Key Points
A gene expression assay that accurately assigns B-cell–associated gene signatures to DLBCL with prognostic impact.
Robust performance irrespective of sample preparation method and GEP platform.
Abstract
Gene expression profiling (GEP) by microarrays of diffuse large B-cell lymphoma (DLBCL) has enabled the categorization of DLBCL into activated B-cell–like and germinal center B-cell–like subclasses. However, as this does not fully embrace the great diversity of B-cell subtypes, we recently developed a gene expression assay for B-cell–associated gene signature (BAGS) classification. To facilitate quick and easy-to-use BAGS profiling, we developed in this study the NanoString-based BAGS2Clinic assay. Microarray data from 4 different cohorts (n = 970) were used to select genes and train the assay. The locked assay was validated in an independent cohort of 88 sample biopsies. The assay showed good correspondence with the original BAGS classifier, with an overall accuracy of 84% (95% confidence interval, 72% to 93%) and a subtype-specific accuracy ranging between 80% and 99%. BAGS classification has the potential to provide valuable insight into tumor biology as well as differences in resistance to immuno- and chemotherapy that can lead to novel treatment strategies for DLBCL patients. BAGS2Clinic can facilitate this and the implementation of BAGS classification as a routine clinical tool to improve prognosis and treatment guidance for DLBCL patients.
Introduction
Determining the cell of origin (COO) in diffuse large B-cell lymphoma (DLBCL) would give caregivers a more specific prognosis and possibly treatment of B-cell tumors. This has motivated the categorization of DLBCL into activated B-cell–like (ABC) and germinal center B-cell-like (GCB) subclasses,1 each with distinct biology and pathogenesis as well as outcome after treatment.2,3 However, this classification agglomerates several naturally occurring B-cell subsets into 2 classes: (1) germinal center cells, which contain both centroblasts and centrocytes, and (2) in vitro activated B cells, including memory cells and plasmablasts.1 We have recently developed a B-cell–associated gene signature (BAGS) that reflects the natural B-cell hierarchy by gene expression profiling (GEP) on microarray.4 Tonsils from healthy donors were sorted by fluorescence-activated cell sorting into 5 distinct B-cell subsets: naive, germinal centrocytes and centroblasts, and post–germinal memory B cells and plasmablasts. By GEP, Dybkær et al trained BAGS classifier for each subtype, which were applied on online available GEP data to associate primary tumors at time of diagnosis to normal B-cell counterparts. In a large metastudy of microarray-based GEP from 5 different clinical cohorts, BAGS provided independent prognostic information to the ABC/GCB subclasses and the international prognostic index.4 BAGS classification has also been associated with drug resistance by GEP of B-cell cancer cell lines subjected to various forms of chemotherapy used in routine clinical treatment.5 The resulting resistance gene signature classifiers were tested in a metastudy of GEP from 3 clinical cohorts, establishing different predicted responses to drugs dependent on BAGS subtype. In addition, BAGS subtypes had distinct genetic profiles and differentially activated signaling pathways.4,5 However, the use of microarrays to acquire GEP data to perform BAGS classification is expensive and labor intensive and requires cumbersome data analysis. We therefore turned to recent technological advances in quick and easy-to-use quantitative gene expression profiling6 to develop a simplistic and parsimonious gene expression assay that accurately and robustly assigns COO in DLBCL according to the natural differentiation hierarchy of B cells independent of sample storage method and GEP platform.
