Key Points
iPSC-derived definitive erythroid cells display a globin expression profile corresponding to yolk sac erythromyeloid progenitors.
iPSC-derived erythroblasts resemble their postnatal counterparts in terms of gene expression and essential biological processes.
Abstract
Robust β-globin expression in erythroid cells derived from induced pluripotent stem cells (iPSCs) increases the resolution with which red blood cell disorders such as sickle cell disease and β thalassemia can be modeled in vitro. To better quantify efforts in augmenting β-globin expression, we report the creation of a β-globin reporter iPSC line that allows for the mapping of β-globin expression throughout human erythropoietic development in real time at single-cell resolution. Coupling this tool with single-cell RNA sequencing (scRNAseq) identified features that distinguish β-globin–expressing cells and allowed for the dissection of the developmental and maturational statuses of iPSC-derived erythroid lineage cells. Coexpression of embryonic, fetal, and adult globins in individual cells indicated that these cells correspond to a yolk sac erythromyeloid progenitor program of hematopoietic development, representing the onset of definitive erythropoiesis. Within this developmental program, scRNAseq analysis identified a gradient of erythroid maturation, with β-globin–expressing cells showing increased maturation. Compared with other cells, β-globin–expressing cells showed a reduction in transcripts coding for ribosomal proteins, increased expression of members of the ubiquitin-proteasome system recently identified to be involved in remodeling of the erythroid proteome, and upregulation of genes involved in the dynamic translational control of red blood cell maturation. These findings emphasize that definitively patterned iPSC-derived erythroblasts resemble their postnatal counterparts in terms of gene expression and essential biological processes, confirming their potential for disease modeling and regenerative medicine applications.
Introduction
Induced pluripotent stem cells (iPSCs) offer opportunities for disease modeling and cell-based therapeutics. Although derivation of patient-specific iPSC lines is now routine, a remaining challenge is the differentiation of PSCs into progeny that accurately resemble the postnatal cell of interest. Decades of research have suggested that in vitro hematopoietic differentiation from PSCs closely mimics in vivo development.1-5 Although resultant erythroid lineage cells are similar to their adult counterparts in several features, adult β-globin expression in iPSC-derived erythroblasts does not reach the levels produced in postnatal erythroid cells,5-7 indicating that PSC-derived erythroid cells, like most other cell types differentiated from PSCs,8 represent a prenatal stage of development. The exact positioning of these cells in human development is still under debate. In the developing embryo, successive hematopoietic programs give rise to hematopoietic progenitors with increasing lineage potential. A first transient wave of hematopoiesis arises in the yolk sac at mouse embryonic day 7 (E7), where it produces primitive erythrocytes, megakaryocytes, and macrophages.9,10 Shortly after, at E8.25, the yolk sac produces erythromyeloid progenitors (EMPs) that give rise to definitive erythroid cells, megakaryocytes, and most myeloid cells.9,11,12 At approximately E9, lymphoid potential can be detected in the yolk sac and paraaortic splanchnopleura as a result of lymphoid-primed multipotent progenitor hematopoiesis.13-15 Hematopoietic ontogeny culminates in the emergence of hematopoietic stem cells (HSCs) in the aorta-gonad-mesonephros (AGM) region at E10.5 that display adult repopulating potential and can sustain lifelong hematopoiesis through their ability to produce all definitive blood cells.16-20
Early hematopoietic differentiation protocols mainly described the production of cells with primitive erythroid characteristics.21-24 More recently, the signaling pathways underlying primitive and definitive hematopoietic specification in vitro have been unraveled, revealing that manipulation of Wnt and activin/nodal signaling can be used to skew differentiating cells toward a definitive rather than primitive fate.25,26 Whether this patterning results in an AGM-type definitive program that generates HSCs and resultant definitive blood cells or a more limited EMP yolk sac program capable of definitive erythropoiesis is unclear. The definitive character of erythroid cells produced using more recent differentiation protocols is reflected in their ability to express enhanced levels of β-globin.5-7,27-29 Although β-globin expression can be assessed via a variety of methods including quantitative reverse transcription polymerase chain reaction (qRT-PCR), high-performance liquid chromatography, mass spectrometry, and western blot analysis, these techniques do not provide information at the single-cell level. Fluorescence-activated cell sorting (FACS) analysis can provide these data; however, this procedure is beholden to β-globin antibody specificity as well as the requirement for permeabilization and fixation, negating live-cell studies and complicating downstream analysis of the transcriptome.
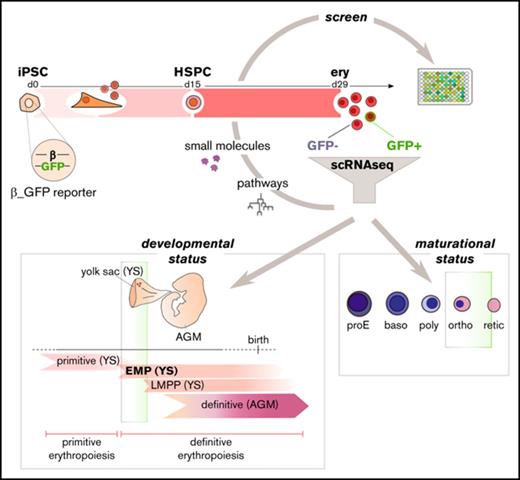
To overcome these limitations and enable the quantification and mapping of β-globin expression in iPSC-derived erythroid cultures, we generated a β-globin reporter iPSC line. This reporter was created through the insertion of a promoterless green fluorescent protein (GFP) cassette after the endogenous β-globin promoter, allowing for the tracking of β-globin expression throughout erythroid development in real time at single-cell resolution. This tool also enables sorting of live β-globin–expressing GFP+ cells, which can then be directly compared with their syngeneic GFP− counterparts by single-cell RNA sequencing (scRNAseq), providing insights into the developmental and maturational identities of these cells, how they compare with their postnatal counterparts, and the features that distinguish β-globin–expressing cells.
Methods
iPSC generation and maintenance
iPSC lines (BU6 and BS31 [BR-SP-31-1]) were generated through hSTEMCCA lentiviral transduction of human peripheral blood mononuclear cells as previously described and met stringent quality control parameters for pluripotency and functionality.30-32 iPSCs were maintained in mTESR (StemCell Technologies) on matrigel (Corning Matrigel hESC-qualified Matrix; #354277) using ReLeSR (StemCell Technologies) for passaging.
Targeting of the β-globin locus
Transcription activator-like effector nucleases (TALENs) targeting the β-globin locus (bL4 and bR4) were a kind gift from Matthew Porteus.33 The targeting vector was adapted from Voit et al33 by replacing the MGMT P140K drug selection cassette with a PGK-PURO cassette. Two million iPSCs were nucleofected with 9 μg of targeting vector and 3 μg of TALEN DNA using Primary Cell Nucleofector Solution P3 (Lonza) and program DC100 of the 4D-Nucleofector System (Lonza). Single resistant colonies were picked and expanded after puromycin selection (500 ng/mL) to establish clonal cell lines.
