Key Points
The first crystal structure of human plasma β-FXIIa in its active state is presented. The conformational lability of FXIIa is discussed.
These novel structural data provide molecular insight into β-FXIIa interaction with its substrates and inhibitors.
Abstract
Activated factor XIIa (FXIIa) is a serine protease that has received a great deal of interest in recent years as a potential target for the development of new antithrombotics. Despite the strong interest in obtaining structural information, only the structure of the FXIIa catalytic domain in its zymogen conformation is available. In this work, reproducible experimental conditions found for the crystallization of human plasma β-FXIIa and crystal growth optimization have led to determination of the first structure of the active form of the enzyme. Two crystal structures of human plasma β-FXIIa complexed with small molecule inhibitors are presented herein. The first is the noncovalent inhibitor benzamidine. The second is an aminoisoquinoline containing a boronic acid–reactive group that targets the catalytic serine. Both benzamidine and the aminoisoquinoline bind in a canonical fashion typical of synthetic serine protease inhibitors, and the protease domain adopts a typical chymotrypsin-like serine protease active conformation. This novel structural data explains the basis of the FXII activation, provides insights into the enzymatic properties of β-FXIIa, and is a great aid toward the further design of protease inhibitors for human FXIIa.
Introduction
Human factor XIIa (FXIIa; Hageman factor, EC 3.4.21.38), a multidomain serine protease of the trypsin-like family, initiates the intrinsic coagulation cascade by contact activation in a reaction involving high-molecular-weight kininogen (HMWK) and plasma prekallikrein (PPK).1 This activation requires proteolytic conversion of plasma FXII zymogen to active protease FXIIa on negatively charged surfaces where FXII undergoes conformational changes and small amounts of active FXIIa are formed.2 At the same time, HMWK bound to the same surface presents PPK to FXIIa for activation. The resulting active plasma kallikrein (PK) reciprocally activates additional FXIIa in a positive feedback loop.3 In the next steps of the intrinsic pathway, FXIIa cleaves its substrate FXI to generate active FXIa, which in turn activates FIX to FIXa.4,5 This series of reactions eventually drives thrombin generation and fibrin formation in the final steps of coagulation. FXII deficiency in humans and animals is not associated with excessive bleeding, demonstrating that FXIIa activation of FXI is not essential for hemostasis.6,7 Except for procoagulant activity, the FXIIa-driven contact system has proinflammatory activity via the kallikrein-kinin system, which liberates the inflammatory mediator bradykinin from HMWK via PK.8,9 FXIIa activity in plasma is mainly regulated by its cognate serpin C1 esterase inhibitor (C1INH).10 Plasma antithrombin (AT) and plasminogen activator inhibitor 1 (PAI-1) also have some minor FXIIa inhibitory activity.11
Thus, recent data have made FXIIa an attractive target for designing safe anticoagulants that inhibit thrombosis without the influence of hemostasis. Currently available antithrombotic agents such as low-molecular-weight heparins, warfarin, and antiplatelet therapies are associated with a high risk of severe bleeding complications because they target components of the blood-clotting mechanism such as thrombin, FVIIa, FIXa, FXa, and FXIa.12 Therefore, designing new drugs against FXIIa, which is involved in the development of pathological thrombus formation while having limited effect on physiological homeostasis, may make antithrombotic therapy safer. However, this strategy is currently limited by the absence of structural data for active FXIIa. Recently, the crystal structure of the catalytic domain of recombinant human FXII (residues 354-596, FXII mature protein sequencing, FXIIc) was determined.13 The structure revealed the zymogen conformation of the enzyme catalytic domain and did not provide a suitable platform for a structure-based drug design approach.
The inactive zymogen form of FXII is secreted as a single-chain polypeptide chain of 596 amino acid residues with a molecular weight of 80 kDa. Upon contact system activation, surface-bound FXII proenzyme is cleaved at the Arg353-Val354 peptide bond by PK, generating α-FXIIa consisting of a 50-kDa heavy chain and a 28-kDa light chain held together by the Cys340-Cys467 disulfide bond. Further proteolytic cleavages of α-FXIIa heavy chain at the Arg334 and Arg343 C termini by PK yields β-FXIIa, consisting of 2 polypeptide chains of a 2-kDa heavy chain remnant and a 28-kDa catalytic domain covalently bonded together by the same disulfide bond.14
In this work, we crystallized and solved, for the first time, structures of human plasma β-FXIIa in complex with 2 different inhibitors. These include the noncovalent inhibitor benzamidine, a classical inhibitor for serine proteases, and a covalent synthetic small molecule inhibitor containing a surrogate of the basic group. For convenience in structural comparison with other members of the trypsin-like serine protease family, we used the chymotrypsinogen residue numbering shown in italics (supplemental Figure 1).14 The catalytic domain of β-FXIIa adopts the same fold typical of other active serine proteases. Structural analysis of surface electrostatic potentials revealed a plausible exosite, which may be crucial for the interactions of cofactors, substrates, and inhibitors with β-FXIIa. Also, the structures of FXIIa-inhibitor complexes provide important information about the geometry of the binding site, which is a critical asset for understanding the determinants for selectivity and specificity in FXIIa-ligand interactions.
Methods
Materials
Human plasma β-FXIIa and PK were purchased from Molecular Innovations (Novi, MI). The purity of the commercial β-FXIIa sample was 95% as assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis (Molecular Innovations) and electrospray mass spectroscopy (supplemental Figure 2). FXIa and Lys-plasmin were obtained from Hematologic Technology (Burlington, VT), and bovine trypsin was obtained from Worthington Biochemical Corporation (Lakewood, NJ). The chromogenic substrates D-Pro-Phe-ArgpNA (S-2302) and N-Z-D-Arg-Gly-Arg-pNA (S-2765) were obtained from Aniara (Westchester, OH); tosyl-Gly-Pro-Lys-4-nitranilide (Chromozyme PL) was obtained from Sigma-Aldrich (St. Louis, MO). Synthesis of compound 1 (3-(1-aminoisoquinolin-6-yl)phenyl)boronic acid), the protease inhibition assay, and the modeling of β-FXIIa interactions with its inhibitor and substrate are described in detail in supplemental Methods.