Methods
A total of 970 DLBCL patients originating from 4 online available data sets3,7,8 spanning different geographical regions and time eras were used in combination as the training cohort. All cases in the training cohort were profiled using the GeneChip Human Genome U133 Plus 2.0 (U133+2) microarray (Affymetrix). The independent validation cohort consisted of 88 de novo DLBCLs diagnosed from 1997 to 2011 at Aalborg University Hospital, Denmark. Each case was individually diagnosed by an expert pathologists, and no cases with additional lymphoid malignancies were included in this study. Patients were treated with either cyclophosphamide, doxorubicin, vincristine, and prednisone or rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. Only patients treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone were used for survival analysis. Samples were stored as either formalin-fixed paraffin-embedded tissue (FFPET; n = 34) or snap-frozen optimal cutting tissue (OCT; n = 54). Digital-multiplexed gene expression (DMGE) profiling was performed with the nCounter (NanoString Technologies) platform, using 300 ng purified RNA for FFPET tissue and 200 ng purified RNA for OCT tissue, respectively. Candidate genes for the assay were identified from the training cohort, which previously has been assigned BAGS subtype according to the gold standard.4 To facilitate the transfer between GEP platforms, a pilot study of 14 OCT DLBCL samples independent of the training and validation cohorts were profiled in parallel on the U133+2 microarray and the nCounter platform to assess the correlation between microarray and DMGE measurements. Highly correlated genes were then included in a multinomial regression model, which calculated gene weights and BAGS-subtype probability scores from the training cohort. To correlate differences in gene expression between BAGS subtypes, differential gene expression analysis was performed according to the DESeq2 workflow.9 This was done on the raw NanoString count data with P values adjusted according to the Benjamini-Hochberg procedure.10 To test the robustness toward one-by-one classification of single cases in the validation cohort, normalization was simulated by calculating the median and variance used for standardization on all samples except one.11 The left-out sample was subsequently standardized with the calculated median and variance before being classified into BAGS subtype. This procedure was repeated for all samples, and the classification was compared with the classification resulting from normalizing all cases together. A detailed description of the gene-selection procedure, model building, and assessment procedure is available in the supplemental Methods. All source code and classifier algorithms are freely available upon request.
Results
The robustness of the gene-selection procedure was validated by bootstrapping, achieving high accuracies ranging from 82% to 92% for the 4 contrasts on the out-of-bag samples. Of the 182 genes initially profiled in the pilot study, 128 genes with a correlation >0.8 between microarray and DMGE measurements were selected to construct a multinomial regression model (Figure 1A). The model was evaluated by bootstrapping, with an overall accuracy of 82.5% (95% CI, 82.4% to 82.6%) estimated from the out-of-bag samples across 1000 iterations. In addition, the subtype-specific sensitivity and specificity were consistently good (supplemental Table 3). The cut of the probability scores for the BAGS-subtype assignment originating from the multinomial regression was set to 0.90, allowing 3% of samples to be left as unclassified in the training cohort. This locked model with established gene weights, probability scores, and cutoff values constitutes the BAGS2Clinic assay.
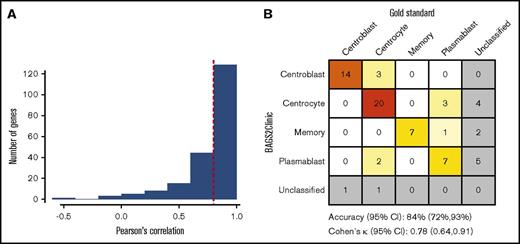
Interplatform performance of the BAGS2Clinic assay. (A) Histogram of correlations between gene expression obtained using nCounter (NanoString Technologies) or the Human Genome U133 Plus 2.0 microarray (Affymetrix) from a pilot study of 14 OCT DLBCL samples; the 0.8 cutoff value was used to select highly correlated genes (indicated by a vertical red dotted line). (B) Confusion matrix of the assignment of DLBCL to BAGS subtypes by the gold standard (1) vs the BAGS2Clinic assay on a subset of the independent validation cohort (n = 70).
Interplatform performance of the BAGS2Clinic assay. (A) Histogram of correlations between gene expression obtained using nCounter (NanoString Technologies) or the Human Genome U133 Plus 2.0 microarray (Affymetrix) from a pilot study of 14 OCT DLBCL samples; the 0.8 cutoff value was used to select highly correlated genes (indicated by a vertical red dotted line). (B) Confusion matrix of the assignment of DLBCL to BAGS subtypes by the gold standard (1) vs the BAGS2Clinic assay on a subset of the independent validation cohort (n = 70).
