Key Points
Basal intracellular Ca2+ levels and migration increase with higher CD38 expression in CLL cells.
Rap1 and the Rap1 guanine-nucleotide exchange factor RasGRP2 are required for CLL migration and regulated by CD38 levels.
Abstract
CD38 is a transmembrane exoenzyme that is associated with poor prognosis in chronic lymphocytic leukemia (CLL). High CD38 levels in CLL cells are linked to increased cell migration, but the molecular basis is unknown. CD38 produces nicotinic acid adenine dinucleotide phosphate and adenosine 5′-diphosphate-ribose, both of which can act to increase intracellular Ca2+ levels. Here we show that CD38 expression increases basal intracellular Ca2+ levels and stimulates CLL cell migration both with and without chemokine stimulation. We find that CD38 acts via intracellular Ca2+ to increase the activity of the Ras family GTPase Rap1, which is in turn regulated by the Ca2+-sensitive Rap1 guanine-nucleotide exchange factor RasGRP2. Both Rap1 and RasGRP2 are required for CLL cell migration, and RasGRP2 is polarized in primary CLL cells with high CD38 levels. These results indicate that CD38 promotes RasGRP2/Rap1-mediated CLL cell adhesion and migration by increasing intracellular Ca2+ levels.
Introduction
Chronic lymphocytic leukemia (CLL) is a cancer of B cells, and one of the most common leukemias in adults. CLL is highly heterogeneous: some patients present with an indolent form, whereas others progress rapidly despite aggressive therapy.1 Disease progression is associated with an increase in CLL cell infiltration of secondary lymphoid tissues and bone marrow, leading to immune dysfunction and bone marrow failure. Within lymphoid niches, but not in the peripheral blood, B-cell receptor (BCR) signaling and microenvironmental stimuli induce CLL cell proliferation.2,3 CLL cell trafficking to and retention within lymphoid niches may therefore play a key role in disease progression. Notably, clinically successful BCR signaling inhibitors, such as the Btk inhibitor ibrutinib and PI-3-kinase-δ inhibitor idelalisib, alter CLL cell trafficking, leading to a decrease in CLL cells in lymphoid tissues and accumulation in the blood.4-7
Several prognostic markers for CLL are implicated in cell adhesion and migration, including the ecto-enzyme CD38 and the tyrosine kinase ZAP70.8,9 Other proteins involved in cell adhesion and migration are also associated with disease progression, including the integrin α4/CD49d, the matrix metalloprotease MMP9, and the adhesion molecule CD44.10-14
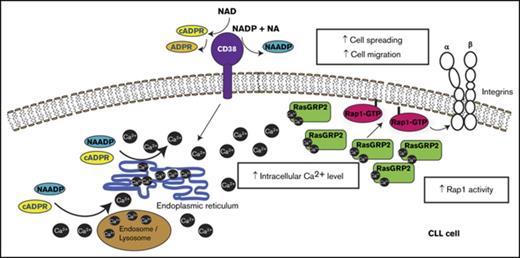
CD38 is a type II transmembrane protein of the adenosine 5′-diphosphate-ribosyl transferase family. The C-terminal extracellular domain of CD38 is an enzyme that converts nicotinamide adenine dinucleotide to adenosine 5′-diphosphate-ribose (ADPR) and cyclic ADP-ribose (cADPR), and nicotinamide adenine dinucleotide phosphate to nicotinic acid adenine dinucleotide phosphate (NAADP).15-17 These products can induce an increase in intracellular Ca2+. CD38 is considered a potential therapeutic target in patients with CLL, either using neutralizing antibodies or enzyme inhibitors.18,19 Indeed, an enzymatically inactive CD38 is unable to support disease progression in a xenograft model for CLL.20
Increasing evidence indicates that CD38 is involved in CLL cell trafficking. For example, higher CD38 levels correlate with increased chemotaxis of CLL cells toward chemokines such as CCL21 and CXCL12, which are present in lymph nodes and likely to regulate CLL cell accumulation in lymphoid niches.20,21 In addition, increased CD38 expression correlates with higher integrin-mediated adhesion to VCAM-1.22 In the human CLL cell line MEC1, overexpression of wild-type but not enzymatically inactive CD38 increases cell migration.20 Together, these results suggest that the catalytic function of CD38 modulates CLL cell adhesion and motility, but the signaling pathways underlying these processes have not been elucidated so far.