Crystallization and structure determination of human plasma β-FXIIa
Human plasma β-FXIIa protein with the presence of 8 mM benzamidine was concentrated to 10 mg/mL using Amicon centrifugal filters with a 10-kDa molecular-weight cutoff (Millipore) in 30 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) buffer, pH 7.0. Initial crystals of the β-FXIIa–benzamidine complex were grown at 18°C from drops containing 0.3 μL of the protein sample and 0.3 μL of reservoir solution (0.2 M ammonium iodide, 2.2 M ammonium sulfate). Protein crystals of the complex suitable for x-ray analysis were obtained after several cycles of microseeding under similar crystallization conditions. SDS-PAGE analysis (supplemental Figure 2B) of protein samples obtained by dissolving the crystals in SDS buffer did not reveal any degradation products and confirmed the presence of only β-FXIIa protein in the crystals used for the data collection.
A soaking approach was used to obtain the protein crystals of the compound 1–β-FXIIa complex. The benzamidine crystals were transferred to a drop of mother liquor containing a saturated solution of compound 1 ligand. The drop was further incubated for 18 hours at 18°C over the reservoir solution before the data collection.
For data collection, crystals were harvested with 20% (vol/vol) glycerol in the reservoir solution. Diffraction data were collected from a single flash-frozen crystal on the LS-CAT ID beamline (APS; Argonne National Laboratory). Data were indexed and processed with HKL-2000.15 The crystals belonged to tetragonal space group I41 and contained 1 molecule of β-FXIIa per asymmetric unit.
The structure of β-FXIIa was solved by molecular replacement using the PHASER program from the CCP4 software suite, with the structure of bovine trypsin (Protein Data Bank [PDB] code 3MFJ) as a search model.16,17 The final structure of β-FXIIa was obtained by carrying out several cycles consisting of manual model building using COOT, followed by structure refinement with REFMAC from the CCP4 software suite.18,19 Coordinates have been deposited in the PDB (codes 6B74 and 6B77).
Results
Overall structure of human plasma β-FXIIa
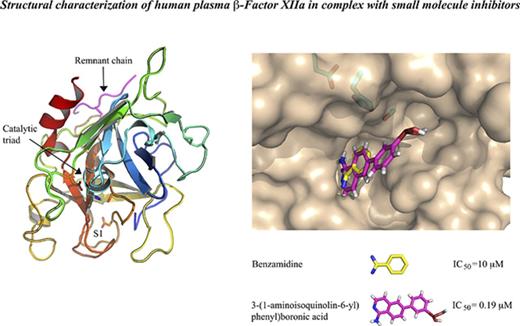
The structures of human plasma β-FXIIa in complex with the small molecule compound 1 (3-(1-aminoisoquinolin-6-yl)phenyl)boronic acid) and benzamidine were solved at a resolution of 2.4 Å and 2.3 Å, respectively (Table 1). For both structures, all 9 residues (residues 335-343, Asn5-Arg4) of the heavy chain remnant and 252 residues (residues 354-596, Val16-Ser244) of the FXIIa light chain, with the exception of a few side chains, were traceable in the final electron-density map (Figure 1). These 2 crystal structures are very similar to one another with an overall root mean square deviation (RMSD) of 0.25 Å for all Cα pairs (Figure 1C). The protease domain of β-FXIIa displays strong structural similarity with other trypsin-like active serine proteases, consisting of 2 β-barrels forming the substrate-binding site and the catalytic triad located at the cleft between 2 barrels in a typical arrangement (Figure 1A). The short remnant heavy chain is positioned on the molecular surface opposite to the active site entrance, and is covalently linked to the protease domain through disulfide bridge Cys1-Cys122. The remnant chain also makes contacts from both the backbone and side chains to residues in the protease domain. These include a hydrogen bond from the backbone carbonyl of Ser-1 to the backbone amino group of Arg205C, a hydrogen bond from the carbonyl of Gly2 to the backbone amino group of Glu205B, and a hydrogen bond from the backbone amino group of Arg4 to the carbonyl oxygen atom of Ala205. Finally, the side chain of Arg4 forms a hydrogen bond with the backbone carbonyl of Gln204 and the NH1 group of Arg205C with the carbonyl of Leu-2. It is interesting to note that the remnant chain of β-FXIIa is organized in a conformation different to that observed for human tissue-type plasminogen activator (tPA), human urokinase-type plasminogen activator (uPA), thrombin, or human hepatocyte growth factor activator (HGFA), although this chain is also linked to the protease domain by a homologous disulfide bridge, but has a much shorter length at its C terminus (supplemental Figure 1). Optimal superposition of the FXIIa catalytic domain with corresponding HGFA, tPA, and uPA homologous domains results in 237, 232, and 231 equivalent Cα atoms with the RMSD values of 1.26 Å, 1.45 Å, and 1.50 Å, respectively.
