7900 HT real-time PCR system (Applied Biosystems). Expression relative to hypoxanthine phosphoribosyltransferase was determined using the cycle threshold value. Primer details are given in supplemental Table 1.
Western blot
Proteins were extracted from sorted cells using 1× radioimmunoprecipitation assay buffer (Cell Signaling Technology) supplemented with protease inhibitor cocktail (Millipore) and 1 mM phenylmethane sulfonyl fluoride. Protein amount was estimated using the bicinchoninic acid assay. Cell extract was denatured in Laemmli buffer containing 1 mM β-mercaptoethanol, loaded to Novex 4% to 12% Bis-Tris protein gel (Life Technologies), and run with NuPAGE 3-(N-morpholino)propanesulfonic acid buffer (Life Technologies). Proteins were transferred to nitrocellulose membrane (iBlot Transfer Stack; Invitrogen). Membrane was blocked in 5% bovine serum albumin in Tris-buffered saline Tween 20 and incubated with antibodies for HMGA2 (Cell Signaling Technology). Goat anti-rabbit–horseradish peroxidase-conjugated secondary antibodies were used at a concentration of 1:15 000 (Santa Cruz Biotechnology). Details for the antibodies are given in supplemental Table 2. Blot was visualized using a Bio-Rad imager.
RNA sequencing and analysis
RNA was extracted from control or HMGA2-overexpressing CB CD34+ cells (n = 4 for each condition, RNeasy kit; Qiagen), and the RNA was amplified using an Ovation RNA-Seq System V2 kit (Nugen). Illumina library construction and sequencing were performed by BGI (Shenzen, China). Raw reads were aligned to the human genome (hg19 build) using TopHat2,9 and differential expression was determined using DEseq.10 Genes were considered differentially expressed when the adjusted P > .05. Ranked gene lists from DEseq were further analyzed using gene-set enrichment analysis (GSEA) using population-specific expression signatures generated for human hematopoietic subpopulations in previous studies.11,12 Pathway analyses were performed using Ingenuity Pathway Analysis software (Qiagen).
Statistical analysis
Unless otherwise stated, the 2-tailed unpaired Student t test was used to determine significance, and mean plus or minus standard error of the mean values are reported in the graphs.
Results
HMGA2 is preferentially expressed in the most immature fractions of human HSPCs
To assess the importance of HMGA2 in human HSPCs, we first quantified the mRNA expression levels in fluorescence-activated cell sorting (FACS)–purified hematopoietic subpopulations from different stages of ontogeny. Cells were isolated from fetal liver, umbilical CB, and adult BM (supplemental Figure 1). The most primitive hematopoietic fractions, that is, HSCs and multipotent progenitors (MPPs), showed higher expression of HMGA2 compared with more committed progenitors across all 3 developmental stages (Figure 1A-C). Moreover, we found that HSCs derived from CB and fetal liver had significantly higher expression of HMGA2 compared with HSCs isolated from adult BM (Figure 1D). This is in agreement with the fetal stage–specific expression of Hmga2 observed previously in the murine hematopoietic system.6 Because HMGA2 is known to be posttranscriptionally regulated by Let7 microRNAs,6 we also validated the expression pattern at the protein level, and confirmed that HMGA2 is robustly expressed in CB-derived HSCs and MPPs, but is almost undetectable in more mature cells (Figure 1E). This protein analysis further indicated that HSCs had substantially higher expression of HMGA2 compared with their immediate downstream progeny (MPPs) that lack self-renewal potential (Figure 1E), indicating a role of HMGA2 in regulating HSC function.
Preferential expression of HMGA2 within the primitive compartment of human hematopoietic cells. (A-C) HMGA2 expression measured by quantitative PCR in FACS-purified populations, HSCs (CD34+CD38−CD45RA−CD90+), MPPs (CD34+CD38−CD45RA−CD90−), MLP (CD34+CD38−CD45RA+CD90−), and committed progenitors (CP; CD34+CD38+) obtained from fetal liver (FL) (A), CB (B), and adult BM (C). (D) Relative expression of HMGA2 mRNA in purified HSCs from different developmental sources. (E) HMGA2 protein expression was assessed by western blot in FACS-purified CB populations. *P < .05; **P < .01; ***P < .001; ****P < .0001. ns, not significant.