Here we investigate the molecular basis for the effects of CD38 on CLL cell migration. We show that CD38 expression stimulates basal as well as chemokine-driven migration. CD38 increases basal intracellular Ca2+ levels, which in turn activates the small GTPase Rap1 via a guanine-nucleotide exchange factor (GEF) for Rap1, RasGRP2, which is likely to be Ca2+-regulated.23 Rap1 is known to stimulate integrin activation,24,25 and hence this pathway could provide a new therapeutic strategy to inhibit trafficking of CLL cells into lymphoid niches.
Methods
Cell culture and patient samples
Blood samples from patients with a confirmed CLL diagnosis were collected after informed consent and in accordance with the Declaration of Helsinki (see supplemental Table 1 for patient characteristics). Ethical approval was obtained from the United Kingdom National Research Ethics Service (08/H0906/94); all patients provided informed written consent. Peripheral blood mononuclear cells were isolated by Ficoll density gradient centrifugation and cryopreserved in aliquots. Thawed cells were cultured in RPMI-1640 containing 10% heat-inactivated fetal calf serum (FCS) and 1% bovine serum albumin (BSA). CD38 expression and B-cell markers were assessed by flow cytometry with anti-CD5-fluorescein isothiocyanate, anti-CD19-phycoerythrin, and anti-CD38-phycoerythrin-Cy5 (Beckman Coulter).
The human MEC1 cell line (kind gift from John Gribben, Queen Mary University of London, London, United Kingdom) was cultured in IMDM containing 2 mM glutamine and 10% FCS. HEK293T cells were cultured in DMEM containing 10% FCS. All media contained penicillin (100 U/mL) and streptomycin (100 μg/mL).
Generation of MEC1 cell populations
Lentiviral particles were obtained by co-transfecting HEK293T cells with 3 vectors: pΔ8.91, pMDG, and vectors expressing CD38 or GFP (pLentiS38W or pLentiSEW, respectively26 ). Lentiviral particles were added to MEC1 cells, and cells were centrifuged at 200g for 1 hour. CD38+ cells, labeled with phycoerythrin-conjugated anti-CD38 antibody (clone HB7; BD Bioscience), and GFP+ cells were sorted with a FACSAria II Cell Sorter (BD Bioscience). Cell populations were used for experiments at least 3 passages after sorting.
siRNA transfection
The siRNA oligonucleotides (Dharmacon) used were RasGRP2 (siRNA1:GUGCAAGGAUCGCCUGUCA; siRNA2: CCAAUUCCCUGCAGGUGAA), RasGRP3 (siRNA1: GGAGAAAGCUGCAAUGAAU; siRNA2: GAAUGCCUCUCACCACUUA), Rap1A (siRNA1: GAUAGAAGAUUCCUACAGA; siRNA2: CAAUAAAUGUGACCUGGAA), Rap1B (siRNA1: GAACAACUGUGCAUUCUUA, siRNA2: CAAUGAUUCUUGUUGGUAA), and control nontargeting siRNA (D-001810-02). MEC1 cells (5 × 106) were transfected with 1.2 μM siRNA in 100 μL human B-cell Nucleofector Kit (Lonza), using an Amaxa nucleofector. Experiments were performed 72 hours after siRNA nucleofection.
Migration assay
MEC1 (3 × 105) or primary CLL cells (5 × 105) were added to VCAM-1-coated (5 µg/mL) 6.5-mm diameter transwells (5-μm pore), and 600 μL medium with or without 100 ng/mL CCL21 (R&D Systems) was added to bottom chambers. MEC1 cells in bottom chambers were counted after 2 hours, using a Casy cell counter (Schärfe System). The migration index for MEC1 cells was calculated by normalizing the values to control MEC1-GFP cells without chemokine or transfected with control siRNA. For primary CLL cells, 300-μL aliquots were harvested from bottom chambers after 16 hours, and the absolute number of migrated cells was determined with CountBright beads for flow cytometry (Molecular Probes) according to the manufacturer’s instructions. Cells in the remaining 300 μL of the bottom chambers were labeled with anti-CD19-fluorescein isothiocyanate antibody (BD-Bioscience) and analyzed using a FACS CantoII (BD-Bioscience) to determine the percentage of CD19+ CLL cells within the total migrated cells.