Data collection and refinement statistics
| . | Benzamidine complex . | Compound 1 complex . |
|---|---|---|
| PDB code . | 6B74 . | 6B77 . |
| Data collection | ||
| Wavelength, Å | 0.9795 | 0.9787 |
| Resolution range, Å | 50.00-2.32 (2.40-2.32) | 50.00-2.37 (2.47-2.37) |
| Space group | I 41 | I 41 |
| Unit cell: a, b, c, Å | 80.289, 80.289, 121.731 | 79.402, 79.402, 122.246 |
| Total reflections | 102 197 | 95 803 |
| Unique reflections | 16 576 (1 632) | 15 282 (1 524) |
| Multiplicity | 6.2 (6.4) | 6.3 (6.4) |
| Completeness, % | 99.9 (100.0) | 99.8 (100.0) |
| <I>/σ(I) | 18.19 (2.98) | 21.4 (3.4) |
| Rmerge (%)* | 8.4 (84.9) | 7.9 (86.7) |
| Refinement | ||
| Resolution range, Å | 36.22-2.32 (2.41-2.32) | 41.35-2.37 (2.45-2.37) |
| Rwork (%)/Rfree (%)† | 19.0 (29.6)/22.2 (32.7) | 17.4 (24.4)/21.8 (32.2) |
| No. of nonhydrogen atoms‡ | 1952 | 2015 |
| Protein | 1849 | 1848 |
| Ligands | 64 | 70 |
| Waters | 51 | 97 |
| RMS bonds, Å | 0.004 | 0.012 |
| RMS angles, ° | 0.832 | 1.360 |
| Ramachandran favored, % | 96.7 | 95.6 |
| Ramachandran allowed, % | 2.5 | 3.6 |
| Ramachandran outliers, % | 0.8 | 0.8 |
| Average B factor, Å2 | 71.8 | 58.0 |
| Macromolecules | 71.2 | 57.4 |
| Ligands | 92.5 | 73.2 |
| Waters | 67.4 | 58.1 |
| . | Benzamidine complex . | Compound 1 complex . |
|---|---|---|
| PDB code . | 6B74 . | 6B77 . |
| Data collection | ||
| Wavelength, Å | 0.9795 | 0.9787 |
| Resolution range, Å | 50.00-2.32 (2.40-2.32) | 50.00-2.37 (2.47-2.37) |
| Space group | I 41 | I 41 |
| Unit cell: a, b, c, Å | 80.289, 80.289, 121.731 | 79.402, 79.402, 122.246 |
| Total reflections | 102 197 | 95 803 |
| Unique reflections | 16 576 (1 632) | 15 282 (1 524) |
| Multiplicity | 6.2 (6.4) | 6.3 (6.4) |
| Completeness, % | 99.9 (100.0) | 99.8 (100.0) |
| <I>/σ(I) | 18.19 (2.98) | 21.4 (3.4) |
| Rmerge (%)* | 8.4 (84.9) | 7.9 (86.7) |
| Refinement | ||
| Resolution range, Å | 36.22-2.32 (2.41-2.32) | 41.35-2.37 (2.45-2.37) |
| Rwork (%)/Rfree (%)† | 19.0 (29.6)/22.2 (32.7) | 17.4 (24.4)/21.8 (32.2) |
| No. of nonhydrogen atoms‡ | 1952 | 2015 |
| Protein | 1849 | 1848 |
| Ligands | 64 | 70 |
| Waters | 51 | 97 |
| RMS bonds, Å | 0.004 | 0.012 |
| RMS angles, ° | 0.832 | 1.360 |
| Ramachandran favored, % | 96.7 | 95.6 |
| Ramachandran allowed, % | 2.5 | 3.6 |
| Ramachandran outliers, % | 0.8 | 0.8 |
| Average B factor, Å2 | 71.8 | 58.0 |
| Macromolecules | 71.2 | 57.4 |
| Ligands | 92.5 | 73.2 |
| Waters | 67.4 | 58.1 |
Statistics for the highest-resolution shell are shown in parentheses.
RMS, root mean square deviation from ideal values (crystallography).
Rmerge = 100Σ(h)Σ(i)|I(i)–<I>|/ Σ(h)Σ(i)I(i), where I(i) is the ith intensity measurement of reflection h, and <I> is the average intensity from multiple observations.
Rfactor = Σ||Fobs|−|Fcalc||/ Σ|Fobs|. Where Fobs and Fcalc are the structure factor amplitudes from the data and the model, respectively; 10% reflections were used to calculate Rfree values.
Per asymmetric unit.
Crystal structure of human plasma β-FXIIa. (A) Stereo ribbon diagram (cyan) of β-FXIIa in complex with covalent compound 1 inhibitor in “standard” orientation. Compound 1 is shown in green sticks. The side chains of key β-FXIIa residues are shown as magenta sticks: His57, Asp102, and Ser195 of the catalytic triads, Asp189 at the base of the S1 specificity pocket. The side chains of N-glycosylated Asn74 and NacGlc-disaccharide are shown in cyan sticks. Some surface loops discussed in the text are labeled and shown in pink. The heavy chain remnant of β-FXIIa is shown in red. Hydrogen bonds are indicated by black dotted lines. (B) The structure of β-FXIIa in complex with noncovalent benzamidine inhibitor is represented as in panel A. Six disulfide bonds are indicated and are shown in yellow. (C) Superposition of the β-FXIIa (benzamidine [magenta] and β-FXIIa) compound 1 (cyan) crystal structures. The catalytic domains are superimposed by aligning the Cα atoms of residues 16-244 and are presented as cartoon diagrams. Benzamidine (orange), compound 1 (green), and residues of the catalytic triad are shown in a stick representation.
Crystal structure of human plasma β-FXIIa. (A) Stereo ribbon diagram (cyan) of β-FXIIa in complex with covalent compound 1 inhibitor in “standard” orientation. Compound 1 is shown in green sticks. The side chains of key β-FXIIa residues are shown as magenta sticks: His57, Asp102, and Ser195 of the catalytic triads, Asp189 at the base of the S1 specificity pocket. The side chains of N-glycosylated Asn74 and NacGlc-disaccharide are shown in cyan sticks. Some surface loops discussed in the text are labeled and shown in pink. The heavy chain remnant of β-FXIIa is shown in red. Hydrogen bonds are indicated by black dotted lines. (B) The structure of β-FXIIa in complex with noncovalent benzamidine inhibitor is represented as in panel A. Six disulfide bonds are indicated and are shown in yellow. (C) Superposition of the β-FXIIa (benzamidine [magenta] and β-FXIIa) compound 1 (cyan) crystal structures. The catalytic domains are superimposed by aligning the Cα atoms of residues 16-244 and are presented as cartoon diagrams. Benzamidine (orange), compound 1 (green), and residues of the catalytic triad are shown in a stick representation.