Preferential expression of HMGA2 within the primitive compartment of human hematopoietic cells. (A-C) HMGA2 expression measured by quantitative PCR in FACS-purified populations, HSCs (CD34+CD38−CD45RA−CD90+), MPPs (CD34+CD38−CD45RA−CD90−), MLP (CD34+CD38−CD45RA+CD90−), and committed progenitors (CP; CD34+CD38+) obtained from fetal liver (FL) (A), CB (B), and adult BM (C). (D) Relative expression of HMGA2 mRNA in purified HSCs from different developmental sources. (E) HMGA2 protein expression was assessed by western blot in FACS-purified CB populations. *P < .05; **P < .01; ***P < .001; ****P < .0001. ns, not significant.
HMGA2 is required to maintain human HSPCs in vitro
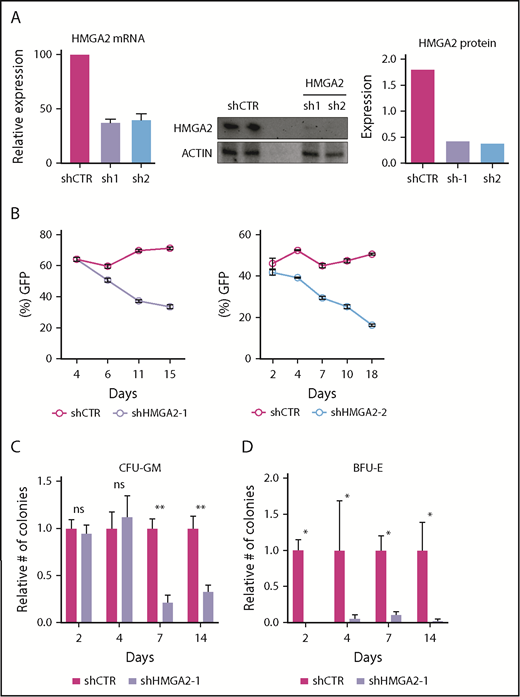
To evaluate the functional role of HMGA2 in human HSPCs, we targeted CB CD34+ cells with lentiviral vectors expressing shRNAs against HMGA2. Two independent shRNAs (sh1 and sh2) showed efficient knockdown of HMGA2 both at mRNA and protein levels (Figure 2A). We first assessed the consequences of HMGA2 knockdown by monitoring the frequency of cultured GFP+-transduced cells over time. We observed a steady decrease of GFP+CD34+ cells during culture of sh1- and sh2-transduced cells compared with cells transduced with a scrambled control shRNA (shCTR) (Figure 2B). Moreover, the propagation of both HSC-enriched CD34+CD90+ cells (supplemental Figure 2), as well as progenitor cells with colony-forming potential (Figure 2C-D), was markedly reduced over 14 days of culture of HMGA2-deficient cells. However, colony assays performed 2 and 4 days after transduction showed normal myeloid colony numbers (colony-forming units granulocyte-macrophage [CFU-GM]) from HMGA2-deficient cells (Figure 2C), indicating that, although HMGA2 is required to maintain production of progenitor cells over time in culture, it is dispensable for the normal maturation and differentiation of myeloid progenitors. By contrast, formation of erythroid colonies (burst-forming units erythroid [BFU-E]) was severely inhibited from cells plated already at day 2 (Figure 2D), suggesting that HMGA2 is critical not only for the propagation of erythroid precursors but also for their maturation.