DMGE profiles were obtained from 94 samples in the validation cohort, with four FFPET and two OCT samples failing to fulfill the quality control criteria (see supplementary material for detail). The concordance between the gold standard and BAGS2Clinic was tested on a subset of the validation cohort (n = 70) with matched U133+2 microarray GEP data available (Figure 1B). Of the 57 cases that were BAGS classified according to the gold standard the BAGS2Clinic assay misclassified 9 cases, obtaining an accuracy of 84% (95% confidence interval [CI], 72% to 93%). No significant differences in frequencies of the BAGS subtype assignment were observed between the training and validation cohorts (χ2 = 10.6, degrees of freedom = 9, and P = .31). The assay was robust irrespective of biopsy storage protocol and achieved a high classification accuracy of 79% (95% CI, 49% to 95%) and 86% (95% CI, 72% to 95%) within the FFPET (n = 16) and OCT (n = 54) subsets, respectively. The intrasubtype accuracy, sensitivity, and specificity calculated using a one-versus-all approach were good, with a range of 80% to 99%, 64% to 100%, and 93% to 98%, respectively (Table 1). Furthermore, the robustness toward classifying single cases revealed a classification accuracy of 83% by leave-one-out cross validation, which is comparable to similar studies assessing normalization and classification of single cases.11 To further test the robustness of the 90% classification probability value, cutoff values were also evaluated at 80%, 85%, 90%, and 95%, all with the same classification accuracy of 84% (supplemental Figure 11). The biological concordance with the original BAGS classification was assessed by a differential gene expression analysis performed on the NanoString GEP data from the validation cohort and compared with previous studies.4 This confirmed the downregulation of LMO2, which is associated with poor survival,12 in the centroblast subtype compared with the centrocyte subtype (fold change = 0.46, P = .014). Similarly, the centroblast subtype had high expression of TCL1A (fold change = 1.9, P = .003) and low expression of CCR7 (fold change = 0.45, P = .014), which is known to be suppressed in CD30+ DLBCL with favorable prognosis.13 The gene BCL6 was suppressed in the memory subtype relative to the plasmablast subtype (fold change = 0.45, P < .001) and is a known oncoprotein associated with poor outcome.14 Similarly, downregulation of MYBL1, which was suppressed in memory cells (fold change = 0.14, P < .001), is associated with poor outcome.15 A heatmap of the gene expressions for all samples is provided in supplemental Figure 9.
Performance of the BAGS2Clinic assay in the validation cohort
| . | Centroblast . | Centrocyte . | Memory . | Plasmablast . |
|---|---|---|---|---|
| Sensitivity, % | 100 | 80 | 100 | 64 |
| Specificity, % | 93 | 91 | 98 | 96 |
| Balanced accuracy, % | 97 | 85 | 99 | 80 |
| . | Centroblast . | Centrocyte . | Memory . | Plasmablast . |
|---|---|---|---|---|
| Sensitivity, % | 100 | 80 | 100 | 64 |
| Specificity, % | 93 | 91 | 98 | 96 |
| Balanced accuracy, % | 97 | 85 | 99 | 80 |
The performance for each subtype was assessed by a one-versus-all approach. The balanced accuracy is calculated as (sensitivity + specificity)/2.
Overall survival (OS) and progression-free survival (PFS) relative to the COO assignment by the BAGS2Clinic assay were inspected in the validation cohort to assess the prognostic stratification identified previously4 (Figure 2). This indicated a shorter survival for the memory subtype compared with the plasmablast subtype in both OS (hazard ratio [HR], 3.46; 95% CI, 1.17-10.23) and PFS (HR, 2.75; 95% CI, 0.99-7.69). Furthermore, the centroblast subtype had an inferior prognosis in PFS (HR, 1.54; 95% CI, 0.60-3.90) compared with the centrocyte subtype, although not observed in OS (HR, 1.08; 95% CI, 0.39-2.97) (Figure 2).
