Key Points
Tissue-specific minor histocompatibility antigens can be predicted through computational analysis of donor and recipient genotyping data.
Targeted mass spectrometry and tetramer analysis confirmed a computationally predicted, public leukemia antigen derived from GRK4.
Abstract
T-cell responses to minor histocompatibility antigens (mHAs) mediate both antitumor immunity (graft-versus-leukemia [GVL]) and graft-versus-host disease (GVHD) in allogeneic stem cell transplant. Identifying mHAs with high allele frequency, tight binding affinity to common HLA molecules, and narrow tissue restriction could enhance immunotherapy against leukemia. Genotyping and HLA allele data from 101 HLA-matched donor-recipient pairs (DRPs) were computationally analyzed to predict both class I and class II mHAs likely to induce either GVL or GVHD. Roughly twice as many mHAs were predicted in HLA-matched unrelated donor (MUD) stem cell transplantation (SCT) compared with HLA-matched related transplants, an expected result given greater genetic disparity in MUD SCT. Computational analysis predicted 14 of 18 previously identified mHAs, with 2 minor antigen mismatches not being contained in the patient cohort, 1 missed mHA resulting from a noncanonical translation of the peptide antigen, and 1 case of poor binding prediction. A predicted peptide epitope derived from GRK4, a protein expressed in acute myeloid leukemia and testis, was confirmed by targeted differential ion mobility spectrometry-tandem mass spectrometry. T cells specific to UNC-GRK4-V were identified by tetramer analysis both in DRPs where a minor antigen mismatch was predicted and in DRPs where the donor contained the allele encoding UNC-GRK4-V, suggesting that this antigen could be both an mHA and a cancer-testis antigen. Computational analysis of genomic and transcriptomic data can reliably predict leukemia-associated mHA and can be used to guide targeted mHA discovery.
Introduction
Minor histocompatibility antigens (mHAs) are peptide epitopes derived from genetic differences (usually single-nucleotide polymorphisms [SNPs]) expressed by a transplant recipient, but not the donor, which are presented by recipient major histocompatibility complex (MHC) molecules and recognized by T cells bearing cognate T cell receptors.1-3 These antigens are key targets of T cell responses following HLA-matched allogeneic stem cell transplantation (allo-SCT), playing a role in the pathogenesis of acute graft-versus-host disease (GVHD) and prevention of disease relapse via the graft-versus-leukemia (GVL) effect.4,5 Multiple mHA have been discovered and several clinical trials of mHA vaccines or cellular immunotherapies have been performed.6-8 Identification of leukemia or hematopoietic tissue-restricted mHA is especially promising as they may drive a beneficial GVL effect without GVHD.
We report a computational mHA prediction method that combines donor/recipient genotyping data with RNA sequencing data from reference human tissue and leukemia samples to predict mHAs with high binding affinity to HLA that are expressed in specific tissues. We used the method to predict tissue-restricted mHAs in a cohort of 101 patients who had undergone allo-SCT for myeloid neoplasms and had been genotyped for 13 917 nonsynonymous coding single-nucleotide polymorphisms (cSNPs). We tested our method’s ability to confirm known mHAs and predict novel GVL mHAs. We discovered a new leukemia-associated antigen by performing targeted mass spectrometry coupled to differential ion mobility spectrometry (DIMS-MS), followed by detection of antigen-specific T-cell populations using peptide/MHC tetramers.
Materials and methods
Patient cohort and determination of genetic variation in allo-SCT recipients
A cohort of 101 myeloid leukemia (acute myeloid leukemia [AML], chronic myeloid leukemia [CML], myelodysplastic syndrome [MDS], and myeloproliferative neoplasms [MPN]) patients who underwent HLA-matched allo-SCT at MD Anderson Cancer Center was analyzed under an institutional review board–approved protocol (LAB99-062).9 Patients and donors were genotyped at 13 917 common cSNPs using Illumina NS-12 microarrays. The cSNP genotyping data are hosted by the Vincent laboratory and are publically available at https://unclineberger.org/vincent/resources. Genetic predictions of a minor mismatch (gMMs) were defined for every cSNP in a DRP where the recipient possessed an allele not found in the corresponding donor DNA. Minor antigen mismatches were defined as predicted peptides present in the recipient, but not the donor. Overall survival (OS) was defined as survival time from time of allo-SCT. GVHD positive was defined as acute GVHD grades II to IV or extensive chronic GVHD. Relapse was defined as recurrent disease following allo-SCT regardless of subsequent therapy.
Isolation of protein-coding differences and enumeration of potentially immunogenic peptides
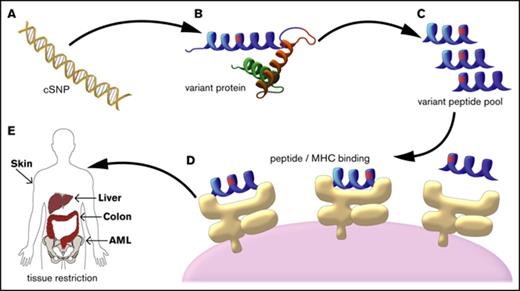
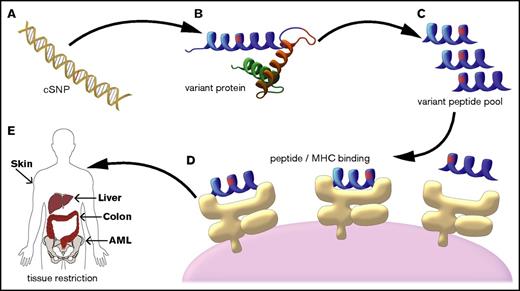
The ENSEMBL variant effect predictor was used in combination with the UCSC Genome Browser database (GRCh37, UCSC hg19) to map all cSNPs with allelic amino acid changes to their associated open reading frames. The resulting 11 172 (out of 13 917 cSNPs on the NS-12 array) possible common cSNPs were translated into all possible 8-, 9-, 10-, 11- (for class I HLA peptide epitope prediction), 16-, 20-, and 24-mer (for class II HLA peptide epitope prediction) peptides on all applicable transcripts using custom software (Figure 1).