The protease domain of human FXIIa has 6 disulfide bonds (Figure 1B). Three of them, Cys42-Cys58, Cys168-Cys182, and Cys191-Cys220 are well conserved among trypsin-like proteases (supplemental Figure 1). Two other disulfide bonds, Cys50-Cys111 and Cys136-Cys201, are less observed. The first of these 2 bridges is found in the structures of HGFA, tPA, and uPA. The second bridge, homologous to Cys136-Cys201, occurs in the protease domains of HGFA, tPA, and uPA, and also in trypsin and chymotrypsinogen. Finally, there is 1 disulfide bond, Cys77-Cys80, which is characteristic for human plasma FXIIa.
There is 1 potential N-glycosylation site in the molecule of β-FXIIa and mass spectroscopic analysis of the commercial sample confirmed the presence of 2911-Da oligosaccharides covalently linked to the protein moiety (supplemental Figure 2A). This N-glycosylation site at Asn74 is located on the surface loop, which is spatially removed from the active site cleft and the substrate-binding site (Figure 1A-B), and, most likely, the glycosylation should not affect FXIIa catalytic properties, as observed for other glycosylated active serine proteases such as PK and FXIa.20,21
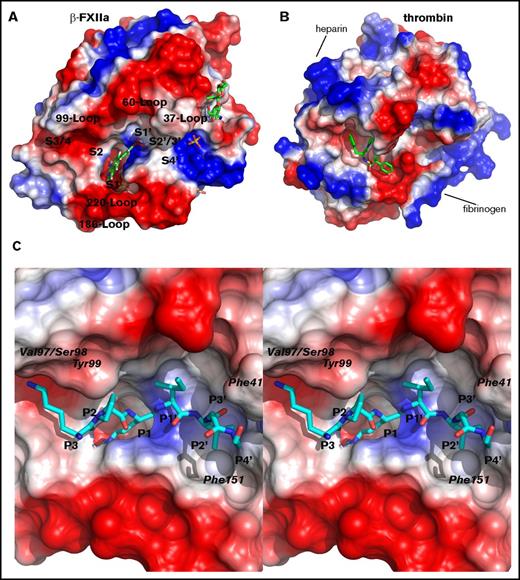
Human plasma β-FXIIa contains 26 negatively charged residues and 15 positively charged residues (and 9 histidine residues), and is a relatively acidic protein, with a calculated isoelectric point value of 5.2, among trypsin-like serine proteases. Analysis of surface charge calculated from the crystal structure of the covalent complex between β-FXIIa and compound 1 revealed several negative surface patches surrounding the active site cleft (Figure 2A). One of these negative potentials spreads along the top rim of the active site starting from the area of the 99 and 60 loops, where side chains of 4 acidic residues (Glu92, Asp60D, Glu62, and Asp63) are exposed to the solvent, toward the 110 loop. Another notable surface patch of negatively charged side chains (Glu146, Glu149, and Glu150) protrudes along the activation loop below the active site cleft and close to the 186 loop (Figure 2A). These acidic residues (except Asp60D, which exists in both β-FXIIa and uPA) are not conserved among trypsin-like proteases and, therefore, the distribution of negatively charged residues along the FXIIa protease domain surface is unique for this enzyme. For example, in thrombin, the former patch is quite positive where Arg60B, Arg86, and Arg89 are surrounded by other alkaline residues, Arg107, Arg129, Arg167, and Arg239, to form the heparin-binding site (Figure 2B).22 Also, the fibrinogen-binding site of the thrombin catalytic domain, which is positively charged, spatially overlays the latter negative site of the FXIIa protease domain.
Surface charge of the β-FXIIa protease domain. (A) Solid surface colored according to the electrostatic potential calculated from the crystal structure of β-FXIIa in complex with compound 1. Positive surface is shown in blue and negative is in red. Some surface loops surrounding the active site cleft and positions of the substrate-binding sites (S4-S4′) are labeled. Noncovalent inhibitors, sugar residues, and sulfate ions are shown as sticks. (B) Thrombin surface charge was calculated from the crystal structure (PDB code 4UFD) of the enzyme in complex with the benzamidine-containing moiety. The inhibitor is shown in green sticks and the view of the thrombin molecule is that of the β-FXIIa molecule orientation shown in panel A. (C) Close-up stereo view of the FXIIa substrate from P3 to P4′ (ball and stick in cyan) interacting with the surface of the substrate-binding area. The residues P3 to P4′ modeled into the active site (see “Materials and methods”) according to the canonical conformation are Lys-Pro-Arg-Ile-Val-Gly-Gly from the FXI amino acid sequence (residues 385-391; the scissile peptide bond is between the underlined residues). The positions of several FXIIa catalytic domain residues, which can form hypothetical interactions with the substrate in the encounter complex are indicated.