GTPase activity assay
MEC1 (3 × 106) and primary CLL cells (1 × 107) were lysed in ice-cold pull-down buffer containing 50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 0.5% deoxycholic acid, 0.1% SDS, 2.5 mM EGTA, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, and protease inhibitors (Complete, Roche Applied Science). Aliquots of clarified lysates were used to determine total protein levels. GST-fusion proteins containing RalGDS-RBD or PAK-PBD, purified from Escherichia coli as described,27 were immobilized onto glutathione-Sepharose beads (Amersham Bioscience) and used to capture active Rap1-GTP or Rac1-GTP, respectively, from cell lysates. Proteins were eluted from beads using Laemmli's sample buffer, and analyzed by western blotting.
Western blotting
Cell lysates were separated on 4% to 12% Bis-Tris polyacrylamide gels (Invitrogen) and transferred to nitrocellulose membranes. Membranes were incubated in blocking solution (5% nonfat dried milk or 5% BSA in TBS containing 0.1% Tween-20) before protein detection with the following antibodies (diluted 1:250 to 1:10,000 in blocking solution): anti-Rap1a/Rap1b (26B4, Cell Signaling), anti-Rac1 (23A8, Upstate-Millipore), anti-Rap1a (Santa Cruz), anti-Rap1b (36E1, Cell Signaling), anti-RasGRP2 (ThermoScientific), anti-RasGRP3 (Cell Signaling), and anti-GAPDH (Upstate-Millipore). Densitometry analysis was performed using ImageJ software.
Intracellular Ca2+ depletion
For Rap1 activity assays, MEC1 and primary CLL cells were resuspended in PBS without CaCl2 or MgCl2 for 10 minutes before incubating with 0.5 μM ionomycin for 5 minutes. Control cells were resuspended in PBS containing 0.9 mM CaCl2 and 0.5 mM MgCl2 and treated with dimethyl sulfoxide instead of ionomycin. For migration assays, MEC1 cells were treated with 0.5 μM ionomycin in low Ca2+ medium (Ca2+-free DMEM, 10% FCS, and 5 mM EDTA) for 5 minutes and then resuspended in low-Ca2+ medium. Control cells were treated with dimethyl sulfoxide instead of ionomycin in standard medium (DMEM with Ca2+ and 10% FCS).
Intracellular Ca2+ detection and F-actin content measurement
To measure intracellular Ca2+ levels, MEC1 cells or primary CLL cells (3 × 106/mL) were incubated with 1 μM Indo1-AM (Life Technologies) for 1 hour at 37°C. Cells were washed, resuspended in fresh medium, and incubated at 37°C for 30 minutes and then stored on ice before being warmed to 37°C for 5 minutes for experiments. Ca2+-free and Ca2+-bound Indo-1 emission wavelengths (420 and 510 nm) were recorded on a BD LSRFortessa flow cytometer (BD Biosciences) in unstimulated cells for up to 1 minute before stimulation with CCL21 (100 ng/mL) for 2 minutes. Ionomycin (1 μM) was used to induce Ca2+ influx as a positive control. The ratio between the 2 Indo1 emission wavelengths (420 nm/510 nm, termed Ratio Indo1) was measured using FlowJo software (Tree Star) and represents the intracellular free Ca2+ levels.
To measure F-actin levels, MEC1 cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS. Cells were incubated in blocking solution (3% BSA in PBS) for 1 hour and then stained with Alexa Fluor 647–conjugated phalloidin (2 units/1 × 105 cells in blocking solution) for 1 hour and analyzed by flow cytometry (FACS CantoII).