Erythroid differentiation from iPSCs
Hematopoietic differentiation from iPSCs to HSCs and progenitor cells (HSPCs) was induced according to Leung et al.29 To induce erythroid differentiation from HSPCs on day 15, 2 × 105 viable cells were plated in individual 35-mm dishes (StemCell Technologies; #27150) in 1.5 mL of methylcellulose-based medium with erythropoietin (EPO) for human cells (StemCell Technologies; MethoCult H4330) per dish. Burst-forming unit-erythroid (BFU-E) colonies were picked after 13 days of culture at 37°C in normoxic, 5% carbon dioxide conditions and cultured in StemSpan Serum-Free Expansion Medium II (StemCell Technologies) supplemented with 4 U/mL of human EPO (hEPO) for analysis the day after (day 29). Alternatively, day-15 HSPCs were specified using a 2-step suspension culture system consisting of Serum-Free Expansion Medium II, 2 mM of l-glutamine, and 100 μg/mL of primocin at 37°C in normoxic, 5% carbon dioxide conditions. Between days 15 and 20, this base was supplemented with 100 ng/mL of human stem cell factor, 40 ng/mL of IGF1, 5 × 10−7 M of dexamethasone, and 0.5 U/mL of hEPO and between days 20 and 25 with 4 U/mL of hEPO.
RNA extraction and quantitative PCR
RNA was extracted using the RNAeasy kit (Qiagen) and DNase treated using a DNA-free kit (Ambion). Complementary DNA (cDNA) was generated using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Predesigned TaqMan primers (Applied Biosystems) were used in conjunction with Taqman Universial Master Mix II (Applied Biosystems; #4440038) for quantitative reverse transcription PCR analysis on the StepOne/QuantStudio 6 Flex Real Time PCR Systems (Applied Biosystems). Fold expression was calculated as 2−(ΔCt), with ΔCt = Ct(gene of interest) − Ct(β-actin). These normalized fold expression values for all time points were then divided by the day-0 values to obtain relative fold expression compared with day 0. The housekeeping gene β-actin was used as an endogenous control.
FACS analysis
Cells were stained on ice for 25 minutes using the following antibodies: CD235a-PE (BD; #555570), CD71-APC (BD; #341028), CD49d-APC (Beckman Coulter; #B01682), CD36-APC (Beckman Coulter; #A87786), Band3 (CD233)-PE (American Research Products; #08-9439-4), and CKIT (CD117)-PE (ebioscience; #12-1178-41). Flow cytometry was conducted on a Stratedigm S1000EXI. FlowJo v8.7 (FlowJo, LLC) software was used for analysis, and FACS plots shown represent live erythroid cells based on side-scatter/forward-scatter gating.
Wright-Giemsa staining
Wright-Giemsa staining was performed using the Hema3 stain set (Fischer Scientific), and images were captured on a Zeiss Axio Imager.A1 microscope equipped with an IS1000 10.0-MP CMOS camera (Tucsen) using a 40× objective.
scRNAseq and analysis
GFP+ and GFP− cells were sorted and entered into the Fluidigm C1 HT workflow for capture, lysis, and reverse transcription of individual cells and library preparation.34 Sequencing was performed on a Nextseq 500 (Illumina) using a high-output kit, resulting in a total of 475 million, paired-end, 75-bp reads. Reads were aligned to the human genome (GRCh38) and quantified using the STAR aligner.35 Outlier removal was performed (>500 genes detected, <3 median absolute deviations away from median total reads, mitochondrial counts), resulting in 242 of a possible 800 cells available for analysis, with a mean of 441 145 aligned reads per cell. Size factors were computed via the Scran Bioconductor package, which uses pool-based scaling factors and deconvolution, followed by log-transformation normalization using the Scater package. Lun et al36 provide more detail regarding the normalization workflow. Analysis of variance was used to determine significantly (false discovery rate [FDR], <0.05) differentially expressed genes between GFP+ and GFP− cells. A heatmap was generated using Ward’s hierarchical clustering method37 on the row-scaled expression values of genes selected from supplemental Figure 4 and genes coding for ribosomal proteins. For pseudotime analysis, the GFP− fraction was divided into HBBlow and HBBintermediate based on whether expression was below or above mean HBB expression. Differential expression between the 3 conditions (q value < 0.01) was performed using the Monocle R package (v2.4), and these differentially expressed genes were used to perform supervised dimensionality reduction and subsequent cell ordering. scRNAseq raw data can be accessed from the Gene Expression Omnibus at GSE111860.
Statistical analysis
Results are presented as mean ± standard deviation. Statistical significance was confirmed using the Student t test.
Results
Creation of a β-globin reporter iPSC line
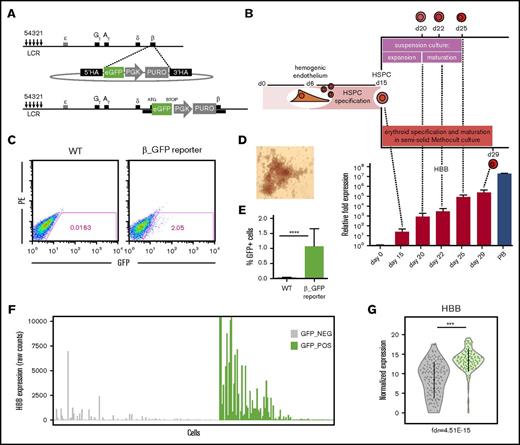
To map β-globin expression throughout erythroid development in real time, a promoterless GFP cassette was fused in frame to the first codon of the β-globin gene (HBB) using a TALEN pair targeting the β-globin locus (Figure 1A). This strategy ensures that the regulatory context controlling the developmentally programmed expression of the β-globin gene cluster remains intact and allows for the visualization of β-globin expression at single-cell resolution via GFP expression. After targeting and puromycin selection of hiPSCs, site-specific integration of the GFP cassette and the absence of random integrations were confirmed by PCR (supplemental Figure 1A). G banding confirmed the retention of a normal karyotype (supplemental Figure 1B). Notably, a twofold reduction in β-globin transcripts (but not ε-globin or γ-globin) was observed in the reporter iPSC line compared with the untargeted wild-type line after erythroid differentiation. This result is consistent with a monoallelic targeting scenario where β-globin transcription from 1 allele is blunted by insertion of the GFP cassette, with the alternate β-globin allele unaltered (supplemental Figure 1C).
Characterization and validation of the β-globin reporter human iPSC (hiPSC) line. (A) Targeting schematic depicting the introduction of a promoterless enhanced GFP (eGFP) cassette after the β-globin promoter. (B) Differentiation schematic and corresponding changes in β-globin transcript levels (quantitative reverse transcription PCR) during erythroid specification ± standard deviation (SD; 2 ≤ n ≤ 9). β-actin was used as a reference gene. CD34+ cells isolated from peripheral blood and specified toward the erythroid lineage (PB) were included as a primary control. (C-E) Characterization of wild-type (WT) or reporter (β_GFP) hiPSC-derived erythroid cells after 29 days of differentiation: representative FACS plots of GFP readout (C), bright field microscopy showing BFU-E colony morphology (×10) (D), and quantification of percentage of GFP+ cells ± SD (n = 9) (E). (F) HBB expression in individual GFP− (gray) and GFP+ (green) cells based on scRNAseq counts. (G) Violin plot demonstrating a significant enrichment for HBB transcripts in the GFP+ fraction (green) compared with the GFP− (gray) fraction based on normalized counts. ***FDR ≤ 0.0005, ****P ≤ .001. HA, homology arm; LCR, locus control region; PE, phycoerythrin; PGK, phosphoglycerate kinase promoter; PURO, puromycin resistance gene.