Surface charge of the β-FXIIa protease domain. (A) Solid surface colored according to the electrostatic potential calculated from the crystal structure of β-FXIIa in complex with compound 1. Positive surface is shown in blue and negative is in red. Some surface loops surrounding the active site cleft and positions of the substrate-binding sites (S4-S4′) are labeled. Noncovalent inhibitors, sugar residues, and sulfate ions are shown as sticks. (B) Thrombin surface charge was calculated from the crystal structure (PDB code 4UFD) of the enzyme in complex with the benzamidine-containing moiety. The inhibitor is shown in green sticks and the view of the thrombin molecule is that of the β-FXIIa molecule orientation shown in panel A. (C) Close-up stereo view of the FXIIa substrate from P3 to P4′ (ball and stick in cyan) interacting with the surface of the substrate-binding area. The residues P3 to P4′ modeled into the active site (see “Materials and methods”) according to the canonical conformation are Lys-Pro-Arg-Ile-Val-Gly-Gly from the FXI amino acid sequence (residues 385-391; the scissile peptide bond is between the underlined residues). The positions of several FXIIa catalytic domain residues, which can form hypothetical interactions with the substrate in the encounter complex are indicated.
Surface loops surrounding the active site of trypsin-like serine proteases are well known to be involved in many specific interactions with their cognate inhibitors, substrates, and cofactors.22-24 All main-chain atoms and most side chains of the surface loops are well defined by electron density in the crystal structures of human plasma β-FXIIa in complex with inhibitors. One of these loops, the β-FXIIa 37-surface loop, although having the same length of polypeptide segment as in trypsin and HGFA, adopts a quite different conformation and does not make any contact with the 60 loop, unlike hydrophobic interactions between side chains of Ile35 and Phe59 in HGFA or salt bridges between side chains of Lys60 and Ser34/Tyr39 in trypsin. Simple modeling revealed 1 possible interaction between the carbonyl atom of Phe41 of β-FXIIa and its substrate at the P3′ site (Figure 2C), which is also observed in the complex between HGFA and the inhibitory domain KD1 from inhibitor-1B.25
The 60-insertion loop resides directly adjacent to the 37 loop (Figures 1A and 2A) and has the same length as in HGFA, tPA, and uPA, is 4 residues longer than in trypsin and trypsinogen, and is 5 residues shorter than in thrombin (supplemental Figure 1). Despite the difference in the sequence, the 60 loop in FXIIa exhibits a main-chain conformation similar to the corresponding loops in HGFA, tPA, and uPA.
The 99 loop of β-FXIIa has a β-hairpin conformation like other related serine proteases, and bears the greatest resemblance to the corresponding loops with the same length in trypsin and tPA. However, this loop is more hydrophobic than in trypsin and tPA and, at its most exposed part, there are Ser95-Pro96-Val97 residues in FXIIa instead of Asn95-Ser96-Asn97 in trypsin or Asp95-Asp96-Asp97 in tPA. The side chain of Phe99 points toward the active site, and similar to trypsin and FXa, this residue could partially block the S2 subsite.26,27 Based on simple modeling using the structure noncovalent complex between active HGFA and the KD1 domain from its cognate HAI-1 inhibitor, the P3 Lys of the FXIIa substrate (in this case, we used the FXI sequence from P3 to P4′; Table 2) projects toward the 99 loop of β-FXIIa and its side chain could form hydrogen bonds with the carbonyl atoms of Val97 and Ser98 (Figure 2C).
Sequences around the scissile peptide bonds of the β-FXIIa substrates FXI and kallikrein and of the β-FXIIa endogenous inhibitors serpin C1, antithrombin, and PAI-1
| . | P4 . | P3 . | P2 . | P1 . | P1′ . | P2′ . | P3′ . | P4′ . | P5′ . |
|---|---|---|---|---|---|---|---|---|---|
| FXI | Ile | Lys | Pro | Arg | Ile | Val | Gly | Gly | Thr |
| Plasma kallikrein | Thr | Ser | Thr | Arg | Ile | Val | Gly | Gly | Thr |
| Serpin C1 | Ser | Val | Ala | Arg | Thr | Leu | Leu | Val | Phe |
| Antithrombin | Ile | Ala | Gly | Arg | Ser | Leu | Asn | Pro | Asn |
| PAI-1 | Val | Ser | Ala | Arg | Met | Ala | Pro | Glu | Glu |
| . | P4 . | P3 . | P2 . | P1 . | P1′ . | P2′ . | P3′ . | P4′ . | P5′ . |
|---|---|---|---|---|---|---|---|---|---|
| FXI | Ile | Lys | Pro | Arg | Ile | Val | Gly | Gly | Thr |
| Plasma kallikrein | Thr | Ser | Thr | Arg | Ile | Val | Gly | Gly | Thr |
| Serpin C1 | Ser | Val | Ala | Arg | Thr | Leu | Leu | Val | Phe |
| Antithrombin | Ile | Ala | Gly | Arg | Ser | Leu | Asn | Pro | Asn |
| PAI-1 | Val | Ser | Ala | Arg | Met | Ala | Pro | Glu | Glu |
All atoms of the 140-autolysis loop, residues 143-152 except the side-chain atoms of Glu150, are well defined in the final 2Fo-Fc density map. As in HGFA and t-PA, this loop is exposed to the solvent, and its main- and side-chain atoms have a relatively high average temperature factor of 88 Å2 (the mean isotropic B factor for all atoms of the protein molecule is 58.0 Å2 for the compound 1–β-FXIIa complex; Table 1), suggesting that the 3 acidic residues Glu146, Glu149, and Glu150 might be involved in specific interactions with substrates, inhibitors, and cofactors. The side chain of Tyr151, similar to HGFA, trypsin, and t-PA, protrudes into the S2′ site (Figure 2C).
The 186 loop (Gly184-Gly188) of FXIIa is close to the entrance frame of the S1 pocket (Gly221-Pro225) and has the same length as in HGFA, trypsin, and uPA, 2 residues longer than in chymotrypsinogen, and 6 residues shorter than in tPA (supplemental Figure 1). The conformations of the loop main-chain residues 184-188 in human FXIIa, HGFA, and bovine trypsin are well superimposed within the RMSD value of 1.0 Å for their Cα residues. At the C terminus of the loop, there are main-chain hydrogen bonds between Asp189-Ala190 and Val16-Val17 located at the N terminus of the β-FXIIa protease domain.