shRNA-mediated knockdown of HMGA2 impairs the in vitro propagation of HSPCs. (A) Knockdown efficiency of shRNAs targeting HMGA2 as measured in CD34+CD38−GFP+ cells 2 days after transduction by quantitative PCR, and in CD34+GFP+ cells 4 days after transduction by western blot. (B) Frequency of GFP in CB CD34+ cells during in vitro culture for 2 shRNAs targeting HMGA2. (C-D) Relative numbers of colony-forming units granulocyte-macrophage (CFU-GM) (C) and burst-forming units erythroid (BFU-E) (D) after 2, 4, 7, and 14 days of culture following shRNA transduction. Numbers are normalized to the control cells. *P < .05; **P < .01.
shRNA-mediated knockdown of HMGA2 impairs the in vitro propagation of HSPCs. (A) Knockdown efficiency of shRNAs targeting HMGA2 as measured in CD34+CD38−GFP+ cells 2 days after transduction by quantitative PCR, and in CD34+GFP+ cells 4 days after transduction by western blot. (B) Frequency of GFP in CB CD34+ cells during in vitro culture for 2 shRNAs targeting HMGA2. (C-D) Relative numbers of colony-forming units granulocyte-macrophage (CFU-GM) (C) and burst-forming units erythroid (BFU-E) (D) after 2, 4, 7, and 14 days of culture following shRNA transduction. Numbers are normalized to the control cells. *P < .05; **P < .01.
HMGA2 is required to maintain the human HSC compartment in vivo following transplantation
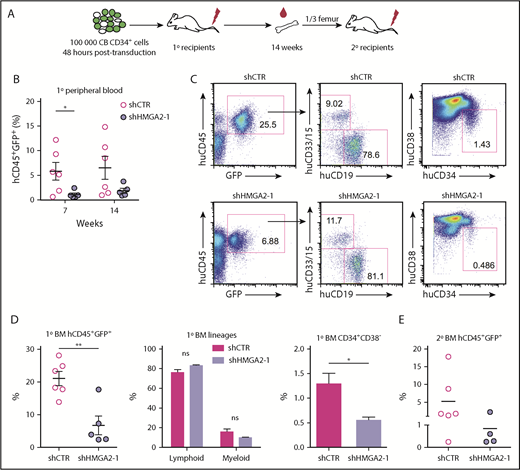
Next, we analyzed the effect of HMGA2 knockdown in vivo by transplanting shRNA-transduced CD34+ cells to sublethally irradiated NOD-SCID/IL2Rgnull (NSG) mice (Figure 3A). The cells were not sorted prior to transplantation allowing green fluorescent protein–negative (GFP−) cells to serve as internal controls (the initial transduction efficiencies are shown in supplemental Figure 3A). Peripheral blood analysis after 7 and 14 weeks showed an impaired hematopoietic reconstitution from GFP+ HMGA2-deficient cells (Figure 3B). By contrast, we observed no reduction in peripheral blood reconstitution from the untransduced GFP− cells (supplemental Figure 3B). Similarly, BM analysis after 14 weeks showed significantly lower reconstitution from GFP+ HMGA2-deficient cells (Figure 3C-D). Despite this marked reduction in long-term reconstitution, we found that the relative distribution between lymphoid and myeloid cells remained intact (Figure 3C-D), indicating a normal maturation potential. Staining for HSPC markers showed a specific reduction of the HSC-enriched CD34+CD38lo/− compartment within the engrafted GFP+ cells (Figure 3D). Accordingly, the reconstitution defect observed in the primary recipient mice could also be propagated to secondary NSG recipients (Figure 3E; supplemental Figure 3C). Overall, these findings show that HMGA2 is critically required to sustain the human HSC compartment in vivo following transplantation.
Knockdown of HMGA2 impairs the in vivo reconstitution potential of CB-derived CD34+cells. (A) Overview of the in vivo transplantation assay for HMGA2-deficient cells. (B) Human CD45+GFP+ chimerism in mice transplanted with shHMGA2-1–transduced cells as analyzed in peripheral blood after 7 and 14 weeks. (C) Representative FACS plots showing BM engraftment of primary recipient NSG mice 14 weeks after transplantation. (D) Accumulated data for huCD45+GFP+ cell engraftment, myeloid (CD15/CD33+), and lymphoid (CD19+) lineage distribution, as well as levels of HSC engraftment (CD34+CD38− cells) measured in huCD45+GFP+ graft in BM of primary recipient NSG. (E) Human CD45+GFP+ engraftment in secondary recipients analyzed 20 weeks posttransplantation. *P < .05; **P < .01.