Characterization and validation of the β-globin reporter human iPSC (hiPSC) line. (A) Targeting schematic depicting the introduction of a promoterless enhanced GFP (eGFP) cassette after the β-globin promoter. (B) Differentiation schematic and corresponding changes in β-globin transcript levels (quantitative reverse transcription PCR) during erythroid specification ± standard deviation (SD; 2 ≤ n ≤ 9). β-actin was used as a reference gene. CD34+ cells isolated from peripheral blood and specified toward the erythroid lineage (PB) were included as a primary control. (C-E) Characterization of wild-type (WT) or reporter (β_GFP) hiPSC-derived erythroid cells after 29 days of differentiation: representative FACS plots of GFP readout (C), bright field microscopy showing BFU-E colony morphology (×10) (D), and quantification of percentage of GFP+ cells ± SD (n = 9) (E). (F) HBB expression in individual GFP− (gray) and GFP+ (green) cells based on scRNAseq counts. (G) Violin plot demonstrating a significant enrichment for HBB transcripts in the GFP+ fraction (green) compared with the GFP− (gray) fraction based on normalized counts. ***FDR ≤ 0.0005, ****P ≤ .001. HA, homology arm; LCR, locus control region; PE, phycoerythrin; PGK, phosphoglycerate kinase promoter; PURO, puromycin resistance gene.
Mapping β-globin expression throughout human erythroid development
To map β-globin expression throughout erythropoiesis, the β-globin reporter iPSC line was differentiated toward an HSPC fate, followed by commitment to and maturation along the erythroid lineage (Figures 1B and 2). Similar to our previous work,29 the differentiation protocol used in this study harnesses Wnt activation during mesodermal specification to initiate definitive patterning25,26 and focuses on the generation of robust numbers of multipotent HSPCs from a hemogenic endothelium intermediate before cells are specified to the erythroid lineage. This approach results in a significant increase in β-globin expression29 as compared with our earlier work,24 which recapitulated primitive hematopoietic and erythropoietic development. Here, to induce erythroid specification, day-15 iPSC-derived HSPCs were embedded in a semisolid extracellular matrix mimetic (Methocult), yielding BFU-E colonies at day 29 of differentiation (Figure 1B,D). Alternatively, day-15 HSPCs were also specified using a 2-step suspension culture system that allows for more convenient sampling of intermediate differentiation stages (Figure 1B). Using this optimized differentiation approach, we routinely observed a discreet population of GFP+ cells after Methocult specification at day 29, when β-globin transcript levels in the bulk cultures are at their peak (Figure 1B-C). GFP+ cells could also sporadically be detected at day 25 using the suspension culture system, but because Methocult specification yielded a more robust GFP readout, we focused on day-29 Methocult-specified cells for additional transcriptomic analyses.
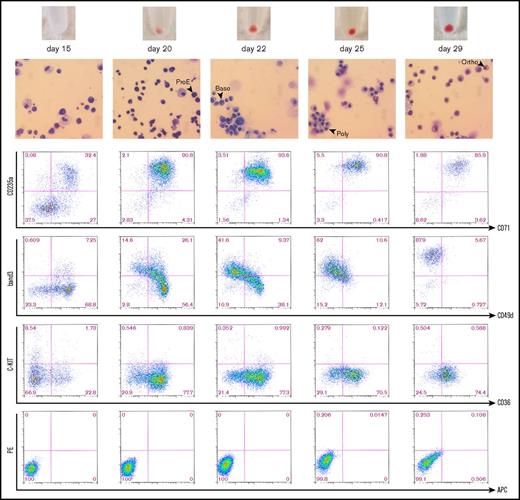
Characterization of erythroid differentiation. Time course including bright field microscopy images (×40) of Wright-Giemsa–stained cytospins and cell pellets (top) and FACS plots showing cell surface marker expression (bottom) at successive stages during erythroid differentiation. The time points shown correspond to the time points illustrated in Figure 1B. Day-15 cells represent HSPCs; suspension cultures were sampled at days 20, 22, and 25 to obtain progenitors at intermediate stages of erythroid specification; and day-29 cells represent BFU-E cells picked from Methocult cultures after 14 days of erythroid specification. Black arrows indicate examples of proerythroblasts (ProE), basophilic erythroblasts (Baso), polychromatic erythroblasts (Poly), and orthochromatic erythroblasts. FACS plot showing unstained cells is included as a reference below. Uniform gating based on day-29 cells was maintained throughout the figure.
Characterization of erythroid differentiation. Time course including bright field microscopy images (×40) of Wright-Giemsa–stained cytospins and cell pellets (top) and FACS plots showing cell surface marker expression (bottom) at successive stages during erythroid differentiation. The time points shown correspond to the time points illustrated in Figure 1B. Day-15 cells represent HSPCs; suspension cultures were sampled at days 20, 22, and 25 to obtain progenitors at intermediate stages of erythroid specification; and day-29 cells represent BFU-E cells picked from Methocult cultures after 14 days of erythroid specification. Black arrows indicate examples of proerythroblasts (ProE), basophilic erythroblasts (Baso), polychromatic erythroblasts (Poly), and orthochromatic erythroblasts. FACS plot showing unstained cells is included as a reference below. Uniform gating based on day-29 cells was maintained throughout the figure.
GFP expression marks cells with increased levels of β-globin transcripts
Interestingly, despite dramatic increases in β-globin transcripts over time in differentiation (Figure 1B), only ∼1% of cells exceeded the GFP detection threshold at the most mature stage of differentiation (Figure 1C,E). Similar results were obtained using multiple iPSC clones representing different genetic backgrounds, and the ability of our targeting approach to visualize β-globin–expressing cells, as well as the correlation between HBB and GFP levels, was validated in postnatally derived HUDEP2 cells capable of robust β-globin protein expression38 (supplemental Figures 2 and 3). This result suggested that the several log-fold increase in β-globin transcripts seen at the population level as the cells progressed through erythroid specification (Figure 1B) could be the result of high levels of β-globin transcription in a small fraction of cells. Using the GFP readout of the reporter line to classify cells based on their β-globin protein expression status, we sorted GFP− and GFP+ cells at day 29 of differentiation. scRNAseq was used to define the correlation between β-globin transcripts and protein-level expression. Comparing the transcriptomes of 143 GFP− and 99 GFP+ cells, we found significantly greater levels of β-globin transcripts in the GFP+ fraction (FDR, 4.51e−15; Figure 1F-G), suggesting an important contribution of this fraction to overall β-globin expression. Moreover, the noted enrichment for β-globin transcripts in the GFP+-sorted fraction further validated our reporter iPSC line as a tool to visualize and enrich for β-globin–expressing cells.