There is a deletion at position 218, and an insertion behind Cys220 (Asp221A) in the β-FXIIa molecule, in common with trypsin, tPA, and HGFA, so this surface 210-220 segment is exposed to the solvent and is part of the top edge of the active site (Figure 1B). In contrast to trypsin, tPA and HGFA, but similar to coagulation factor Xa and thrombin, there is an insertion of 3 residues (205A-205B-205C) behind residue 205; this results in a bulged surface loop located opposite to the active site and in salt bridges between this loop and a few residues of the heavy chain remnant.
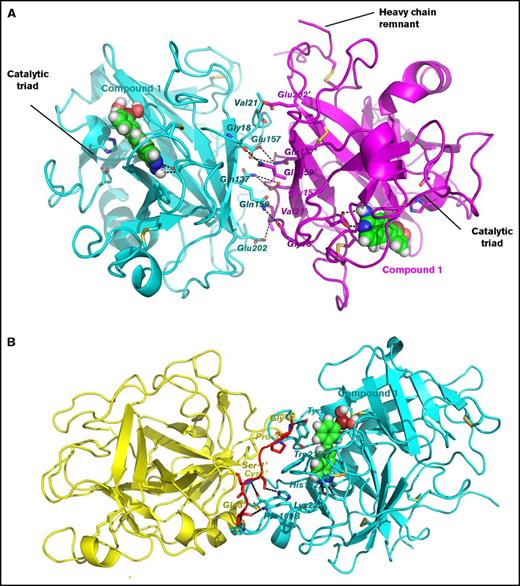
Although β-FXIIa exists as a monomer in solution under physiological conditions, extensive intermolecular contacts between human FXIIa protease domains in the crystal packing were observed (Figure 3). However, there is no biological or biochemical evidence that dimer or multimer formations have potential relevance for FXIIa functions.
Two distinct intermolecular contacts between symmetry related molecules of β-FXIIa protease domain in the crystal packing. (A) First extensive interface. Ribbon representation of 2 protease domains shown in cyan and magenta. Contacting residues are labeled and shown as sticks. The residues of the neighboring molecule are denoted by the “′” mark. Salt bridges and hydrogen bonds are depicted by black dotted lines. Compound 1, buried in the active site, is shown as balls. Positions of the catalytic triad for both molecules and the heavy chain remnant for one molecule (on the right) are indicated. (B) Second contact region. The same as in panel A except the heavy chain remnant of the β-FXIIa molecule on the left (yellow cartoon) is shown in red.
Two distinct intermolecular contacts between symmetry related molecules of β-FXIIa protease domain in the crystal packing. (A) First extensive interface. Ribbon representation of 2 protease domains shown in cyan and magenta. Contacting residues are labeled and shown as sticks. The residues of the neighboring molecule are denoted by the “′” mark. Salt bridges and hydrogen bonds are depicted by black dotted lines. Compound 1, buried in the active site, is shown as balls. Positions of the catalytic triad for both molecules and the heavy chain remnant for one molecule (on the right) are indicated. (B) Second contact region. The same as in panel A except the heavy chain remnant of the β-FXIIa molecule on the left (yellow cartoon) is shown in red.
Inhibitor binding
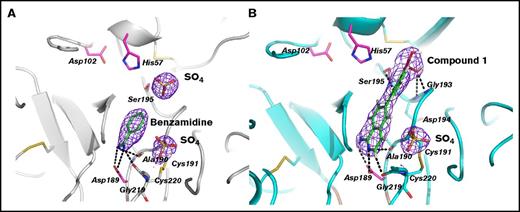
The β-FXIIa specificity pocket bordered by segments Ile213-Cys220, Asp189-Ser195, Pro225-Thr229, and disulfide bond Cys191-Cys220 is practically identical to that of other active trypsin-like serine proteases. Two different small compound inhibitors in the crystal complex structures solved in this work are clearly defined by electron density in the enzyme primary site, with Asp189 positioned at the bottom of the pocket to allow salt bridge formation with the basic group of the P1 moiety. The basic benzamidine molecule in the β-FXIIa–benzamidine complex is sandwiched between main-chain segments Trp215-Gly216 and Cys191-Gln192, which is very similar, for instance, to the corresponding trypsin complex (PDB code 3MFJ), except for a 22° rotation around its long molecular axis (Figure 4A).27 The amidinium group of the inhibitor makes a symmetric salt bridge with Asp189, also forming 2 salt bridges to the carbonyl oxygens of Cly219 and Ala190.
Binding interactions of small molecule inhibitors to β-FXIIa. (A) Close-up view of the inhibitor binding area in the benzamidine complex (gray ribbon). The benzamidine molecule (green sticks) bound to the S1 site, the residues defining the interaction with the inhibitors, the residues of the catalytic triad, and the bound phosphate ions (yellow) are shown as sticks. The electron density maps (blue) around the bound inhibitor and the ion are countered at 1σ. (B) The compound 1–β-FXIIa complex. The same view as in panel A.
Binding interactions of small molecule inhibitors to β-FXIIa. (A) Close-up view of the inhibitor binding area in the benzamidine complex (gray ribbon). The benzamidine molecule (green sticks) bound to the S1 site, the residues defining the interaction with the inhibitors, the residues of the catalytic triad, and the bound phosphate ions (yellow) are shown as sticks. The electron density maps (blue) around the bound inhibitor and the ion are countered at 1σ. (B) The compound 1–β-FXIIa complex. The same view as in panel A.