Knockdown of HMGA2 impairs the in vivo reconstitution potential of CB-derived CD34+cells. (A) Overview of the in vivo transplantation assay for HMGA2-deficient cells. (B) Human CD45+GFP+ chimerism in mice transplanted with shHMGA2-1–transduced cells as analyzed in peripheral blood after 7 and 14 weeks. (C) Representative FACS plots showing BM engraftment of primary recipient NSG mice 14 weeks after transplantation. (D) Accumulated data for huCD45+GFP+ cell engraftment, myeloid (CD15/CD33+), and lymphoid (CD19+) lineage distribution, as well as levels of HSC engraftment (CD34+CD38− cells) measured in huCD45+GFP+ graft in BM of primary recipient NSG. (E) Human CD45+GFP+ engraftment in secondary recipients analyzed 20 weeks posttransplantation. *P < .05; **P < .01.
HMGA2 overexpression enhances the propagation of HSPCs in vitro and in vivo
To assess how HMGA2 regulates HSPC function in more detail, we overexpressed the gene using a lentiviral vector (supplemental Figure 4A). Overexpression of HMGA2 in transduced CB HSPCs was confirmed both at the mRNA and protein levels (supplemental Figure 4B-C). CD34+ cells targeted with the HMGA2 vector showed a gradual increase in the frequency of ZsGreen+ cells during 5 weeks of culture compared with control cells targeted with the same vector lacking the HMGA2 gene (supplemental Figure 4D). Similarly, the propagation of immature CD34+CD90+ cells gradually increased over 3 weeks of culture (Figure 4A) and the numbers of myeloid colony-forming cells showed an increase over control after 2 weeks (Figure 4B). There was further a trend toward increasing numbers of erythroid colony-forming cells in the HMGA2-transduced cultures (Figure 4C). Together, this suggests that enforced HMGA2 expression promotes the long-term maintenance of HSPCs ex vivo. However, we did not observe any significant differences in cell-cycle status or in the levels of apoptotic cells among the cultured CD34+ cells (supplemental Figure 5).
HMGA2 overexpression increases engraftment levels of CB CD34+in serially transplanted immunodeficient mice. CB CD34+ cells were transduced with a lentiviral vector overexpressing HMGA2 and assayed in vitro and in vivo by transplantation into NSG mice. (A) Relative numbers of CD34+CD90+ cells over 21 days of culture following shRNA transduction. Numbers are normalized to the control cells. (B-C) Relative numbers of CFU-GM (B) and BFU-E (C) after 2, 7, and 14 days of culture following shRNA transduction. Numbers are normalized to the control cells. (D) Overview of in vivo transplantation assay for HMGA2-overexpressing cells. (E) Chimerism of huCD45+ZsGreen+ cells in primary recipient mice transplanted with HMGA2-overexpressing cells as analyzed in peripheral blood after 6 to 7 and 14 weeks and (F) in BM after 14 weeks. (G) Levels of HSC engraftment (CD34+CD38− cells) measured in BM of primary recipients 14 weeks after transplantation. Representative FACS plots gated on huCD45+ZsGreen+ cells and the accumulated data are shown. (H) Unfractionated BM equivalent of one-third of femur from primary mice were transplanted in secondary mice. Chimerism of huCD45+ZsGreen+ cells in BM of secondary NSG recipients. Numbers on the top show the positively engrafted mice (CD45+ZsGreen+ cells ≥0.1%). *P < .05; **P < .01; ***P < .001.