iPSC-derived erythroid cells recapitulate primary human erythropoiesis
scRNAseq analysis resulted in a list of 7644 genes expressed in day-29 iPSC-derived erythroid cells isolated from Methocult cultures when we combined both GFP− and GFP+ cells. Enrichr analysis39,40 (http://amp.pharm.mssm.edu/Enrichr) was used to identify annotated gene sets enriched in the 200 most highly expressed genes and resulted in a CD71+ early erythroid signature (FDR, 1.895e−23; Table 1, gene fraction B), matching immunostaining data validating the erythroblast staging of these cells (Figure 2). Pathways enriched in the top 200 expressed genes at this time point include heme biosynthesis (FDR, 1.82e−3) and hemoglobin’s chaperone (FDR, 6.46e−10) consistent with hemoglobin production in these cells (Table 1, gene fraction A). Moreover, comparison of these data with transcriptomic analyses performed on human cord blood CD34+ HSPCs during terminal erythroid differentiation showed a considerable overlap between the most highly expressed genes in iPSC-derived erythroid cells at the most mature stage of differentiation and the top 25 expressed genes at the orthochromatic erythroblast stage identified in maturing primary erythroid cell cultures (supplemental Table 1).41 The presence of orthochromatic-staged erythroblasts in day-29 differentiation cultures was also confirmed by Wright-Giemsa staining (Figure 2). Together, these findings indicate that iPSC-derived erythroid cells display a transcriptional signature matching their immunophenotype and that these cells resemble their postnatally derived counterparts in terms of gene expression and the biological processes essential for the maturation of red blood cells.
Enrichr analysis of scRNAseq data
| Gene fraction . | Enriched terms* . | Adjusted P . | Source† . |
|---|---|---|---|
| A | Processes/pathways | ||
| Top 200 expressed genes (all cells) | Hemoglobin’s chaperone_Homo sapiens_h_ahspPathway | 6.46E−10 | BioCarta 2016 |
| Heme biosynthesis_Homo sapiens_PWY-5920 | 1.82E−03 | HumanCyc 2016 | |
| Heme biosynthesis_Homo sapiens_P02746 | 1.39E−01 | Panther 2016 | |
| Hypoxic and oxygen homeostasis regulation of HIF-1-alpha_Homo sapiens_4c0f3584-6193-11e5-8ac5-06603eb7f303 | 5.54E−02 | NCI-Nature 2016 | |
| Ribosome_Homo sapiens_hsa03010 | 7.22E−41 | KEGG 2016 | |
| HIF-1 signaling pathway_Homo sapiens_hsa04066 | 2.75E−02 | KEGG 2016 | |
| Upregulated in GFP+ | Hemoglobin’s chaperone_Homo sapiens_h_ahspPathway | 8.26E−07 | BioCarta 2016 |
| Heme biosynthesis_Homo sapiens_PWY-5920 | 4.54E−02 | HumanCyc 2016 | |
| Heme biosynthesis_Homo sapiens_P02746 | 7.41E−02 | Panther 2016 | |
| Ubiquitin protein ligase binding (GO:0031625) | 4.20E−02 | GO molecular function 2017 | |
| Oxygen transport (GO:0015671) | 1.29E−04 | GO biological process 2015 | |
| Oxidative stress_Homo sapiens_WP408 | 1.12E−02 | WikiPathways 2016 | |
| Cellular responses to stress_Homo sapiens_R-HSA-2262752 | 2.82E−04 | Reactome 2016 | |
| HIF-1-α transcription factor network_Homo sapiens_20ef2b81-6193-11e5-8ac5-06603eb7f303 | 5.57E−04 | NCI-Nature 2016 | |
| Downregulated in GFP+ | Ribosome_Homo sapiens_hsa03010 | 4.75E−29 | KEGG 2016 |
| tRNA binding (GO:0000049) | 6.01E−16 | GO molecular function 2017b | |
| Peptide chain elongation_Homo sapiens_R-HSA-156902 | 1.52E−31 | Reactome 2016 | |
| Eukaryotic Translation Elongation_Homo sapiens_R-HSA-156842 | 1.71E−31 | Reactome 2016 | |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC)_Homo sapiens_R-HSA-975956 | 1.71E−31 | Reactome 2016 | |
| 3′ -UTR-mediated translational regulation_Homo sapiens_R-HSA-157279 | 5.82E−30 | Reactome 2016 | |
| B | Cell type | ||
| Top 200 expressed genes (all cells) | CD71+_EarlyErythroid | 1.90E−23 | Human Gene Atlas |
| Bone marrow (bulk tissue) | 2.98E−25 | ARCHS4 Tissues | |
| Cord blood | 1.60E−23 | ARCHS4 Tissues | |
| Erythroblast | 1.07E−03 | ARCHS4 Tissues | |
| Upregulated in GFP+ | CD71+_EarlyErythroid | 4.25E−49 | Human Gene Atlas |
| Erythroid_cell | 6.60E−30 | Jensen Tissues | |
| Blood | 7.61E−13 | Jensen Tissues | |
| Erythroblast | 1.96E−12 | ARCHS4 Tissues | |
| C | Transcription factor targets | ||
| Upregulated in GFP+ | KLF1 | 5.196E−32 | Enrichr Submissions TF-Gene Coocurrence |
| KLF1_20508144_ChIP-Seq_FETAL-LIVER-ERYTHROID_Mouse | 1.490E−06 | ChEA 2016 | |
| GATA1_CHEA | 4.279E−15 | ENCODE and ChEA | |
| GATA1_erythroblast_hg19 | 7.311E−9 | Consensus TFs from ChIP-X | |
| GATA1_19941826_ChIP-Seq_K562_Human | 4.508E−8 | ChEA 2016 | |
| TAL1_20566737_ChIP-Seq_PRIMARY_FETAL_LIVER_ ERYTHROID_Mouse | 5.805E−12 | ChEA 2016 | |
| TAL1_erythroblast_mm9 | 4.769E−10 | ENCODE TF ChIP-seq 2015 |
| Gene fraction . | Enriched terms* . | Adjusted P . | Source† . |
|---|---|---|---|
| A | Processes/pathways | ||
| Top 200 expressed genes (all cells) | Hemoglobin’s chaperone_Homo sapiens_h_ahspPathway | 6.46E−10 | BioCarta 2016 |