Compound 1 binds to the FXIIa primary site in an extended conformation and its aminoisoquinoline group interacts productively with Asp189 at the bottom of the S1 pocket (distances of 2.9 Å and 3.5 Å between 2 carboxylate oxygens and the amino group; Figure 4B). In addition, the other electrostatic interactions in the S1 pocket include the hydrogen bonds between the carbonyl oxygens of Gly219 and Ala190 and the amino group of the isoquinolone, but the Cly219-Cys220 peptide bond flips by 60° compared with the benzamidine structure (Figure 4A). At the top of the enzyme primary site, compound 1 displaces the sulfate ion found in the benzamidine structure; the acid group of the inhibitor forms 2 salt bridges with the main-chain nitrogens of Gly193 and Asp194. Continuous electron density links the inhibitor boron atom to the γ-O of Ser195, indicating the presence of a covalent linkage between the inhibitor and the enzyme.
Discussion
The discovery that both the zymogen and the activated form of FXII play significant physiological functions in vivo, such as the FXII growth factor activity or its essential contribution to thrombosis demonstrated in the f12 knockout mouse model, has generated great interest in the protein because it makes this coagulation factor an attractive target for new anticoagulants.6-9 However, structural data for the activated form of FXII have been unavailable. In this study, we present the first structural characterization of human plasma β-FXIIa in complex with 2 different inhibitors: benzamidine, the canonical basic inhibitor of serine proteases, and a small synthetic inhibitor that contains a boronic acid. The primary specificity site of β-FXIIa is practically identical to that of HGFA, tPA, FXa, and thrombin, and there are only 2 differences in the FXIIa primary pocket compared with uPA, chymotrypsin, and trypsin where residues Ala190 and Ile213 are replaced by Ser and Val, respectively (supplemental Figure 1). Therefore, like HGFA, thrombin, tPA, and the other related coagulation protease FXa, the primary pocket of β-FXIIa is slightly less polar. This could explain the FXIIa preference for substrate and cognate inhibitors with P1 Arg, as had been previously observed.28,29 Another structural feature of β-FXIIa includes the restriction of the S2 subsite by the imposing side chain of Tyr99 similar to thrombin and FXa (Figure 2C), explaining the preference of β-FXIIa for small residues at the P2. This argument is in agreement with the solution-phase fluorogenic peptide microarrays and single substrate studies that report the preference for Thr, Ser, and Gly in the P2 position, Met and Gln in the P3, and disfavor Glu at the P3.30,31 These data are also consistent with the physiological substrates of FXIIa, FXI zymogen, and PPK, where the sequences around the scissile bonds are Lys-Pro-Arg-Ile and Ser-Thr-Arg-Ile, respectively (Table 2).
It is assumed that serpins, inhibitors of serine proteases, interact with their cognate proteases via their reactive center loops (RCLs), which adopt a canonical conformation from P4 to P3′ and make extensive contacts from both backbone and side chains to residues in proteases from P5 to P6′.32 With the exception of RCL interactions, there may be extensive interactions between serpin exosites and surface loops surrounding the enzyme active site in the encounter serpin-protease complexes.33 Serpin C1 esterase inhibitor is cognate inhibitor of β-FXIIa, and accounts for 92% of FXIIa inhibition in plasma by a serpin.10 There are several structural models available for this serpin where the RCL is not ordered, making these C1INH structures unusable for modeling. PAI-1 is a trivial inhibitor of activated FXII.34 Therefore, it could be possible to obtain a reliable structural model for polypeptide substrate-binding interactions based on a structural comparison between the crystal structure of β-FXIIa solved in this work and the available tPA–PAI-1 encounter complex structure.35 Our simple modeling results provide a suitable model for this substrate interaction geometry of β-FXIIa (supplemental Figure 3A-B). There are also interactions between serpin exosites and protease surface loops; side chains of Thr205, Lys207, and Arg271 of PAI-1 might form salt bridges with Gln192, Gln60, and Asp60A of β-FXIIa, respectively, similar to those in the tPA-PAI-1 encounter complex (supplemental Figure 3B).35
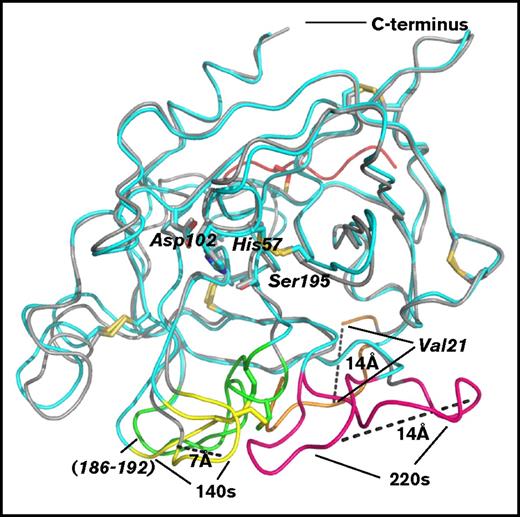
Figure 5 represents optimal superposition of the crystal structure of β-FXIIa onto that of the FXIIa zymogen13 and can provide some insight into an examination of the FXII activation mechanism. The structure of the apo-form (nonbound with any ligands) of human FXII light chain (FXIIc, PDB code 4XDE, Val16-Ser244) lacking 9 residues of the heavy chain remnant revealed a typical zymogen conformation for the protease.13 The largest conformational differences between 2 catalytic domains are found in the autolysis loop (residues 16-26), in the activation domain (residues 186-192), and in the adjacent surface 140 and 220 loops; in particular, the first ordered N-terminal residue Val21 in the zymogen structure is 14 Å away from its position in the activated enzyme, the Val16-Leu20 segment is completely missing in the zymogen, which results in the lack of an oxyanion hole, whereas the largest differences between the corresponding C-α atoms are 7 Å and 14 Å for the residues in the 140 and 220 loops, respectively (Figure 5). The main-chain conformations of other regions and the residues of the catalytic triad superimpose well with the RMSD value of 0.9 Å for 207 C-α atoms of the catalytic domains, excluding the above-mentioned labile regions.