Prognostic impact of the BAGS2Clinic assay in an independent validation cohort. OS (A) and PFS (B) evaluated by Kaplan-Meier survival curves indicating the prognostic impact of BAGS2Clinic on the validation cohort (n = 77). HRs were computed by Cox regression. All unclassified samples were excluded from the survival analysis. CB, centroblast; CC, centrocyte; M, memory; PB, plasmablast.
Prognostic impact of the BAGS2Clinic assay in an independent validation cohort. OS (A) and PFS (B) evaluated by Kaplan-Meier survival curves indicating the prognostic impact of BAGS2Clinic on the validation cohort (n = 77). HRs were computed by Cox regression. All unclassified samples were excluded from the survival analysis. CB, centroblast; CC, centrocyte; M, memory; PB, plasmablast.
Discussion
The results of this study supports the hypothesis of BAGS being a prognostic marker, although the sample sizes of BAGS subsets in the validation cohort do not provide the statistical power needed for establishing significant results. The assay has demonstrated concordant results in COO assignment according to the gold standard, irrespective of GEP platform and tissue storage method, which have been validated in an independent cohort of DLBCL. It has also demonstrated a good classification performance when evaluated on single cases, and the classification probability cutoff is robust. The introduction of the BAGS2Clinic assay provides a more detailed stratification of DLBCL than dichotomous ABC/GCB classification. Stratification of DLBCL by ABC/GCB classification and molecular features is required according to the latest World Health Organization guidelines.16 This includes the distinction of high-grade B-cell lymphomas from DLBCL by the presence of MYC and BCL2 and/or BCL6 rearrangements referred to as double- or triple-hit lymphomas,17 previously constituting a subgroup of GCB DLBCLs with inferior prognosis.18,19 To what extent the BAGS-derived subgroups correlate with the biomarkers defined by the World Health Organization guidelines is the subject of ongoing research testing the BAGS2Clinic assay in a large cohort of DLBCL patients. This will also clarify the prognostic relevance independent of ABC/GCB classification and the international prognostic index. Nevertheless, dividing DLBCL into more subtle subgroups reflecting naturally occurring B cells may provide the level of detail needed to link observed biomarkers of clinical relevance with B-cell biology. The stratification of DLBCL is particularly relevant for the development of targeted therapies, which is an area of intense research with multiple drugs currently being tested.20 The correlation of BAGS subtypes to molecular targets and resistance gene signatures may provide an actionable path forward, developing tools for predicting subtype-dependent response to treatment.5 This may not be limited to DLBCL. A recent study demonstrated the prognostic value of BAGS in chronic lymphocytic leukemia, with elevated mutation frequencies of specific subtypes and distinct predicted responses to treatment depending on subtype.21 In other B-cell–like malignancies, such as follicular lymphoma and Burkitt lymphoma, the BAGS subtypes may provide additional important information to explain clinical heterogeneity. The BAGS2Clinic makes this information accessible for a low price and short turnaround time to help explain some of the molecular and prognostic heterogeneity observed in DLBCL.4 This is particularly relevant for clinical applications as a novel routine tool for diagnostic and prognostic assessment as well as insight into the stages of clonal differentiation and oncogenesis.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors would like to thank laboratory technicians Dana Germer and Helle Høholt for laboratory preparation and analysis of patient samples.
Authorship
Contribution: M.B. and T.Y.M. designed the study, performed research, and wrote the manuscript; J.R., and K.D designed the study, collected data and performed research; T.Y.M. conducted the statistical analysis and collected data; M.B., R.F.B, and T.Y.M designed the statistical analysis; W.K., H.E.J., and M.A. provided guidance on sample collection, data interpretation, and biotechnological questions; and all authors read and approved the final version of the manuscript before submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martin Bøgsted, Sdr Skovvej 15, DK-9000, Aalborg, Denmark; e-mail: martin.boegsted@rn.dk.