Microscopy and image analysis
MEC1 and primary CLL cells were seeded on VCAM-1-coated coverslips (5 μg/mL; R&D Systems) or 0.01% poly-l-lysine. Cells were then fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with 3% BSA, and incubated with anti-RasGRP2 (1:200; Thermo Scientific, PA5-28865) or anti-CD38 (1:100; Abcam, ab33535) antibody followed by Alexa Fluor 546– or 488–conjugated secondary antibody (Life Technologies). F-actin was visualized with Alexa Fluor 488- or 633-conjugated phalloidin (Invitrogen). Cells were imaged with a LSM510 confocal microscope, using Zen software (Zeiss).
RasGRP2 distribution was quantified with the Oval Profile plug-in of ImageJ. A circle was drawn around each cell, and a Radial Sum profile plot was generated, representing the interpolated pixel intensities (from the circle center to the periphery) along 360 radii. The 360 values obtained per cell were then sorted from the lowest to the highest to calculate the slope of the curve, which reflects the variation of fluorescence around the cell. A slope score of 0 (straight horizontal line) represents no variation within the 360 radii; hence, a uniformly distributed fluorescence signal (low polarization) and a high positive slope score represent a high variation of the fluorescence localization along the radii (high polarization).
Interference reflection microscopy (IRM) was used to produce images containing only the regions of close contact between the cell and the adhesive surface.28 Bright field and IRM images were obtained simultaneously on a LSM510 confocal microscope. The cell attachment area was quantified from IRM images using ImageJ.
Statistical analysis
Statistical significance was determined with 2-tailed Student t test using IBM SPSS Statistic or GraphPad Prism software.
Results
CD38 expression promotes CLL cell migration and cell spreading
To investigate the effects of CD38 expression on cell motility, we transfected the CLL-derived MEC1 cell line, which does not express endogenous surface CD38,20 with a lentivirus encoding CD38 or a control GFP-encoding lentivirus. Stably expressing CD38+ (termed MEC1-CD38H) and GFP+ (termed MEC1-GFP) cells were sorted by FACS, and CD38 expression was monitored by flow cytometry (supplemental Figure 1A).
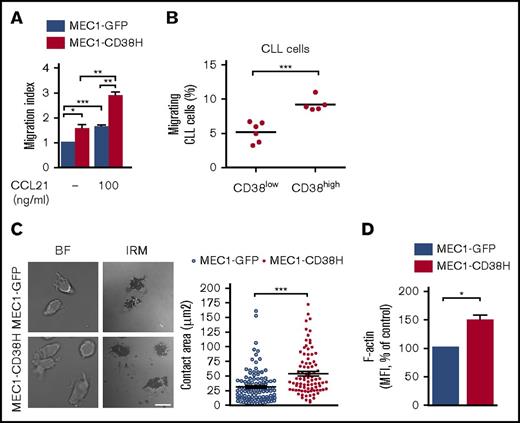
Previous studies have shown that expression of CD38 in CLL cells is associated with enhanced cell migration in response to a chemokine gradient.20,21 We sought to assess whether CD38 expression is specifically linked to chemokine-directed cell migration or whether its expression also alters basal cell motility. In transwell assays, MEC1-CD38H cells not only exhibited higher chemotaxis toward CCL21 compared with MEC1-GFP cells, in agreement with previous observations,20,21 but also showed increased migration in the absence of chemokine (Figure 1A). The higher migration toward CCL21 was not a result of different levels of its receptor CCR7 between MEC1-GFP and MEC1-CD38H cells (supplemental Figure 1B), and is instead likely to be a result of the increased basal migration.