| Heme biosynthesis_Homo sapiens_PWY-5920 | 1.82E−03 | HumanCyc 2016 | |
| Heme biosynthesis_Homo sapiens_P02746 | 1.39E−01 | Panther 2016 | |
| Hypoxic and oxygen homeostasis regulation of HIF-1-alpha_Homo sapiens_4c0f3584-6193-11e5-8ac5-06603eb7f303 | 5.54E−02 | NCI-Nature 2016 | |
| Ribosome_Homo sapiens_hsa03010 | 7.22E−41 | KEGG 2016 | |
| HIF-1 signaling pathway_Homo sapiens_hsa04066 | 2.75E−02 | KEGG 2016 | |
| Upregulated in GFP+ | Hemoglobin’s chaperone_Homo sapiens_h_ahspPathway | 8.26E−07 | BioCarta 2016 |
| Heme biosynthesis_Homo sapiens_PWY-5920 | 4.54E−02 | HumanCyc 2016 | |
| Heme biosynthesis_Homo sapiens_P02746 | 7.41E−02 | Panther 2016 | |
| Ubiquitin protein ligase binding (GO:0031625) | 4.20E−02 | GO molecular function 2017 | |
| Oxygen transport (GO:0015671) | 1.29E−04 | GO biological process 2015 | |
| Oxidative stress_Homo sapiens_WP408 | 1.12E−02 | WikiPathways 2016 | |
| Cellular responses to stress_Homo sapiens_R-HSA-2262752 | 2.82E−04 | Reactome 2016 | |
| HIF-1-α transcription factor network_Homo sapiens_20ef2b81-6193-11e5-8ac5-06603eb7f303 | 5.57E−04 | NCI-Nature 2016 | |
| Downregulated in GFP+ | Ribosome_Homo sapiens_hsa03010 | 4.75E−29 | KEGG 2016 |
| tRNA binding (GO:0000049) | 6.01E−16 | GO molecular function 2017b | |
| Peptide chain elongation_Homo sapiens_R-HSA-156902 | 1.52E−31 | Reactome 2016 | |
| Eukaryotic Translation Elongation_Homo sapiens_R-HSA-156842 | 1.71E−31 | Reactome 2016 | |
| Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC)_Homo sapiens_R-HSA-975956 | 1.71E−31 | Reactome 2016 | |
| 3′ -UTR-mediated translational regulation_Homo sapiens_R-HSA-157279 | 5.82E−30 | Reactome 2016 | |
| B | Cell type | ||
| Top 200 expressed genes (all cells) | CD71+_EarlyErythroid | 1.90E−23 | Human Gene Atlas |
| Bone marrow (bulk tissue) | 2.98E−25 | ARCHS4 Tissues | |
| Cord blood | 1.60E−23 | ARCHS4 Tissues | |
| Erythroblast | 1.07E−03 | ARCHS4 Tissues | |
| Upregulated in GFP+ | CD71+_EarlyErythroid | 4.25E−49 | Human Gene Atlas |
| Erythroid_cell | 6.60E−30 | Jensen Tissues | |
| Blood | 7.61E−13 | Jensen Tissues | |
| Erythroblast | 1.96E−12 | ARCHS4 Tissues | |
| C | Transcription factor targets | ||
| Upregulated in GFP+ | KLF1 | 5.196E−32 | Enrichr Submissions TF-Gene Coocurrence |
| KLF1_20508144_ChIP-Seq_FETAL-LIVER-ERYTHROID_Mouse | 1.490E−06 | ChEA 2016 | |
| GATA1_CHEA | 4.279E−15 | ENCODE and ChEA | |
| GATA1_erythroblast_hg19 | 7.311E−9 | Consensus TFs from ChIP-X | |
| GATA1_19941826_ChIP-Seq_K562_Human | 4.508E−8 | ChEA 2016 | |
| TAL1_20566737_ChIP-Seq_PRIMARY_FETAL_LIVER_ ERYTHROID_Mouse | 5.805E−12 | ChEA 2016 | |
| TAL1_erythroblast_mm9 | 4.769E−10 | ENCODE TF ChIP-seq 2015 |
Enriched in different fractions of day-29 β_GFP reporter hiPSC-derived erythroid cells according to Enrichr analysis of scRNAseq data.
Gene set libraries were accessed through the Enrichr platform (http://amp.pharm.mssm.edu/Enrichr/).
GFP+ β-globin–expressing iPSC-derived erythroid cells display advanced maturation
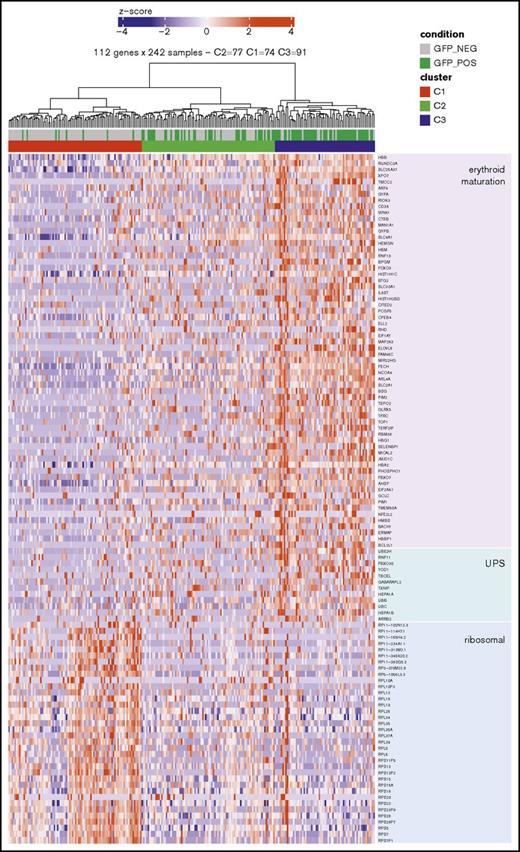
After sorting and scRNAseq of day-29 iPSC-derived erythroid cells, 219 genes were found differentially expressed between GFP+ and GFP− fractions with an FDR of <5% (FDR, <0.05). Of these genes, 74 were downregulated and 145 were upregulated in the GFP+ fraction. Enrichr analysis39,40 was used to identify annotated gene sets upregulated in the GFP+ compared with the GFP− fraction and resulted in CD71+ early erythroid (FDR, 4.2514e−49), erythroblast (FDR, 1.96e−12), or erythroid cell (FDR, 6.60e−30) signatures (Table 1, gene fraction B). Oxygen transport (FDR, 1.29e−4), heme biosynthesis (FDR, 4.54e−2), and hemoglobin’s chaperone (FDR, 8.26e−07) pathways were enriched in GFP+ cells (Table 1, gene fraction A). These results indicate that key signature genes important for erythroid-specific identity and biological processes are expressed at higher levels in GFP+ cells. Among these genes, we found typical erythroid cell surface markers (GYPA [CD235a], SLC4A1 [band3], CD36), genes involved in heme biosynthesis (FECH, SLC25a37, TFRC [CD71]), hemoglobin chaperones (AHSP), transcription factors (FOXO3, HEMGN, NFE2L2, XPO7), and other genes linked to erythroid maturation (Figure 3).28,41-46 Although master erythroid regulators such as KLF1 and GATA1 and TAL1 were not found differentially expressed, their targets made up a majority of the differentially expressed genes (Table 1, gene fraction C). Moreover, gene set enrichment analysis demonstrated enrichment for genes that are typically upregulated during the terminal erythroid maturation of postnatal cells in the GFP+ fraction (supplemental Figure 5). Similarly, genes that were downregulated in the GFP+ fraction corresponded to genes that are typically downregulated during erythroid maturation, suggesting increased maturation in the GFP+ β-globin–expressing fraction compared with the GFP− cells.