mHA prediction algorithm schema. (A) The alleles at cSNPs for all 101 DRPs in the patient cohort were determined using the Illumina NS-12 array platform (n = 13 917 cSNPs). (B) Each allele for each cSNP was mapped to the reference coding transcriptome. (C) The peptide sequences for all 8-, 9-, 10-, 11-, 16-, 20-, and 24-mer amino acid sequences that contained the allele of interest were computationally generated (n = 2 189 712 amino acid sequences covering both alleles for all 11 172 cSNPs). (D) All peptides that could be mHAs for a given DRP (ie, one allele was present in the recipient, but not the donor) were tested for predicted binding affinity to the DRP’s HLA types. (E) All predicted mHAs were classified as GVL or GVHD associated based upon the tissue expression of the source protein for the mHA.
mHA prediction algorithm schema. (A) The alleles at cSNPs for all 101 DRPs in the patient cohort were determined using the Illumina NS-12 array platform (n = 13 917 cSNPs). (B) Each allele for each cSNP was mapped to the reference coding transcriptome. (C) The peptide sequences for all 8-, 9-, 10-, 11-, 16-, 20-, and 24-mer amino acid sequences that contained the allele of interest were computationally generated (n = 2 189 712 amino acid sequences covering both alleles for all 11 172 cSNPs). (D) All peptides that could be mHAs for a given DRP (ie, one allele was present in the recipient, but not the donor) were tested for predicted binding affinity to the DRP’s HLA types. (E) All predicted mHAs were classified as GVL or GVHD associated based upon the tissue expression of the source protein for the mHA.
RNA-seq of AML samples
Total RNA was isolated using QIAGEN RNeasy kits, and complementary DNA sequencing libraries were prepared using the TruSeq mRNA-Seq Sample Preparation Protocol (Illumina). Paired-end sequencing (76 cycles/end) was performed on an Illumina HiSeq 2000 at the UNC High Throughput Sequencing Facility. RNA-sequencing (RNA-seq) data were processed as previously described.10-12
Determination of the allo-SCT donor and recipient HLA types
Donor and recipient HLA typing was performed at the MD Anderson HLA-typing Laboratory. For patients with missing allele-level data at an HLA locus for subtype (55% of loci) or type (1% of loci), the data were imputed as the most common subtype or type for that locus, respectively. Global HLA frequencies were determined from 1000 Genomes Project data.13 Published 1000 Genomes Project sample HLA types (n = 1277) were confirmed and updated with PHLAT calls for all 1000 Genomes Project samples with paired-end Illumina sequencing runs meeting PHLAT requirements of average coverage depth >50 (n = 1916).14
Identification of peptide fragments with strong HLA binding and GVL/GVH mHA prediction based on tissue expression
All possible 8- to 11-, 16-, 20-, and 24-mer peptides resulting from genetic variation in each allo-SCT recipient elaborated by crossing genotype data with the human reference transcriptome (Gencode GRCh37.p13) were screened in silico for predicted peptide/HLA dissociation constants (Kd) <500 nM against each patient’s individual HLA alleles (A, B, C, DRB1) using NetMHCpan and NetMHCIIpan.15,16 GVL mHAs were defined as peptides predicted to bind any patient HLA alleles with peptide source protein messenger RNA (mRNA) expression >50 transcripts per million (TPM) in AML, normal bone marrow or testis. GVL restricted mHA were defined as GVL mHAs with < 5 TPM in skin, hepatobiliary and colonic tissues. Gene expression in testis was included to broaden filtering parameters to include possible cancer-testis antigens (CTAs). Graft-versus-host (GVH) mHAs were defined as peptides predicted to bind any patient HLA alleles with peptide source protein mRNA expression >50 TPM in normal skin, hepatobiliary, or colonic tissues. mRNA transcript expression levels for mean AML samples (n = 8) were determined via RNA-seq, and mean mRNA expression levels for normal bone marrow, testis, skin, hepatobiliary, and colonic tissues (n = 4/7/3/3/7, respectively) were taken from Human Protein Atlas RNA-seq data. Fragments per kilobase of exon per million reads (FPKM) values were converted to TPM by dividing each FPKM value by the FPKM sum for all genes expressed in that tissue.
Tissue expression of GRK4 mRNA
Tissue-specific RNA was purchased (Ambion, Applied Stem Cell) or purified from de-identified primary human AML cells obtained from the UNC Hematologic Malignancies Tissue Procurement Facility using the QIAGEN RNeasy kit. Reverse-transcription polymerase chain reaction (RT-PCR) was performed on 100 ng of each RNA sample using the SuperScript III One-Step RT-PCR kit (Ambion).
Protein expression of GRK4
Human testis protein lysate was purchased from Abcam, and protein lysates were prepared from primary human AML cells using RIPA buffer. 50 μg of each lysate was loaded onto polyacrylamide gels (Bio-Rad) and run in Tris/glycine/sodium dodecyl sulfate buffer. Proteins were transferred to a PVDF membrane. GRK4 was probed using mouse anti-GRK4 (clone A-5, Santa Cruz Biotechnology) and detected using ECL. β-Actin was probed using anti-β-actin (Sigma).