CD38 expression increases CLL cell migration and cell spreading. (A) MEC1-GFP and MEC1-CD38H cells were added to VCAM-1-coated transwell filters in the absence or presence of 100 ng/mL CCL21 in the bottom chamber. The migration index was obtained by normalizing the values to the MEC1-GFP control cells. Data shown are the mean of 3 independent experiments ± SEM. (B) Peripheral blood mononuclear cells from 11 patients with CLL were added to VCAM-1-coated transwell filters; values represent the percentage of CD19+ migrated cells divided by the total number of CD19+ cells added to the filter. Horizontal bars indicate mean values of migrating cells for CD38low (n = 6) and CD38high (n = 5) CLL samples. (C) Representative images of fixed cells and quantification of the cell attachment area (μm2) of MEC1-GFP and MEC1-CD38H cells seeded on VCAM-1 (n ≥ 100 cells per population from 3 independent experiments). Scale bar, 10 μm. (D) MEC1-GFP and MEC1-CD38H cells were stained with Alexa Fluor 647–conjugated phalloidin and analyzed by flow cytometry. Values were obtained by normalizing the median fluorescent intensity to the MEC1-GFP control cells. Data shown are the mean of 5 independent experiments ± SEM. Horizontal bars indicate mean values ± SEM. *P < .05; **P < .01; *** P < .001 determined by 2-tailed Student t test. BF, bright field; IRM, interference reflection microscopy.
CD38 expression increases CLL cell migration and cell spreading. (A) MEC1-GFP and MEC1-CD38H cells were added to VCAM-1-coated transwell filters in the absence or presence of 100 ng/mL CCL21 in the bottom chamber. The migration index was obtained by normalizing the values to the MEC1-GFP control cells. Data shown are the mean of 3 independent experiments ± SEM. (B) Peripheral blood mononuclear cells from 11 patients with CLL were added to VCAM-1-coated transwell filters; values represent the percentage of CD19+ migrated cells divided by the total number of CD19+ cells added to the filter. Horizontal bars indicate mean values of migrating cells for CD38low (n = 6) and CD38high (n = 5) CLL samples. (C) Representative images of fixed cells and quantification of the cell attachment area (μm2) of MEC1-GFP and MEC1-CD38H cells seeded on VCAM-1 (n ≥ 100 cells per population from 3 independent experiments). Scale bar, 10 μm. (D) MEC1-GFP and MEC1-CD38H cells were stained with Alexa Fluor 647–conjugated phalloidin and analyzed by flow cytometry. Values were obtained by normalizing the median fluorescent intensity to the MEC1-GFP control cells. Data shown are the mean of 5 independent experiments ± SEM. Horizontal bars indicate mean values ± SEM. *P < .05; **P < .01; *** P < .001 determined by 2-tailed Student t test. BF, bright field; IRM, interference reflection microscopy.
Given that CD38 enhanced MEC1 cell basal migration, we tested whether CD38 expression levels affected the basal migration of primary human CLL cells. Samples were categorized into CD38low and CD38high cells, based on a cutoff value of 30% of CD38+ CLL cells, for CD38high. Consistent with our findings with MEC1 cells, high CD38 expression was associated with increased CLL cell basal migration (Figure 1B; supplemental Figure 1C).
To determine how CD38 might regulate cell migration in the absence of added chemokine, we initially evaluated its effect on cell adhesiveness, which is an important component of cell migration.29 We used IRM to measure MEC1-GFP and MEC1-CD38H cell spreading on VCAM-1. MEC1-CD38H cells showed a significantly increased spreading compared with MEC1-GFP cells (Figure 1C). This was not a result of any difference in expression of the VCAM-1 ligands α4- (CD49d) and β1-integrin (CD29) between MEC1-GFP and MEC1-CD38H cells (supplemental Figure 1D-E), but correlated with an increased level of polymerized actin (F-actin) in MEC1-CD38H compared with MEC1-GFP (Figure 1D). Taken together, these results indicate that CD38 directly alters CLL cell motility by enhancing cell spreading and intrinsic cell migration.