Heatmap showing a subset of genes differentially expressed between day-29 GFP−and GFP+sorted fractions. Heatmap illustrating downregulation of ribosomal transcripts, upregulation of a subset of genes involved in the ubiquitin-proteasome system (UPS) pathway and increased expression of genes linked to erythroid maturation in GFP+ vs GFP− cells. In addition to genes coding for ribosomal proteins, this heatmap contains a selection of genes identified in supplemental Figure 4 as linked to erythroid maturation based on comparison of transcriptomic data sets representing terminal erythroid maturation found in the literature.40,44 The UPS-related genes shown represent a subset of these erythroid maturation genes identified in supplemental Figure 4.
Heatmap showing a subset of genes differentially expressed between day-29 GFP−and GFP+sorted fractions. Heatmap illustrating downregulation of ribosomal transcripts, upregulation of a subset of genes involved in the ubiquitin-proteasome system (UPS) pathway and increased expression of genes linked to erythroid maturation in GFP+ vs GFP− cells. In addition to genes coding for ribosomal proteins, this heatmap contains a selection of genes identified in supplemental Figure 4 as linked to erythroid maturation based on comparison of transcriptomic data sets representing terminal erythroid maturation found in the literature.40,44 The UPS-related genes shown represent a subset of these erythroid maturation genes identified in supplemental Figure 4.
Another striking observation is that transcripts coding for ribosomal proteins of the small (RPS) and large (RPL) ribosomal subunits make up a majority of transcripts downregulated in the GFP+ fraction (Figure 3). This finding was corroborated by Enrichr analysis showing enrichment for the ribosome pathway (FDR, 4.75e−29) and other translational processes involving RPS and RPL genes in the list of genes upregulated in the GFP− vs the GFP+ fraction (Table 1, gene fraction A). Similarly, a comprehensive proteomic analysis of human erythropoiesis at successive stages of maturation showed a decrease in most proteins of the translation machinery during erythroid maturation, including RPS and RPL members, indicating that this finding is in agreement with increased maturation of the GFP+ fraction.44
Additionally, we also found increased expression of components of the UPS (UBE2H, RNF11, FBXO30, YOD1, TBCEL, GABARAPL2) in the GFP+ fraction (Figures 2B and 3). These proteasome remodeling factors are induced during late erythroid differentiation to accomplish the selective degradation of a vast set of preexisting proteins, while sparing hemoglobin and other essential components of mature erythroid cells.47
Lastly, coupled with signatures indicating posttranslational remodeling of the erythroid proteome, we also identified factors involved in the posttransriptional control of erythropoiesis. Transcription of MIR22HG, for example, a gene coding for a microRNA controlling erythroid maturation in mice (MIR22),48 was upregulated in the GFP+ fraction. Similarly, RBM38 and CPEB4 were found upregulated in the GFP+ vs GFP− fraction (Figure 3). These 2 factors coordinate the dynamic translational control of red blood cell development, with CPEB4 responsible for the translational silencing of expendable messenger RNAs (mRNAs) and RBM38 concomitantly promoting enhanced decoding of transcripts essential for red blood cell maturation.49,50 Together, these findings strongly suggest an increased maturational status of the GFP+ β-globin–expressing fraction compared with the GFP− fraction.
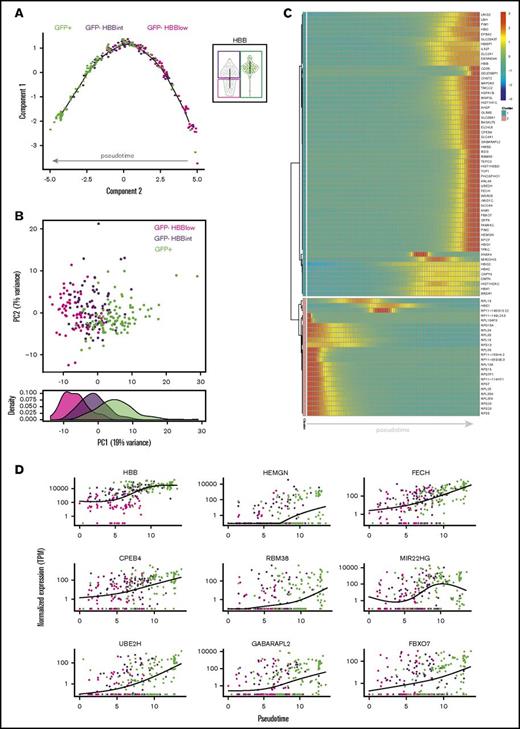
Pseudotime analysis shows a gradual increase in erythroid maturation from GFP− to GFP+ cells
To examine whether these iPSC-derived erythroid lineage cells follow a continuous developmental trajectory or represent distinct populations coexisting at this time point in differentiation, we ordered them according to pseudotime using the differentially expressed genes for the dimensionality reduction step (Monocle R package). For this analysis, we chose to further subdivide the GFP− population into HBBlow and HBBintermediate fractions, because we noticed a bimodal distribution of HBB transcript levels in this population (Figures 1G and 4A) and wanted to gain insight into how these fractions relate to each other and to GFP+ cells in a pseudotemporal manner. The trajectory that was calculated based on these 3 input populations showed a gradual progression from GFP− cells that expressed low levels of β-globin transcripts (GFP−HBBlow) to GFP+ cells expressing high levels of β-globin transcriptionally and also at the protein level (Figure 4A). Interestingly, a GFP−HBBintermediate fraction expressing β-globin transcripts but not protein according to its GFP expression status was identified as an intermediate population based on the pseudotime trajectory. Similarly, principal component analysis positioned this fraction as an intermediate between HBBlow and GFP+ populations (Figure 4B). Gene expression changes over pseudotime again showed downregulation of ribosomal transcripts, upregulation of genes involved in the UPS, and increased expression of genes linked to erythroid maturation as the cells advanced through pseudotime (Figure 4C-D). Taken together, these data suggest a gradual increase in maturation from the GFP− to GFP+ fraction.
Pseudotime analysis of scRNAseq data. (A) Supervised monocle plot displaying the trajectory of day-29 hiPSC-derived erythroid cells along pseudotime indicating a gradation from GFP−HBBlow to GFP+ cells. The 3 input fractions specified when performing this analysis are shown in the violin plot on the right. (B) Principal component (PC) analysis plot constructed using the top 500 differentially expressed genes between the 3 conditions illustrating the separation of day-29 hiPSC-derived erythroid cells (top). Density graph showing the distribution of the 3 subpopulations with respect to PC1 (bottom). (C) Heatmap showing gene expression changes over pseudotime. Vertical columns represent units of pseudotime. (D) Scatter plots illustrating the expression pattern of selected genes over pseudotime. Normalized expression is shown as transcripts per million (TPM).
Pseudotime analysis of scRNAseq data. (A) Supervised monocle plot displaying the trajectory of day-29 hiPSC-derived erythroid cells along pseudotime indicating a gradation from GFP−HBBlow to GFP+ cells. The 3 input fractions specified when performing this analysis are shown in the violin plot on the right. (B) Principal component (PC) analysis plot constructed using the top 500 differentially expressed genes between the 3 conditions illustrating the separation of day-29 hiPSC-derived erythroid cells (top). Density graph showing the distribution of the 3 subpopulations with respect to PC1 (bottom). (C) Heatmap showing gene expression changes over pseudotime. Vertical columns represent units of pseudotime. (D) Scatter plots illustrating the expression pattern of selected genes over pseudotime. Normalized expression is shown as transcripts per million (TPM).