Isolation of class I peptide epitopes
U937 cells transfected with HLA-A*02:01 (U937.A2) were genotyped at rs1801058 and found to be heterozygous. Approximately 2 × 108 U937.A2 cells were lysed and cleared by centrifugation. Anti-class I HLA antibody (W6/32) was added to the lysate and incubated on a rocker at 4°C. Antibody complexes were immunoprecipitated using protein G Sepharose resin. The resin was washed and loaded onto columns, and peptide/HLA complexes were released with 10% acetic acid. Glacial acetic acid was then added drop-wise until pH 2.5. The mixture was filtered through 5-kDa filters to isolate ∼1-kDa peptides.17,18
DIMS-MS/MS targeted MS for UNC-GRK4-V
Pure UNC-GRK4-V peptide was diluted to 100 nM in water/acetonitrile/formic acid (50/50/0.1%). The tandem mass spectrometry (MS/MS) spectrum and EC (compensation voltage) for optimal transmission of UNC-GRK4-V were recorded.19,20 The peptide epitope pool was desalted using a ZipTip (C18, 0.6 μL) and directly nanoelectrosprayed into the DIMS-MS with the EC set for optimal transmission (86 V/cm). The MS/MS spectrum of the UNC-GRK4-V peptide from the epitope pool was compared with that of the pure UNC-GRK4-V peptide, and a Fit score was calculated based on the agreement between the reference and experimental UNC-GRK4-V MS/MS spectra using Bruker DataAnalysis.
Synthesis of UNC-GKR4-V/HLA-A*02:01 tetramers
HLA-A*02:01 containing a biotinylation site and β2 microglobulin were overexpressed in Escherichia coli and purified by fast protein liquid chromatography as previously reported.21 HLA-A*02:01, β2 microglobulin, and UNC-GRK4-V peptide were combined to make UNC-GRK4-V/HLA-A*02:01 monomers. Purified monomers were biotinylated by BirA ligase and complexed with allophycocyanin (APC)/avidin, yielding UNC-GRK4-V/HLA-A*02:01 tetramers.
Detection of UNC-GRK4-V–specific CD8+ T cells in post-SCT samples
Post-SCT peripheral blood mononuclear cell (PBMC) samples from HLA-A*02:01 expressing patients and donors were genotyped for the rs1801058 cSNP under institutional review board–approved protocols (Laboratory 99-062 for MD Anderson samples and LCCC-0824 for UNC samples). PBMCs were incubated with the following fluorescent markers: DAPI live/dead stain, CD4 fluorescein isothiocyanate (FITC), CD14-FITC, CD16-FITC, CD19-FITC (dump channel), CD8 phycoerythrin, and tetramer-APC (either negative tetramer or UNC-GRK4-V/HLA-A*02:01 tetramer). All samples were run on a MacQuant analytical flow cytometer with subsequent analysis using FlowJo software.
Results
Patient characteristics
Characteristics of the 101 patients are given in Table 1. Median patient age was 48 years, and 63% of patients were male. Disease types were AML (61 patients), CML (25 patients), MDS (14 patients), and MPN (1 patient). 48 patients were transplanted either in complete remission (CR) or chronic phase (CP). 76 patients underwent myeloablative conditioning, and 71 grafts were from HLA-matched related donors (MRDs). All 29 matched unrelated donor (MUD) patients received rabbit ATG as part of their conditioning therapy. There were nonstatistically significant differences in 5-year OS rates according to disease type: 58% for CML, 41% for AML and 35% for MDS (Figure 2A). However, OS was different based upon disease status at time of SCT (Figure 2B) and patient age (supplemental Figure 1A). There were no differences in OS based upon donor type, conditioning regimen (Figure 2C-D), SCT source, gender, or gender mismatch (supplemental Figure 1B-D). 38 patients developed grade II to IV acute GVHD (32% of MRD and 52% of MUD), and 33 developed extensive chronic GVHD (31% of MRD and 38% of MUD). 34 patients relapsed after SCT (36% of MRD, 28% of MUD, 31% of MAC, and 40% of reduced-intensity conditioning [RIC]).
Patient characteristics
| Characteristic . | No. of patients (N = 101) . |
|---|---|
| Age (range), y | 48 (21-72) |
| Sex, n (%) | |
| Male | 64 (63) |
| Female | 37 (37) |
| Disease, n (%) | |
| AML | 61 (60) |
| CML | 25 (25) |
| MDS | 14 (14) |
| MPN | 1 (1) |
| Disease stage, n (%) | |
| CR/CP | 48 (48) |
| Not in CR/CP | 53 (52) |
| Conditioning intensity, n (%) | |
| Myeloablative | 71 (71) |
| Reduced intensity | 30 (29) |
| Donor type, n (%) | |
| HLA-matched related donor | 72 (72) |
| HLA-matched unrelated donor | 29 (28) |
| Graft source, n (%) | |
| Peripheral blood stem cells | 68 (67) |
| Bone marrow | 33 (33) |
| Sex mismatch, n (%) | |
| Female donor into male recipient | 28 (28) |
| Male donor into female recipient | 19 (19) |
| Characteristic . | No. of patients (N = 101) . |
|---|---|
| Age (range), y | 48 (21-72) |
| Sex, n (%) | |
| Male | 64 (63) |
| Female | 37 (37) |
| Disease, n (%) | |
| AML | 61 (60) |
| CML | 25 (25) |
| MDS | 14 (14) |
| MPN | 1 (1) |
| Disease stage, n (%) | |
| CR/CP | 48 (48) |
| Not in CR/CP | 53 (52) |
| Conditioning intensity, n (%) | |
| Myeloablative | 71 (71) |
| Reduced intensity | 30 (29) |
| Donor type, n (%) | |
| HLA-matched related donor | 72 (72) |
| HLA-matched unrelated donor | 29 (28) |
| Graft source, n (%) | |
| Peripheral blood stem cells | 68 (67) |
| Bone marrow | 33 (33) |
| Sex mismatch, n (%) | |
| Female donor into male recipient | 28 (28) |
| Male donor into female recipient | 19 (19) |
Patient cohort survival outcomes. (A) Two-year OS was 55%, and there was no significant difference in OS based upon disease type (AML, MDS, or CML). (B) There was a statistically significant (P = .0010) difference in OS based upon disease status at time of SCT (25% vs 65%, not in CR/CP vs CR/CP). (C-D) There were no significant differences in OS based upon conditioning type (myeloablative conditioning [MAC] or RIC) or donor source (MRD or MUD).