HMGA2 overexpression increases engraftment levels of CB CD34+in serially transplanted immunodeficient mice. CB CD34+ cells were transduced with a lentiviral vector overexpressing HMGA2 and assayed in vitro and in vivo by transplantation into NSG mice. (A) Relative numbers of CD34+CD90+ cells over 21 days of culture following shRNA transduction. Numbers are normalized to the control cells. (B-C) Relative numbers of CFU-GM (B) and BFU-E (C) after 2, 7, and 14 days of culture following shRNA transduction. Numbers are normalized to the control cells. (D) Overview of in vivo transplantation assay for HMGA2-overexpressing cells. (E) Chimerism of huCD45+ZsGreen+ cells in primary recipient mice transplanted with HMGA2-overexpressing cells as analyzed in peripheral blood after 6 to 7 and 14 weeks and (F) in BM after 14 weeks. (G) Levels of HSC engraftment (CD34+CD38− cells) measured in BM of primary recipients 14 weeks after transplantation. Representative FACS plots gated on huCD45+ZsGreen+ cells and the accumulated data are shown. (H) Unfractionated BM equivalent of one-third of femur from primary mice were transplanted in secondary mice. Chimerism of huCD45+ZsGreen+ cells in BM of secondary NSG recipients. Numbers on the top show the positively engrafted mice (CD45+ZsGreen+ cells ≥0.1%). *P < .05; **P < .01; ***P < .001.
Next, to evaluate the effect of enforced HMGA2 expression on HSPC renewal and differentiation in vivo, we injected transduced CD34+ cells into sublethally irradiated NSG mice (Figure 4D). Transduction efficiencies between the control and HMGA2 vectors were kept at similar levels to allow a direct comparison of the transduced, ZsGreen+ cell populations (the transduction efficiencies in each experiment are given in supplemental Figure 6A). Evaluation of human cell reconstitution in peripheral blood at 6 to 7 weeks showed equivalent levels of hCD45+ZsGreen+ cells for both groups (Figure 4E), indicating that HMGA2 overexpression did not impact the functional output from short-term repopulating cells. After 14 weeks, we observed a mild increase in human chimerism in the blood and BM of primary recipient mice transplanted with HMGA2-overexpressing cells (Figure 4E-F). Importantly, in the BM, we found more than twofold higher frequencies of HSC-enriched CD34+CD38− cells among the engrafted HGMA2-overexpressing ZsGreen+ cells compared with the control group (Figure 4G). To functionally validate this finding, we performed secondary transplantations and found a progressive increase of human ZsGreen+ cells in peripheral blood in the HMGA2 group (supplemental Figure 6B). BM analysis showed that of 17 of 17 mice in the HMGA2 group were successfully engrafted with human ZsGreen+ cells (≥0.1%), compared with 11 of 16 mice in the control group (Figure 4H). The levels of ZsGreen+ cells, as well as overall levels of human cells, were significantly higher in the mice transplanted with HMGA2-transduced cells (Figure 4H; supplemental Figure 6C). Thus, enforced HMGA2 expression promotes the self-renewal ability of human HSCs in vivo. Additional experiments using transplantation of highly purified HSCs (CD34+CD38−CD90+CD49f+ cells) transduced with HMGA2 showed a similar but more pronounced effect of HMGA2 overexpression in primary recipients compared with the transplantations using bulk CD34+ cells (supplemental Figure 7).
HMGA2 overexpression alters the differentiation pattern of human HSPCs
We next analyzed the distribution of specific lineages in the BM of mice transplanted with HMGA2-transduced cells. We detected an increase in human CD33+/15+ myeloid cells and a reduction in human CD19+ lymphocytes in both the primary and secondary recipient mice (Figure 5A). Similar, but milder, changes were seen in the peripheral blood and spleen (supplemental Figure 8). Additionally, the peripheral blood and spleen analysis showed an increased contribution to mature human CD3+ T cells (supplemental Figure 8). We further assessed the contribution of human erythroid cells in the transplanted mice and observed a threefold increase of human GlyA+ cells in the BM of primary recipients transplanted with HMGA2-overexpressing cells compared with control (Figure 5B). Strikingly, in the secondary recipients, the levels of GlyA+ cells reached above 10%, suggesting that HMGA2 is a strong positive regulator of human erythropoiesis (Figure 5B).