Coexpression of embryonic, fetal, and adult globins in individual iPSC-derived erythroid cells is indicative of a definitive EMP program of hematopoietic development
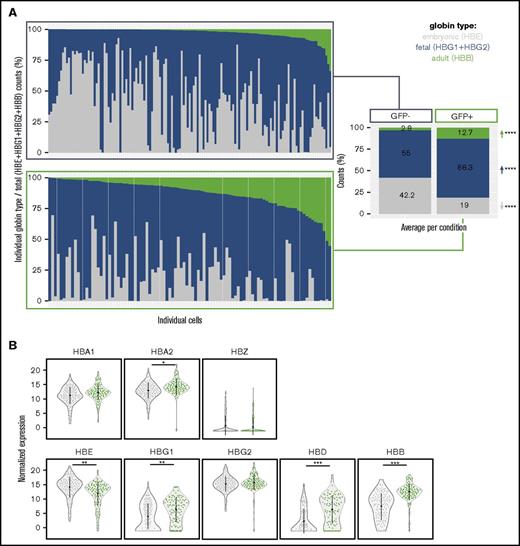
In addition to gaining insight into the maturational status of GFP+ vs GFP− cells, scRNAseq analysis of the BFU-E cells obtained at day 29 of differentiation also shed light on the developmental timing of iPSC-derived erythroid cells. During hematopoietic development, different hematopoietic programs can be distinguished, each with their characteristic globin expression profile.51 Positioning PSC-derived erythroid cells in ontogeny, however, is challenging, because in vitro differentiation cultures lack the spatiotemporal separation of the different hematopoietic waves present in the embryo. Because of the possibility of multiple hematopoietic programs coexisting in 1 well, we used scRNAseq analysis to get an accurate idea of the developmental stage of the cells in our cultures rather than relying on bulk expression analyses, which are inherently confounding in this setting. Looking at the coexpression of different globins in individual day-29 iPSC-derived erythroid cells, we found that a majority of the cells expressed a combination of embryonic (ε-globin; HBE), fetal (γ-globin, HBG1 and HBG2), and adult (β-globin; HBB) globins (Figure 5A), suggesting an EMP stage of hematopoietic development.51 This result is in agreement with the presence of BFU-Es in our differentiation cultures at day 29, given that the emergence of BFU-Es defines the onset of definitive erythropoiesis in the embryo and that the EMP wave arising in the yolk sac is the first hematopoietic wave with definitive erythroid potential.9,11,12
Globin expression in day-29 hiPSC-derived erythroid cells. (A) Coexpression of globin genes (HBE, HBG1, HBG2, HBB) in individual cells visualized as the percentage of transcripts from the individual globin type vs total β-globin gene cluster transcripts (HBE + HBG1 + HBG2 + HBB). Average percentages per globin type are shown on the right for the GFP− and GFP+ fractions. A 2-tailed t test was performed to determine significant differences between GFP− and GFP+ fractions. (B) Violin plots showing normalized expression of globin genes in the GFP−– (gray) vs GFP+–sorted (green) fraction of day-29 hiPSC-derived erythroid cells. *FDR ≤ 0.05, **FDR ≤ 0.005, ***FDR ≤ 0.0005, ****P ≤ .001.
Globin expression in day-29 hiPSC-derived erythroid cells. (A) Coexpression of globin genes (HBE, HBG1, HBG2, HBB) in individual cells visualized as the percentage of transcripts from the individual globin type vs total β-globin gene cluster transcripts (HBE + HBG1 + HBG2 + HBB). Average percentages per globin type are shown on the right for the GFP− and GFP+ fractions. A 2-tailed t test was performed to determine significant differences between GFP− and GFP+ fractions. (B) Violin plots showing normalized expression of globin genes in the GFP−– (gray) vs GFP+–sorted (green) fraction of day-29 hiPSC-derived erythroid cells. *FDR ≤ 0.05, **FDR ≤ 0.005, ***FDR ≤ 0.0005, ****P ≤ .001.
In this study, reads that can be mapped to the HBB gene were detected in nearly all cells (231 of 242 cells). Notably, contribution of HBB transcripts to the sum of transcripts from the β-globin gene cluster (ε+γ+β) in the GFP− fraction was minimal (Figure 5A). This was especially apparent in a subset of GFP− cells predominantly expressing embryonic globin (Figure 5A). Conversely, the GFP+ fraction showed a significant reduction in the contribution of HBE transcripts to the total β-globin gene cluster transcripts. At the same time, HBB transcripts accounted for a larger percentage of this total (ε+γ+β) in the GFP+ fraction, as would be expected based on the enrichment of HBB transcripts here compared with the GFP− fraction (Figure 5B). In addition to a decrease in HBE transcripts and an increase in HBB transcripts, the GFP+ fraction also showed increased expression of HBD, HBG1, and HBA2. No significant differences in expression were noted for HBZ, HBA1, or HBG2 (Figure 5B).
Taken together, scRNAseq analysis showed the coexpression of embryonic, fetal, and adult globins in a vast majority (231 of 242) of sampled cells, suggesting that the iPSC-derived erythroid cells in our cultures correspond to an EMP stage of development described in the yolk sac and representing the onset of definitive erythropoiesis.9,11,12,51 Moreover, we found that embryonic characteristics (HBE transcripts) were gradually lost as cells gained more adult features (HBB transcripts, β-globin protein expression), paralleling the gradual increase in erythroid maturation from the GFP− fraction to the GFP+ fraction suggested by pseudotime analysis. These results indicate that iPSC-derived erythroid cells have the ability to undergo some degree of maturational globin switching.
Discussion
Our creation of a β-globin reporter has expanded the toolbox that can be used to map β-globin expression in human PSC progeny. Combined with scRNAseq, this tool allows for the dissection of the maturational and developmental statuses of iPSC-derived erythroid cells, an especially relevant topic given the prospect of using PSC-derived erythroid cells for disease modeling and regenerative medicine applications.