Patient cohort survival outcomes. (A) Two-year OS was 55%, and there was no significant difference in OS based upon disease type (AML, MDS, or CML). (B) There was a statistically significant (P = .0010) difference in OS based upon disease status at time of SCT (25% vs 65%, not in CR/CP vs CR/CP). (C-D) There were no significant differences in OS based upon conditioning type (myeloablative conditioning [MAC] or RIC) or donor source (MRD or MUD).
Number of predicted mHAs is not associated with patient clinical outcomes
We tested all in silico generated peptides derived from DRP genetic variants for predicted binding to each HLA represented in the patient cohort. Any peptide with a predicted Kd < 500 nM was considered a possible mHA.15,22-28 The number of mHAs for each DRP in the patient cohort was calculated based upon the allelic cSNP differences between the donor and recipients. To avoid bias potentially introduced by inadequate performance of the peptide/MHC-binding prediction algorithm (netMHCpan) for some HLA alleles with insufficient training data, we analyzed the sensitivity and specificity of netMHCpan by HLA allele. Performance measures were highly correlated with the proportion of training data peptides that were “binders” and less so with the amount of training data for each allele (supplemental Figure 2A-D). We considered alleles with sensitivity ≥0.5 and specificity ≥0.7 as “verified” and used only these alleles for further analysis.
For each DRP, we calculated the number of predicted mHAs and normalized this value to the number of gMMs. The predicted number of mHAs per gMM, among class I HLA, ranged from 0.09 per gMM to 1.32 (Figure 3A), while the predicted number of mHAs per gMM for class II HLA ranged from 12.4 to 27.3 (supplemental Figure 6A).
Characteristics of mHAs by HLA type and donor relation. (A) There is a large difference in the number of peptides that can bind with high affinity to class I HLAs (0.09 to 1.32 per gMM). (B) MUD SCT is associated with roughly twice as many class I mHAs compared with MRD SCT. (C) Because of variable frequencies of peptide presentation by different HLA molecules, there is a greater variability in the mHAs compared with gMMs among both MRD and MUD patients. (D) There is a linear association between the number of GVL mHAs and GVHD mHAs. (E) There is no association between the number of GVL mHAs and relapse for MRD or MUD. (F) There is similarly no association between the number of GVH mHAs and GVHD.
Characteristics of mHAs by HLA type and donor relation. (A) There is a large difference in the number of peptides that can bind with high affinity to class I HLAs (0.09 to 1.32 per gMM). (B) MUD SCT is associated with roughly twice as many class I mHAs compared with MRD SCT. (C) Because of variable frequencies of peptide presentation by different HLA molecules, there is a greater variability in the mHAs compared with gMMs among both MRD and MUD patients. (D) There is a linear association between the number of GVL mHAs and GVHD mHAs. (E) There is no association between the number of GVL mHAs and relapse for MRD or MUD. (F) There is similarly no association between the number of GVH mHAs and GVHD.
To accommodate DRPs with different numbers of verified HLAs, numbers of mHAs associated with verified HLAs were divided by the number of verified HLA in the given DRP. Using these normalized values, we compared the number of predicted class I and class II mHAs among all 101 DRPs. There were twice as many class I mHA and 2.5-fold more class II mHAs in the MUD setting relative to MRD SCT (Figure 3B; supplemental Figure 6B). There was greater variability in the number of predicted class I mHAs compared with the variability in the number of gMMs in both MRD and MUD SCT (Figure 3C). The numbers of total of class I GVH mHAs and GVL mHAs were higher in MUD relative to MRD transplants, and there was a linear association between the number of GVL and GVH mHAs (Figure 3D).
We tested for associations between numbers of mHAs and clinical outcomes. Because of the large differences in numbers of mHAs between MRD and MUD SCT, analyses were conducted on these 2 groups separately. There was no association between numbers of HLA-A*02:01 restricted GVL mHAs and relapse (Figure 3E) or numbers of GVH mHAs and GVHD (Figure 3F). The same lack of association was also observed when all class I and class II mHAs were analyzed for the clinical outcomes of relapse, GVHD, and OS (supplemental Figures 5 and 6).
Previously confirmed mHAs are confirmed through bioinformatics analysis
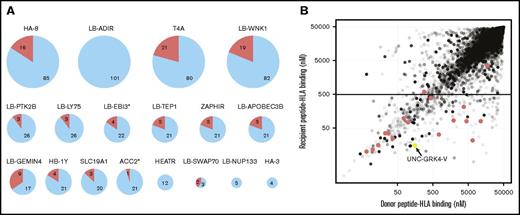
We tested our method’s ability to confirm previously reported mHAs summarized in the recent publication by Oostvogels et al.7 We identified 14 of the 18 mHAs that were potentially confirmable in our SNP array and patient cohort (Figure 4A). Two of the missed mHAs (LB-NUP133 and HA-3) were not identified in the peptide pool because they are expressed on low-prevalence HLAs, and there were no DRPs with the appropriate gMM. The mHA HEATR had a predicted HLA-binding affinity of 589 nM (slightly above our filtering threshold), and the mHA LB-ADIR is derived from an alternate open reading frame that was not represented in the Gencode reference transcriptome.3,29 We compared the predicted binding of the DRP “recipient” vs “donor” allelic variants for every 8- to 11-mer peptide, over all HLA types in the patient cohort, for the 12 cSNPs in the microarray that mapped to known class I mHA (a total of 20 926 DRP peptide pairs). The 12 cSNPs map to 17 mHAs, and of these 17, 15 were found in our peptide list with the recipient allele peptide having a Kd <500 nM (Figure 4B).