HMGA2 overexpression in CB CD34+leads to altered hematopoietic differentiation. (A) Distribution of hematopoietic lineages as measured by expression of CD15/CD33, CD19, and CD3 within the engrafted human CD45+ZsGreen+ in primary and secondary recipient mice. (B) Frequency of GlyA+ cells in the BM of primary and secondary recipients. Representative FACS plots for GlyA+ cells in the BM of secondary recipients are shown to the right. *P < .05; **P < .01.
HMGA2 overexpression in CB CD34+leads to altered hematopoietic differentiation. (A) Distribution of hematopoietic lineages as measured by expression of CD15/CD33, CD19, and CD3 within the engrafted human CD45+ZsGreen+ in primary and secondary recipient mice. (B) Frequency of GlyA+ cells in the BM of primary and secondary recipients. Representative FACS plots for GlyA+ cells in the BM of secondary recipients are shown to the right. *P < .05; **P < .01.
HMGA2-overexpressing HSPCs show gene-expression signatures associated with myeloerythroid differentiation
To obtain a global view of gene-expression changes associated with enforced expression of HMGA2, we performed RNA sequencing (RNA-Seq) on HMGA2-transduced CD34+ cells 72 hours posttransduction. Overall, we found 127 significantly upregulated genes and 106 downregulated genes with a fold change >1.5 compared with control (supplemental Table 3). The data were analyzed by the GSEA software, using gene sets from population-specific expression signatures generated for human hematopoietic subpopulations in previous studies.11,12 The gene signature normally associated with common myeloid progenitors was significantly enriched in HMGA2-overexpressing HSPCs, whereas gene signatures associated with multilymphoid progenitors (MLPs) were not significantly altered (Figure 6A). This is consistent with the myeloid skewing observed in the transplanted animals. Similarly, gene signatures associated with megakaryocytic erythroid progenitors (MEPs) as well as more mature erythroid signatures were strongly enriched in HMGA2-overexpressing cells (Figure 6B). Indeed, several hemoglobin genes and other erythroid-specific genes were among the 20 most upregulated genes in HMGA2-transduced cells (Figure 6C). Thus, HMGA2 overexpression in HSPCs induces transcriptional programs of altered hematopoietic differentiation, which are reflected in the functional phenotypes observed in vivo.
HMGA2-overexpressing HSPCs show gene-expression signatures associated with myeloerythroid differentiation. (A-B) GSEA to identify gene sets enriched or depleted in HMGA2-overexpressing CD34+ cells compared with controls. GSEA plots were generated using population-specific gene signatures described in previous studies for human hematopoietic cells. (C) Top 20 upregulated genes in HMGA2-overexpressing cells. CMP, common myeloid progenitor; EryUP, erythroid upregulated; FDR, false discovery rate; NES, normalized enrichment score.
HMGA2-overexpressing HSPCs show gene-expression signatures associated with myeloerythroid differentiation. (A-B) GSEA to identify gene sets enriched or depleted in HMGA2-overexpressing CD34+ cells compared with controls. GSEA plots were generated using population-specific gene signatures described in previous studies for human hematopoietic cells. (C) Top 20 upregulated genes in HMGA2-overexpressing cells. CMP, common myeloid progenitor; EryUP, erythroid upregulated; FDR, false discovery rate; NES, normalized enrichment score.