Structural comparison of the catalytic domains of β-FXIIa and FXIIc. Superposition of the β-FXIIa–compound 1 (cyan) reported in this study and the zymogen form of the FXII catalytic domain (FXIIc, residues 16-244; PDB code 4XDE, gray cartoon). The greatest conformational changes between 2 structures are indicated by dashed lines.
Structural comparison of the catalytic domains of β-FXIIa and FXIIc. Superposition of the β-FXIIa–compound 1 (cyan) reported in this study and the zymogen form of the FXII catalytic domain (FXIIc, residues 16-244; PDB code 4XDE, gray cartoon). The greatest conformational changes between 2 structures are indicated by dashed lines.
Incompetent conformation of the protease domain of serine proteases was observed for the apo-form of HGFA expressed as a recombinant protein with 2 chains covalently linked together by a disulfide bond, whereas the same 2-chain construct in complex with the Kunitz domain inhibitor was expressed as an active enzyme.25 Such conformational plasticity is also observed for other proteases, including thrombin activity regulation by sodium atoms and partial transformation of the inactive trypsinogen into a catalytically active enzyme in the presence of Ile-Val dipeptides without activation cleavage or conformation transition observed for apo-prostatin and prostatin.36,37
Here, we assume that the zymogen conformation of FXIIc could be explained by such conformational diversity. The presence of the heavy chain remnant in the protein construct seems not to be one of the main requirements for expression of the active form of FXII because other serine proteases, such as trypsin, kallikrein, and FXIa, lacking the short peptide segments covalently linked with the catalytic domain, have been crystallized in the canonical conformation.20,37,38 Moreover, as was discussed earlier in this section, the HGFA construct having the remnant segment has been crystallized in both competent (in complex with an inhibitor) and incompetent (the apo-enzyme with no inhibitor bound) forms.25 Also, the crystal structure of β-FXIIa, where the remnant chain does not make any interactions with the autolysis loop to stabilize the active conformation, supports this argument. Furthermore, in the zymogen-like crystal structure of FXIIc determined by Pathak et al,13 an interaction between side chains of Arg73 and Asp194 was observed. The Asp194 is well known to play a critical role in switching between the activated and inactive forms of trypsin-like serine proteases. Our structural study of β-FXIIa in complex with different inhibitors revealed the canonical interaction between the Asp194 side chain and the N-terminal Val16.
In conclusion, a high-quality structure of human plasma β-FXIIa, which likely contributes to different pathological diseases via its biological functions, is presented. The new structural features of the active site cleft, substrate-binding sites, and the surface loop conformations of β-FXIIa may be a foundation for an efficient structure-based drug design platform targeting the FXIIa-driven plasma contact activities.
The full-text version of this article contains a data supplement.
Acknowledgment
The authors are grateful to the staff at the LS-CAT ID beamline of the Advanced Proton Source, Argonne National Laboratory, Lemont, IL, where data were collected.
Authorship
Contribution: A.D. designed the crystallization experiments, analyzed data, solved crystal structures, and wrote the manuscript; A.S., Z.L., and H.S. conducted biochemical experiments; C.Y. synthesized compound 1; M.T.F. was responsible for oversight of the project; J.R.P. analyzed experimental data and wrote the manuscript; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: A.D. and M.T.F. are Shamrock Structures, LLC employees. A.S., C.Y., Z.L., H.S. and J.R.P. are Global Blood Therapeutics employees.
Correspondence: Alexey Dementiev, Shamrock Structures, 1440 Davey Rd, Woodridge, IL 60517; e-mail: adementiev@shamrockstructures.com; and James R. Partridge, GBT, 400 East Jamie Ct, Suite 101, South San Francisco, CA 94080; e-mail: jpartridge@globalbloodtx.com.

![Figure 1. Crystal structure of human plasma β-FXIIa. (A) Stereo ribbon diagram (cyan) of β-FXIIa in complex with covalent compound 1 inhibitor in “standard” orientation. Compound 1 is shown in green sticks. The side chains of key β-FXIIa residues are shown as magenta sticks: His57, Asp102, and Ser195 of the catalytic triads, Asp189 at the base of the S1 specificity pocket. The side chains of N-glycosylated Asn74 and NacGlc-disaccharide are shown in cyan sticks. Some surface loops discussed in the text are labeled and shown in pink. The heavy chain remnant of β-FXIIa is shown in red. Hydrogen bonds are indicated by black dotted lines. (B) The structure of β-FXIIa in complex with noncovalent benzamidine inhibitor is represented as in panel A. Six disulfide bonds are indicated and are shown in yellow. (C) Superposition of the β-FXIIa (benzamidine [magenta] and β-FXIIa) compound 1 (cyan) crystal structures. The catalytic domains are superimposed by aligning the Cα atoms of residues 16-244 and are presented as cartoon diagrams. Benzamidine (orange), compound 1 (green), and residues of the catalytic triad are shown in a stick representation.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/2/5/10.1182_bloodadvances.2018016337/3/m_advances016337f1.jpeg?Expires=1767698205&Signature=oNa7HncPTABqAu4wPhMi10hO8lH1GZSeo6d4Qf3AtQisSLKSOi2FQ2zWpLnGfHMfM0D6c8KSklqdeyKmbsmf2J26vI7TY-tTzZBFIvmmAkCapNGMNYRbzf6GhL71rZZS6-GYx2R7GOObN2o5O5IN3XWXVe3EoBBpSSFkusdHSXJIjHaHEmjm5Lu~azT26mBMMl-dbXYjLvQH9o0LxSpFg2W-WoOBXkPsv-45vRCQWZK0IS75kgmMWxLJPypdHM0gIEQfMKfV9qgZuWVQjiF5LZ41BjyEO0eUcdZgJWglZRCu58mJL~TKBMUA2GMN8KCRFf853~HunO8HOI4C9IIdTg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)