Confirmation of previously discovered mHAs. (A) Eighteen previously described HLA class I and class II mHAs were potentially identifiable in the patient genotyping data set by having the relevant cSNP contained on the NS-12 array and the appropriate HLA type contained in the patient population. For each pie chart, the total number represents the number of DRPs in the 101-patient cohort that expressed the appropriate HLA for the examined mHA. The red wedges represent the number of DRPs where the actual mHA could be presented (ie, the appropriate gMM existed in the DRP, and the actual peptide was contained in the total peptide pool). Of the 18 mHAs, 14 were successfully identified by the prediction algorithm. Two mHAs (LB-NUP133 and HA-3) were represented on rare HLAs, and no patients in the cohort had the appropriate gMMs to predict the mHA. For the other 2 mHAs not predicted, the peptide epitope from the mHA derived from HEATR has a predicted binding affinity of >500 nM, and the mHA LB-ADIR is derived from an alternative reading frame. (B) Twelve cSNPs from the SNP array data mapped to known class I mHAs that could be contained in the generated peptide data set. The HLA binding affinity for all 8- to 11-mer DRP peptide pairs to all class I HLA types contained in the patient data set for the 12 cSNPs is shown. A total of 20 926 DRP peptide pairs are shown with increasing shades of gray corresponding to the frequency of the peptide pair in the patient cohort. The 12 cSNPs have been shown to yield 17 mHA peptides that are represented by the red dots. Only 2 of the 17 mHAs fall out of our threshold Kd of 500 nM: the HEATR mHA and 1 confirmed mHA derived from LB-APOBEC3B. The UNC-GRK4-V peptide pair (see Figures 5 and 6) is also mapped (yellow dot).
Confirmation of previously discovered mHAs. (A) Eighteen previously described HLA class I and class II mHAs were potentially identifiable in the patient genotyping data set by having the relevant cSNP contained on the NS-12 array and the appropriate HLA type contained in the patient population. For each pie chart, the total number represents the number of DRPs in the 101-patient cohort that expressed the appropriate HLA for the examined mHA. The red wedges represent the number of DRPs where the actual mHA could be presented (ie, the appropriate gMM existed in the DRP, and the actual peptide was contained in the total peptide pool). Of the 18 mHAs, 14 were successfully identified by the prediction algorithm. Two mHAs (LB-NUP133 and HA-3) were represented on rare HLAs, and no patients in the cohort had the appropriate gMMs to predict the mHA. For the other 2 mHAs not predicted, the peptide epitope from the mHA derived from HEATR has a predicted binding affinity of >500 nM, and the mHA LB-ADIR is derived from an alternative reading frame. (B) Twelve cSNPs from the SNP array data mapped to known class I mHAs that could be contained in the generated peptide data set. The HLA binding affinity for all 8- to 11-mer DRP peptide pairs to all class I HLA types contained in the patient data set for the 12 cSNPs is shown. A total of 20 926 DRP peptide pairs are shown with increasing shades of gray corresponding to the frequency of the peptide pair in the patient cohort. The 12 cSNPs have been shown to yield 17 mHA peptides that are represented by the red dots. Only 2 of the 17 mHAs fall out of our threshold Kd of 500 nM: the HEATR mHA and 1 confirmed mHA derived from LB-APOBEC3B. The UNC-GRK4-V peptide pair (see Figures 5 and 6) is also mapped (yellow dot).
Confirmation of a computationally predicted leukemia-associated mHAs derived from GRK4
Our method applied to the patient cohort provided a list of 102 peptides with desirable properties for public, leukemia-associated mHAs: (1) presentation on a common HLAs; (2) high binding affinity; (3) expression in AML, but not in GVHD target organs; and (4) optimal allele frequencies to allow minor mismatches to be common. One of these predicted mHAs was linked to cSNP rs1801058, which is a polymorphism in the G-protein–coupled receptor 4 kinase (GRK4) gene. We predicted a recipient allele peptide, VLDIEQFSV (UNC-GRK4-V, HLA-A*02:01 Kd = 19.6 nM), with the C-terminal valine resulting from the T allele and a “donor” allele peptide, VLDIEQFSA (UNC-GRK4-A, HLA-A*02:01 Kd = 389.1 nM), with the C-terminal alanine resulting from the C allele. Of our 101 patients, 11 had the appropriate alleles for a gMM (ie, the donor was C/C and the recipient was either T/C or TT).
Gene expression databases indicated that GRK4 is highly expressed in testis with little to no expression in other normal tissues.30 RT-PCR on RNA from primary human AML, normal PBMCs, liver, colon, skin, and testis showed an amplicon product in all 3 primary AML samples and testis RNA, but no products in the other samples (Figure 5A). GRK4 was identified in 3 of 4 primary human AML lysates by western blotting (Figure 5B). In addition to AML, analysis of gene expression data from The Cancer Genome Atlas also identified increased GRK4 expression in several other cancers, including glioma and glioblastoma (supplemental Figure 4).
UNC-GRK4-V is expressed in AML. (A) GRK4 has limited tissue expression, with transcripts only detectable by RT-PCR in human AML and testis, but not in PBMCs, liver, colon, or skin. (B) Western blot confirms GRK4 protein expression in testis and 3 of 4 human AML samples. (C) Extracted ion chromograms using DIMS identifies the EC to allow the maximum signal from UNC-GRK4-V (m/z = 1049.5) into the MS using the pure standard (red trace), which was 86 V/cm. Extracted ion chromogram of m/z = 1049.5 in the U937.A2 cell epitope pool (blue trace) shows that other species with m/z = 1049.5 can be detected across a range of EC. (D) The fragmentation pattern for pure UNC-GRK4-V peptide was determined for comparison with results from the peptide pool. (E) Targeted MS was performed on the epitope pool by setting EC = 86 V/cm and fragmenting the parent ion with an m/z = 1049.5. The resulting MS/MS spectrum is virtually identical to that of the pure peptide.