iPSC-derived day-29 erythroid cells resemble their postnatal counterparts in terms of cell surface marker expression, gene expression, and biological processes essential for erythroid maturation.28,41,44,45 Our data agree with a recent study comparing the transcriptome of iPSC-derived erythroid cells and cord blood–derived cells28 and further emphasize that erythroid differentiation from iPSCs closely mirrors primary human erythropoiesis. An outstanding question is whether definitive erythropoiesis from PSCs is representative of an AGM-type definitive or EMP yolk sac program. Although both types of definitive erythroid cells are indistinguishable near the end of prenatal development, a unique globin expression signature distinguishes these programs at the start of erythroid specification.51 In mice transgenic for the human β-globin locus, EMP-derived erythroid cells express a combination of embryonic, fetal, and adult globins, distinguishing them from primitive erythroid cells that lack adult globins and AGM-derived definitive erythroid cells that lack embryonic globin.51 Because multiple hematopoietic programs can coexist in 1 differentiation culture,1,4,25 the developmental identity of iPSC-derived erythroid cells in bulk populations is challenging. Single-cell RNAseq analyses revealed coexpression of embryonic, fetal, and adult globins in a vast majority of sampled cells, suggesting that the iPSC-derived erythroid cells in our cultures correspond to a yolk sac EMP stage of development.51 A similar globin coexpression pattern has also been reported in BFU-Es from 6-week-old human embryos.52 Although an EMP wave has not been formally characterized in human development, BFU-Es, which are considered the first definitive erythroid progenitors, can be detected in the human yolk sac ∼4 to 5 weeks postconception.53 This similarity to their first appearance in mice, which coincides with the onset of EMP hematopoiesis, strongly suggests the emergence of a similar EMP wave in humans at ∼4 to 5 weeks in gestation.11,54 In addition, scRNAseq demonstrated that HBE transcripts are gradually reduced as cells gain more adult HBB transcripts, identical to the inverse correlation between the levels of ε-globin and β-globin described in BFU-Es isolated from human embryos.52 Recapitulation of yolk sac hematopoiesis, including a definitive EMP-like program, has previously been described during hematopoietic differentiation from both mouse and human PSCs.1,4 The high-resolution analysis of globin coexpression in our study provides additional confidence that the unique coexpression pattern characteristic for the EMP wave is present in individual cells and is not the result of pooling cells from different developmental programs. Although this analysis describes the cells produced using our in-house differentiation protocol, we predict that these findings will be expandable to many of the current differentiation protocols making use of Wnt stimulation early on in the differentiation process to inhibit the emergence of primitively patterned cells and instead promote definitive hematopoiesis.
Furthermore, scRNAseq analysis of GFP+ and GFP− sorted cells at day 29 indicated advanced maturation of GFP+ compared with GFP− cells. The pathway analyses performed herein suggest that an iPSC-based system such as that described here can effectively recapitulate human erythropoietic development. Gene set enrichment analysis demonstrated that genes up- or downregulated in the GFP+ fraction corresponded to genes typically up- or downregulated during terminal erythroid maturation of postnatal cells.45 Although many of these genes represent targets of erythroid master regulators like GATA1, KLF1, and TAL1, these transcription factors were not found differentially expressed between GFP− and GFP+ fractions, suggesting other levels of regulation besides their transcript levels. We also noted a decrease in ribosomal transcripts, in line with a comprehensive proteomic analysis of human erythropoiesis showing a decrease in most proteins of the translation machinery during erythroid maturation.44 Moreover, a subset of members of the UPS, recently identified to be involved in remodeling of the erythroid proteome during maturation, was upregulated in the GFP+ fraction, in agreement with their upregulation in primary cells undergoing terminal erythroid maturation.41,47,55 In addition to posttranslational control mechanisms being upregulated in the GFP+ fraction, we also found enrichment for posttranscriptional mechanisms controlling red blood cell development. CPEB4 and RBM38, 2 key genes involved in the dynamic translational control of red blood cell development, were enriched in the GFP+ fraction. As erythroid cells mature and prepare to condense and extrude their nuclei, CPEB4 causes translational silencing of expendable mRNAs, while RBM38 enhances decoding of essential erythroid transcripts.50 This coordinated action reduces the dependence of protein synthesis on active mRNA production, which is downregulated in terminally maturing red blood cells. These findings demonstrate that the produced iPSC-derived populations are extraordinarily similar to their postnatal counterparts in terms of the dynamic developmental processes of human erythroid lineage cells.
Although multiple iPSC lines were targeted with our β-globin reporter construct, only ∼1% of cells exceeded the GFP detection threshold upon erythroid differentiation. This suggests that we are missing key molecular cues for cellular development and maturation and/or we are not effectively recapitulating a necessary niche. Because the pseudotime trajectory identified a transitory population of cells (GFP-HBBintermediate) that express β-globin at the transcript level but not at the protein level, one could hypothesize that these cells lack the posttranscriptional machinery to translate those transcripts efficiently into protein. Differential expression of MIR22HG and factors such as CPEB4 and RBM38 between the GFP− and GFP+ fractions would suggest that that is a possibility. Moreover, it is also likely that the bulk analyses performed in many studies do not account for cellular heterogeneity in iPSC-derived erythroid cultures, which might lead to overestimation of the number of cells achieving robust adult β-globin expression, with relatively few cells accounting for the adult-type signature. Modeling fetal hemoglobin expression suggested that its distribution is variable from person to person and cell to cell, and a subject’s global total fetal hemoglobin expression could be the result of many cells expressing modest amounts of globin or a few cells expressing robust amounts of globin.56
Although the coexistence of a primitive erythropoietic wave in our cultures cannot be completely excluded, it is highly unlikely that the day-29 BFU-E–derived cells are the progeny of different developmental programs. This is supported by principal component analysis presenting the data as a continuum of cells (Figure 4B), whereas a recent study has shown that iPSC-derived primitive erythroid cells cluster separately from both iPSC- and cord blood–derived definitive erythroblasts based on the differential expression of 3149 genes.28 We only found 219 genes differentially expressed between GFP− and GFP+ cells, mostly linked to erythroid maturation. Therefore, coexpression of embryonic, fetal, and adult globins in individual cells is likely to represent a yolk sac EMP program of hematopoietic development, displaying a gradient of erythroid maturation. Importantly, the identification of these cells as definitive erythroid cells also implies that they are all inherently capable of producing β-globin and can undergo more extensive γ-to-β switching once the signals catalyzing this switch are unraveled. That PSC progeny can switch from γ- to β-globin in an in vivo setting has been previously demonstrated after transplantation and maturation of PSC-derived cells in immunodeficient mice.57,58 Making use of this reporter cell line and its GFP readout will enable high-throughput screens for compounds that might facilitate this switch and increase β-globin expression in PSC-derived cells in vitro, in a controlled environment, and improve the resolution with which we can study β-thalassemia and sickle-cell disease.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Brian R. Tilton and Riley M. F. Pihl of the Boston University Flow Cytometry Core Facility for technical assistance and Matthew Porteus for constructs and technical advice.
This work was supported by NextGen Consortium Grant U01HL107443, a collaborative grant between the Murphy and Steinberg Groups (1R01HL133350), and the Training Grant for Hematology (5T32 HL007501), all from the National Heart, Lung, and Blood Institute, National Institutes of Health.
Authorship
Contribution: K.V. performed conception and design, collection and/or assembly of data, data analysis and interpretation, and manuscript writing; T. Matte performed data collection and computational analysis; A.L., Z.H.N., and T. Morrison performed collection and/or assembly of data; D.H.K.C. performed data analysis and interpretation; M.H.S. performed data analysis and interpretation and manuscript writing; and G.J.M. performed conception and design, collection and/or assembly of data, data analysis and interpretation, and manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: George J. Murphy, Center for Regenerative Medicine, Boston University School of Medicine, 670 Albany St, Suite 208, Boston, MA 02118; e-mail: gjmurphy@bu.edu.