HMGA2-overexpressing HSPCs show increased activation of AKT
Ingenuity pathway analysis (IPA) showed that cellular functions associated with proliferation and survival were significantly enriched among both the upregulated and downregulated genes in HMGA2-overexpressing cells (Figure 7A), possibly associated with the enhanced proliferation and maintenance of HSPCs observed in vitro and in vivo. Specifically, mammalian target of rapamycin (mTOR) and p70S6 kinase were among the most significantly perturbed pathways (Figure 7B). As both mTOR and p70S6 are downstream of phosphatidylinositol 3-kinase (PI3K)–AKT, we determined the phosphorylation status of the components of the AKT/mTOR/S6 kinase cascade, and observed increased levels of phosphorylated AKT (p-S473), whereas mTOR (p-S2448), S6 kinase (p-T421/S424), and 4EBP1 (p-T37/46) were unaltered (Figure 7C). Thus, there appears to be selective activation of AKT in response to enforced expression of HMGA2 in HSPCs, which at least in part, may explain the enhanced proliferation and maintenance phenotype.
Increased activation of AKT signaling in HMGA2-overexpressing cells. (A) IPA showing cellular functions associated with upregulated or downregulated genes in HMGA2-overexpressing cells. (B) IPA analysis showing cellular pathways enriched in upregulated genes in HMGA2-overexpressing cells. (C) Western blot for the components of the AKT-mTOR pathways in HMGA2-overexpressing cells. Exp., experiment; NO, nitric oxide; ROS, reactive oxygen species.
Increased activation of AKT signaling in HMGA2-overexpressing cells. (A) IPA showing cellular functions associated with upregulated or downregulated genes in HMGA2-overexpressing cells. (B) IPA analysis showing cellular pathways enriched in upregulated genes in HMGA2-overexpressing cells. (C) Western blot for the components of the AKT-mTOR pathways in HMGA2-overexpressing cells. Exp., experiment; NO, nitric oxide; ROS, reactive oxygen species.
Discussion
In this study, we have used loss-of-function and gain-of-function approaches to characterize the role of HMGA2 in human hematopoiesis and HSPCs. We demonstrate key functions of HMGA2 in regulating both renewal and differentiation of human HSPCs in vitro and in vivo.
Previously, the role of HMGA2 in hematopoiesis has been characterized in mouse models. For the most part, our findings in primary human cells are compatible with the mouse studies. Ikeda et al reported clonal expansion of HSCs in the BM coupled with signs of myeloproliferation upon overexpression of Hmga2.5 Similarly, Copley et al found that overexpression of Hmga2 enhanced HSC self-renewal in serial transplantation models.6 Conversely, the enhanced self-renewal activity seen during early development was compromised by loss of Hmga2.6 Indeed, our findings suggest that HMGA2 promotes self-renewal of human HSCs also, and that enforced expression preferentially drives the propagation of long-term repopulating HSCs compared with more short-lived cells. This is supported by the relatively delayed effect seen here in both the in vitro and in vivo assays when overexpressing HMGA2 in CD34+ cells, as well as the more pronounced repopulation advantage seen when purified HSCs were transplanted, compared with bulk CD34+ cells. Moreover, similar to Copley et al, we did not observe any overt changes in the proliferative state of HMGA2-overexpressing HSPCs, indicating that HMGA2 mainly regulates the balance between renewal and differentiation rather than proliferation.6
In both primary and secondary transplant recipients of cells overexpressing HMGA2, the lineage distribution was skewed toward myelopoiesis and we also observed a striking accumulation of human GlyA+ erythrocytes in the BM. Our RNA-Seq data further provided molecular evidence for a differentiation pattern skewed to the megakaryocytic and erythroid lineages with a particular strong enrichment for the MEP and mature erythroid gene signature. Furthermore, the near complete absence of erythroid colony formation upon HMGA2 knockdown suggests an important and specific role of HMGA2 in regulation of human erythropoiesis. This is supported by the previously reported studies by Ikeda et al, and Rowe et al, which show that ectopic expression of Hmga2 enhances erythroid development in mice.5,8
The enhanced self-renewal capacity and altered differentiation pattern from enforced HMGA2 expression could potentially indicate a role of HMGA2 in malignant development. However, there is little evidence of HMGA2 as a primary driver of leukemogenesis. No recurrent genetic aberrations within the HMGA2 locus have been associated with leukemia. Moreover, even though overexpression of full-length HMGA2 transcripts has been associated with advanced stages of malignancies, this is linked to the repression of let-7, a microRNA that in addition to regulating HMGA2 expression, is known to control the expression of oncogenes such as C-MYC and KRAS.13-15 This suggests that malignant transformation may be dependent on derepression of other let-7 target genes rather than the specific activities of HMGA2. Importantly, we did not observe any incidence of leukemia from cells overexpressing HMGA2 in our transplanted mice. This may indicate that HMGA2 triggers a benign expansion in vivo but is not sufficient to initiate leukemia on its own. The notion of benign expansion effects from HMGA2 is further supported by findings from X-SCID and β-thalassemia gene therapy trials where recurrent events of lentiviral vector integration within the HMGA2 locus resulting in increased HMGA2 expression were observed.16,17 Indeed, it was found that this provided a clonal advantage to HSPCs bearing such specific vector integrations and enhanced the efficiency of gene correction and therapeutic benefits of the gene therapy approach.