UNC-GRK4-V is expressed in AML. (A) GRK4 has limited tissue expression, with transcripts only detectable by RT-PCR in human AML and testis, but not in PBMCs, liver, colon, or skin. (B) Western blot confirms GRK4 protein expression in testis and 3 of 4 human AML samples. (C) Extracted ion chromograms using DIMS identifies the EC to allow the maximum signal from UNC-GRK4-V (m/z = 1049.5) into the MS using the pure standard (red trace), which was 86 V/cm. Extracted ion chromogram of m/z = 1049.5 in the U937.A2 cell epitope pool (blue trace) shows that other species with m/z = 1049.5 can be detected across a range of EC. (D) The fragmentation pattern for pure UNC-GRK4-V peptide was determined for comparison with results from the peptide pool. (E) Targeted MS was performed on the epitope pool by setting EC = 86 V/cm and fragmenting the parent ion with an m/z = 1049.5. The resulting MS/MS spectrum is virtually identical to that of the pure peptide.
To confirm that the UNC-GRK4-V peptide is endogenously processed and presented on the cell surface by HLA-A*02:01, HLA-presented peptides were isolated from U937.A2 cells and analyzed by targeted MS using a custom built planar DIMS device coupled to a Bruker HCT ion trap mass spectrometer.19,20 DIMS is an ion-separation technique based on the principle that different gas phase ion species (even species of the same molecular weight) migrate on different trajectories through a chamber that has a rapidly oscillating electric field. By applying a fixed compensation field (EC), the device can be used as an ion filter to transfer ions of a specific trajectory into the mass spectrometer. We obtained pure UNC-GRK4-V peptide and measured the ion transmission into the mass spectrometer at various ECs (Figure 5C). Maximum transmission of UNC-GRK4-V occurred at an EC of 86 V/cm. The MS/MS spectrum of pure UNC-GRK4-V was obtained so the dominant product ion peaks could be determined (Figure 5D). The peptide epitopes obtained from U937.A2 cells were then directly electrosprayed (ie, no high-performance liquid chromatography [HPLC] was used) into the DIMS-MS with EC = 86 V/cm to filter out the many peptides which transmit at other ECs. An appropriate peak of mass-to-charge (m/z) = 1049.5 was detected with a fragmentation pattern that matched (fit = 927/1000) pure UNC-GRK4-V (Figure 5E; supplemental Methods).
Minor antigen responses to UNC-GRK4-V in post-SCT patients
We produced UNC-GRK4-V/HLA-A*02:01 tetramers and probed for expanded antigen-specific T-cell populations in post-SCT samples obtained from UNC (n = 4) and MD Anderson (n = 8). The alleles of rs1801058 for all donors and recipients were determined, and 3 scenarios were considered: (1) SCT with a gMM (ie, donor homozygous for the C allele and recipient expressing ≥1 copy of the T allele [n = 3]); (2) SCT with both the donor and recipient expressing at least 1 copy of the T allele, consistent with a cancer-testis/self-antigen response [n = 6]; and (3) the recipient not expressing UNC-GRK4-V (n = 3).
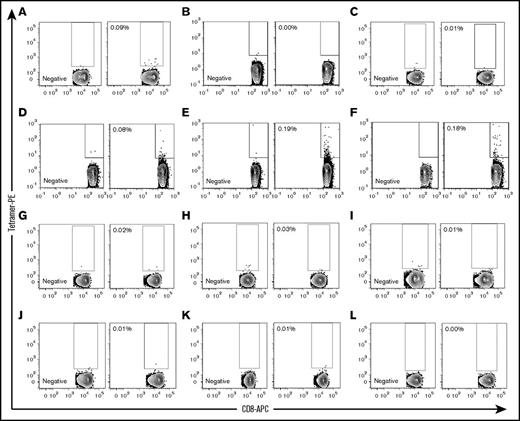
Using a negative tetramer for gating, we identified UNC-GRK4-V/HLA-A*02:01–specific T cells in 1 (Figure 6A) of the 3 predicted gMMs (Figure 6A-C) and 3 (Figure 6D-F) of the 6 cancer-testis/self-antigen samples (Figure 6D-I). None of the 3 UNC-GRK4-V nonexpressing patients showed a UNC-GRK4-V/HLA-A*02:01tetramer response (Figure 6J-L).
UNC-GRK4-V–specific T cells are present post-SCT. UNC-GRK4-V–specific T-cell populations were tested in 12 HLA-A*02:01–expressing AML patients who had undergone allo-SCT. Negative tetramers were used for each sample to define tetramer-positive gates for each patient. (A-C) Three patients in DRPs with gMMs for UNC-GRK4-V were tested, with 1 patient (A, alive 29 months post-SCT with chronic GVHD) showing an expanded tetramer-positive population. (D-I) Six patients in DRPs where both the recipient and donor carried the UNC-GRK4-V allele were tested, and 3 patients (D, alive 75 months post-SCT with mild chronic GVHD; E, died of relapsed AML 21 months post-SCT; F, alive 84 months post-SCT without GVHD) showed evidence of a tetramer-positive population. (J-L) Three patients in DRPs in which the patient did not carry the UNC-GRK4-V allele were tested, and no samples showed a tetramer-positive population.