CD38 stimulates cell migration via the GTPase Rap1
To determine the molecular basis for the effects of CD38 on CLL cell adhesion and migration, we tested the role of Rap and Rac GTPases, because they are known to stimulate leukocyte adhesion, polarization, and migration.30,31
To investigate whether Rap1 or Rac1 is involved in the motility of CD38-expressing cells, we first measured their activity in the 2 MEC1 cell populations. The level of active Rap1-GTP was significantly higher in MEC1-CD38H compared with MEC1-GFP cells, both in basal conditions and after CCL21 stimulation (Figure 2A). Total Rap1 levels were not affected by CD38 expression (Figure 2A). In contrast, Rac1 activity did not differ between MEC1-GFP and MEC1-CD38H cells, and Rac1 activity was not increased by CCL21 stimulation (supplemental Figure 2A-B). These results are in agreement with recent observations showing that in CLL cells, in contrast with normal B cells, Rac1 is not activated by chemokines and does not contribute to CLL cell motility.32
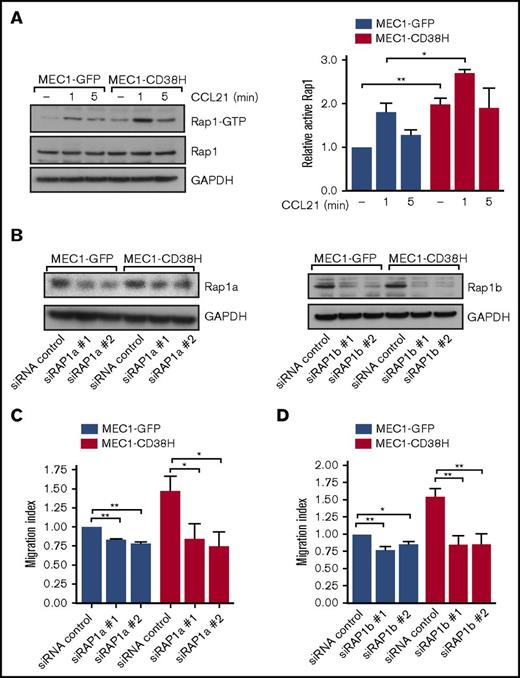
CD38 increases Rap1 activity that is required for cell motility. (A) Representative blot (left) and quantification (right) of active Rap1-GTP levels, assessed by Rap1 pulldown assays, in MEC1-GFP and MEC1-CD38H cells untreated or treated with CCL21 (100 ng/mL) for the indicated times. Graph shows the mean of 3 independent experiments ± SEM. Relative active Rap1 levels were obtained by normalizing each value to the untreated MEC1-GFP control cells. *P < .05; **P < .01 determined by 2-tailed Student t test. (B-D) MEC1-GFP and MEC1-CD38H cells were transfected with siRNAs targeting Rap1a or Rap1b. (B) Representative blots of Rap1a and Rap1b expression levels in MEC1-GFP and MEC1-CD38H cells. (C-D) Migration of MEC1-GFP and MEC1-CD38H cells through transwell filters after Rap1a (C) or Rap1b depletion (D). The migration index was obtained by normalizing the values to the MEC1-GFP control cells. Data shown are the mean of 3 independent experiments ± SEM. *P < .05; **P < .01 determined by 2-tailed Student t test.
CD38 increases Rap1 activity that is required for cell motility. (A) Representative blot (left) and quantification (right) of active Rap1-GTP levels, assessed by Rap1 pulldown assays, in MEC1-GFP and MEC1-CD38H cells untreated or treated with CCL21 (100 ng/mL) for the indicated times. Graph shows the mean of 3 independent experiments ± SEM. Relative active Rap1 levels were obtained by normalizing each value to the untreated MEC1-GFP control cells. *P < .05; **P < .01 determined by 2-tailed Student t test. (B-D) MEC1-GFP and MEC1-CD38H cells were transfected with siRNAs targeting Rap1a or Rap1b. (B) Representative blots of Rap1a and Rap1b expression levels in MEC1-GFP and MEC1-CD38H cells. (C-D) Migration of MEC1-GFP and MEC1-CD38H cells through transwell filters after Rap1a (C) or Rap1b depletion (D). The migration index was obtained by normalizing the values to the MEC1-GFP control cells. Data shown are the mean of 3 independent experiments ± SEM. *P < .05; **P < .01 determined by 2-tailed Student t test.