Mechanistically, we found that genes downstream of PI3K-AKT–mTOR and mTOR complex 1 signaling were differentially expressed in CD34+ HSPCs upon enforced expression of HMGA2, and we observed increased levels of p-AKT. Activation of AKT signaling has been associated with HMGA2 activity in diverse cellular contexts, that is, stromal cells,18 hematopoietic cells,5 and acute myeloid leukemia.19 PI3K/AKT signaling has been extensively studied in hematopoiesis and regulators downstream to this pathway such as mTOR, WNT, and GSK3b are critical in regulating HSC functions.20 In the mouse, knockout of both the isoforms of AKT (AKT1/2) result in compromised HSC competitiveness.21 Increased activation of the pathway, on the other hand, results in increased HSC proliferation, which ultimately may cause exhaustion and impaired hematopoietic reconstitution.22 However, we did not observe any signs of HSC exhaustion over time in our serially transplanted animals and our cell-cycle analysis did not indicate any increased cycling upon HMGA2 overexpression. Similarly, Lechman et al has found that knockdown of miR-126 results in activation of PI3K-AKT/GSK3b that leads to expansion of HSCs in vivo without exhaustion.23 The PI3K/AKT pathway has complex and diverse functions and it is likely that the functional consequences of enhanced activity of this pathway in HSCs are highly dose- and context-dependent. Further studies are required to demonstrate a direct functional role of this pathway in mediating the HMGA2 phenotype in HSCs.
HMGA2 has been shown to be one of the most highly expressed proteins in murine HSCs. Here, we now provide a functional analysis of its role in human hematopoiesis and position it as a central regulator of HSC renewal and differentiation.
The RNA-sequencing data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE107594).
The full-text version of this article contains a data supplement.
Acknowledgments
This work was supported by grants from the Swedish Research Council, the Swedish Cancer Foundation, the Swedish Paediatric Cancer Foundation, and the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 648894) (J.L.); the National Health and Medical Research Council of Australia (J.E.P.); Cancer Australia (D.B.); and the Anthony Rothe Memorial Trust (J.A.I.T. and D.B.). P.K. was supported by a Wenner-Gren postdoctoral fellowship and D.B. was supported by early career fellowships from the National Health and Medical Research Council of Australia and Cancer Institute of New South Wales. This work was further supported by the HematoLinné and StemTherapy programs at Lund University and the Translational Cancer Research Centre at UNSW and UTS Sydney.
Authorship
Contribution: P.K. and J.L. conceived and designed the study; P.K. performed most experiments with assistance from R.G., M.S.T., I.d.J., A.B., and A.S.; D.B., J.A.I.T., A.U., and J.E.P. designed, executed, and analyzed RNA-Seq experiments; and P.K. and J.L. wrote the manuscript with assistance from the other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonas Larsson, Molecular Medicine and Gene Therapy, Lund University, BMC A12, 221 84 Lund, Sweden; e-mail: jonas.larsson@med.lu.se.