UNC-GRK4-V–specific T cells are present post-SCT. UNC-GRK4-V–specific T-cell populations were tested in 12 HLA-A*02:01–expressing AML patients who had undergone allo-SCT. Negative tetramers were used for each sample to define tetramer-positive gates for each patient. (A-C) Three patients in DRPs with gMMs for UNC-GRK4-V were tested, with 1 patient (A, alive 29 months post-SCT with chronic GVHD) showing an expanded tetramer-positive population. (D-I) Six patients in DRPs where both the recipient and donor carried the UNC-GRK4-V allele were tested, and 3 patients (D, alive 75 months post-SCT with mild chronic GVHD; E, died of relapsed AML 21 months post-SCT; F, alive 84 months post-SCT without GVHD) showed evidence of a tetramer-positive population. (J-L) Three patients in DRPs in which the patient did not carry the UNC-GRK4-V allele were tested, and no samples showed a tetramer-positive population.
Discussion
Because mHAs play a central role in both GVL and GVHD their discovery and characterization are crucial in developing strategies to improve SCT outcomes.1,3,7 Classic mHA discovery methods start with the isolation of a T-cell clone with subsequent screening for reactivity against allogeneic cell populations.29,31-39 While this approach has provided the majority of currently discovered mHAs, it is time consuming and has the possibility of yielding mHAs with limited translational use due to disadvantageous features such a low population prevalence or unfavorable tissue restriction.
Because of these limitations, reverse immunologic approaches that start with a list of candidate peptide epitopes and then aim to identify T-cell responses to the epitopes in post-SCT patients have been developed. Candidate peptides have been selected based upon clinical and genomic data as well as through nontargeted MS.9,40-42 We describe here a complete computational model that can use patient genomic information in any format to predict candidate minor antigen epitopes and confirm the mHAs using high-sensitivity targeted MS and standard peptide/HLA tetramer analysis.
Using genotyping data from 101 myeloid leukemia SCT patients, we computationally generated a pool of 1 655 540 potential mHA peptides and analyzed them for tissue expression and predicted binding to HLA types. In agreement with prior analyses, we observed roughly twice as many class I and II mHAs in MUD SCT compared with MRD SCT, which is primarily driven by the larger genetic disparity in MUD DRPs.9,43,44 There was diversity in the numbers of peptide epitopes predicted to bind with high affinity on various HLAs, a featured observed by other research groups.45 We observed no differences in OS, GVHD, or relapse based upon the number of predicted mHAs for each DRP (Figure 3E-F; supplemental Figures 5 and 6), a result consistent with genotyping analyses by Martin et al.44 Initial studies investigating MUD SCT showed higher incidences of GVHD compared with MRD,46,47 and our analysis shows significantly more mHAs in the MUD context. We suspect the reason that GVHD (and other outcomes) were similar between MRD and MUD SCT in this cohort is due to the fact that all MUD SCT patients received T-cell depletion with ATG.48,49 If no patients had received ATG, we suspect that clinical outcomes (particularly GVHD) would differ between MUD and MRD because of the large differences in mHA number between these groups; however, clinical outcomes among MRD patients (or MUD patients) treated as separate groups would not differ substantially based upon mHA number due to the relatively narrow distribution in these subgroups. Taken together, this lack of association implies only a subset of immunodominant mHA elicit robust GVL or GVHD responses.
Our computational screening approach was highly sensitive, capturing from within the candidate epitope pool 14 of 16 mHAs (88%) that could have been represented in our patient cohort based upon cSNP, HLA type, and presence of the appropriate minor mismatch. In addition, it allowed us to perform more targeted bioanalytical and immunologic assays. Peptide sequencing by MS has been the definitive test for class I epitope presentation for >2 decades.34,50 Nontargeted MS can identify mHAs; however, the approach only obtains MS/MS spectra of the most abundant species, which may not be the optimal targets. Furthermore, the computational alignment of peptides to proteomics databases is generally inefficient, with <30% of MS/MS spectra aligning with high confidence to any peptide in a proteomics database.51,52 This problem is further hindered if the database does not contain polymorphic variants (eg, mHAs) or mutations (eg, neoantigens).40,42 In this study, we used a targeted MS strategy capitalizing upon DIMS’s ability to filter out many competing peptides so that we could cleanly confirm the expression of our target mHA based upon 2 complementary features: (1) the concordance of the EC of maximal transmission between the model peptide and the sample, and (2) direct comparison between the model peptide and the sample MS/MS. By using DIMS, we were able to confirm UNC-GRK4-V using only direct nanoelectrospray, avoiding the need for HPLC optimization. We also observed that because of the UNC-GRK4-V peptide’s fragmentation pattern, it could not align to a standard proteomics database, and therefore, would not have been identified by nontargeted HPLC-MS/MS. While targeted MS will not provide coverage for as many peptides as a nontargeted approach, it can still test for tens of to a hundred peptides in a single experiment and is a strategy that could be applied to both mHAs and neoantigen discovery.
The gMM leading to a UNC-GRK4-V mHA is predicted to be common among SCTs because the recipient allele (T) frequency is 0.3, and the donor allele frequency (C) is 0.7. From Hardy-Weinberg equilibrium, we would expect 49% (0.7 × 0.7) of persons to be homozygous for the allele that does not encode GRK4-V and 51% [(0.3 × 0.3) + (2 × 0.3 × 0.7)] of persons to have an appropriate recipient genotype (ie, at least 1 copy of the T allele). In a MUD context, these allele frequencies would result in a ∼25% chance of there being a gMM and somewhat less of a chance in the MRD context because of genetic linkage.
GRK4 is expressed in testis and several cancer types, but not in normal hematopoietic tissue or GVHD target organs. Given its very narrow tissue expression, it was considered as a candidate CTA in addition to an mHA from the allelic variation at rs1801058. We tested for tetramer binding of T cells to UNC-GRK4-V/HLA-A*02:01 in DRPs predicted to have a minor mismatch (gMMs) and in DRPs where the donor also contained the UNC-GRK4-V allele. We observed tetramer-positive populations in 4 of 9 post-SCT patients (1 of 3 mHA context, 3 of 6 CTA context) who were predicted to express the epitope, indicating that while UNC-GRK4-V is an mHA, it can also function as a CTA. This feature is important, because it expands the number of HLA-A*02:01–expressing leukemia patients who could potentially be treated with a UNC-GRK4-V immunotherapeutic. Furthermore, several additional peptides at rs1801058 are predicted to bind tightly to HLA-A*02:01, A*03:01, A*11:01, B*35:01, and B*44:03. Detailed biochemical characterization, like that performed on UNC-GRK4-V, would be needed to confirm if any of these predicted peptides are mHAs.
This study used cSNP array data to generate the computational pool of candidate mHAs, and as such, in this format, it could not identify mHAs derived from other genetic differences such as alternative splicing, gene deletion, frameshifts, or novel open reading frames, all of which can lead to mHAs.29,33,37,53 If different genomics and RNA-seq data were used, however, mHAs derived from these alterations could also be predicted and then confirmed in the same general manner.
In summary, the novel computational mHA prediction method we have developed combined with the targeted confirmatory methods have enabled us to predict 102 novel public GVL mHAs and confirm 1 predicted GVL mHA as a potentially useful immunotherapy target. Extrapolating from our study of 11 172 cSNPs to the roughly 70 000 common cSNPs annotated in the 1000 Genomes Project, we would predict ∼700 public GVL mHAs, with ∼40 public GVL mHAs per DRP. These public GVL-restricted mHAs could be developed into new SCT treatment strategies applicable to virtually all AML patients, regardless of HLA type or unique genotype, where initial T-cell depletion, such as post-SCT cyclophosphamide to reduce GVHD risk, is coupled to later public GVL mHA-targeted immunotherapy, such as post-SCT vaccine or adoptive T-cell transfer, to enhance antileukemia alloreactivity without increasing the risk of GVHD.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the patients who were enrolled in the Laboratory 99-042 and LCCC-0824 studies and contributed specimens. The authors acknowledge Joel Parker with the Lineberger Comprehensive Cancer Bioinformatics Group and Mark Reed and Sandeep Sarangi with UNC Research Computing for assistance with large data file indexing and processing.
This work was supported by National Institutes of Health, National Cancer Institute grant R01 CA201225 (P.M.A.), as well as an ASCO Young Investigator Award (P.M.A.). P.M.A. and B.G.V. were also supported by the North Carolina University Cancer Research Fund. J.L.L. was supported by the Scott Neil Schwirck Fellowship.
Authorship
Contribution: B.G.V., P.M.A., and J.J.M. conceived the project, designed experiments, and interpreted results; J.L.L., S.C., and B.G.V. developed the bioinformatics pipeline for prediction of mHAs; J.L.L. applied the pipeline to data sets in this study and interpreted results; D.S.B. analyzed the performance of netMHCpan. P.M.A., G.A., and J.J.M. provided SNP genotyping, HLA typing, and clinical data, including GVHD incidence, GVHD grade, and relapse incidence; U.D. and E.J.C. synthesized tetramers; U.D. analyzed GRK4 RNA and protein expression and performed tetramer staining; S.A.H. provided SNP genotyping for the samples used for tetramer staining and performed tetramer analysis; J.E.K., I.M.S., and G.L.G. performed DIMS-MS/MS; and all authors critically reviewed and revised the manuscript, providing intellectual content, and approved the manuscript for publication.
Conflict-of-interest disclosure: Bruker Daltonics has licensed, from the University of North Carolina, some intellectual property related to the DIMS technology that was developed in the Glish (G.L.G.) laboratory. The remaining authors declare no competing financial interests.
Correspondence: Paul M. Armistead, Lineberger Comprehensive Cancer Center, 5205 Marsico Hall, Chapel Hill, NC 27599; e-mail: paul_armistead@med.unc.edu; and Benjamin G. Vincent, Lineberger Comprehensive Cancer Center, 5206 Marsico Hall, Chapel Hill, NC 27599, e-mail: benjamin_vincent@med.unc.edu.
REFERENCES
Author notes
P.M.A. and B.G.V. contributed equally to this study.

![Figure 2. Patient cohort survival outcomes. (A) Two-year OS was 55%, and there was no significant difference in OS based upon disease type (AML, MDS, or CML). (B) There was a statistically significant (P = .0010) difference in OS based upon disease status at time of SCT (25% vs 65%, not in CR/CP vs CR/CP). (C-D) There were no significant differences in OS based upon conditioning type (myeloablative conditioning [MAC] or RIC) or donor source (MRD or MUD).](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/2/16/10.1182_bloodadvances.2018022475/3/m_advances022475f2.jpeg?Expires=1763745685&Signature=lLNighLzfzrN5vn3l~9NLFJ5F2cCqZQ43YAOFJt1Rhx8vjC3zmkz7tW6Nh~MjuiPkY2yO64uxWhwLhjCgYafXQf~xyNFlrrpNQXVhQbVGkNNHTgnVZUg8Y-WULLECMC7yIZNI6eTy5sjk23LMobout2whVgzXkCLHxAyYAebS6kPmL4oqL12D48ud1Wa8yl~wcUlca0l0R7tWU9ciUDGFu4QxfytOFYf4sgpkq1ABB3~nTdT22uor5CVKHYWXTh0RjotjxkJ9cE2Fa3J6thn2nIFqPXG21ZfdLWr0dbX2lzcOeTb~Y82damZC4Ycu6INCjoYUkkmDucxALBp41zEwQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)