To explore the effects of the differential Rap1 activity on CLL cell motility, we used siRNAs to knockdown the expression of the 2 Rap1 proteins, Rap1a and Rap1b (Figure 2B; supplemental Figure 2C-D). Downregulation of Rap1a and Rap1b significantly impaired both basal migration and CCL21-stimulated migration (Figure 2C-D; supplemental Figure 2E-F). Notably, MEC1-CD38H cells exhibited a more pronounced decrease in migration compared with MEC1-GFP control cells on Rap1a or Rap1b depletion (supplemental Figure 2G). This indicates that the higher basal Rap1 activity of CD38-expressing MEC1 cells contributes to their increased migration.31
Rap1 activity in CD38-expressing CLL cells is linked to basal intracellular Ca2+ levels
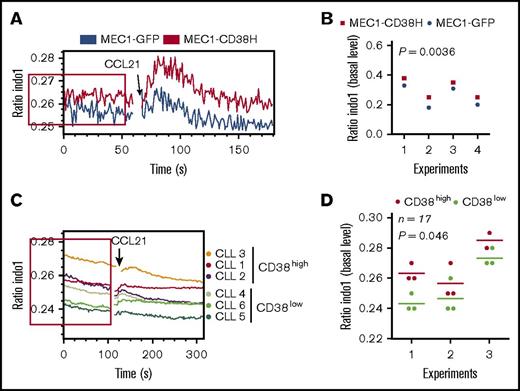
CD38 is an ectoenzyme that catalyzes the synthesis of 3 different products (NAADP, cADPR, and ADPR) involved in increasing intracellular Ca2+ levels.20,33-35 We therefore analyzed the effect of CD38 expression on Ca2+ levels. MEC1-CD38H cells exhibited higher Ca2+ levels both under basal conditions and after CCL21 stimulation compared with MEC1-GFP cells (Figure 3A-B; supplemental Figure 3A). The higher intracellular Ca2+ in MEC1-CD38H cells after CCL21 addition reflects the increased basal Ca2+ levels, as there was no difference between the 2 populations in CCL21-induced Ca2+ levels when the basal levels were normalized to each other (supplemental Figure 3B).
CD38 expression is associated with elevated basal Ca2+levels. MEC1-GFP, MEC1-CD38H, and primary CLL cells with varying CD38 expression levels (n = 17 patient samples, analyzed in 3 independent experiments) were incubated with the ratiometric Ca2+ indicator Indo-1 and analyzed by flow cytometry. The basal fluorescence level was recorded for 60 seconds before stimulation with CCL21 (100 ng/mL) for a further 120 seconds. (A,C) Representative bivariate plot of the median of fluorescent intensity ratio (Indo-1 emission wavelengths: 420/510 nm) against time; red boxes indicate the basal fluorescence level. (B,D) Peak values of the median basal fluorescence intensity were obtained using FlowJo Kinetic statistic tool. Horizontal bars in panel D indicate mean values of the fluorescence intensity peak calculated in each experiment for CD38low (green) and CD38high (red) samples. Significant differences between the 2 experimental groups were determined using a 2-tailed Student t test.
CD38 expression is associated with elevated basal Ca2+levels. MEC1-GFP, MEC1-CD38H, and primary CLL cells with varying CD38 expression levels (n = 17 patient samples, analyzed in 3 independent experiments) were incubated with the ratiometric Ca2+ indicator Indo-1 and analyzed by flow cytometry. The basal fluorescence level was recorded for 60 seconds before stimulation with CCL21 (100 ng/mL) for a further 120 seconds. (A,C) Representative bivariate plot of the median of fluorescent intensity ratio (Indo-1 emission wavelengths: 420/510 nm) against time; red boxes indicate the basal fluorescence level. (B,D) Peak values of the median basal fluorescence intensity were obtained using FlowJo Kinetic statistic tool. Horizontal bars in panel D indicate mean values of the fluorescence intensity peak calculated in each experiment for CD38low (green) and CD38high (red) samples. Significant differences between the 2 experimental groups were determined using a 2-tailed Student t test.
We further investigated the link between CD38 and basal Ca2+ levels by comparing Ca2+ levels between CD38low and CD38high primary CLL samples (n = 17 patients with CLL analyzed in 3 independent experiments). Because of the inherent interexperimental variability, Ca2+ levels of primary CLL cells were compared between samples in the same experiment. CD38high samples showed significantly higher basal Ca2+ levels compared with CD38low samples (Figure 3C-D). No detectable changes in Ca2+ levels were observed after CCL21 stimulation in the samples analyzed (data not shown).
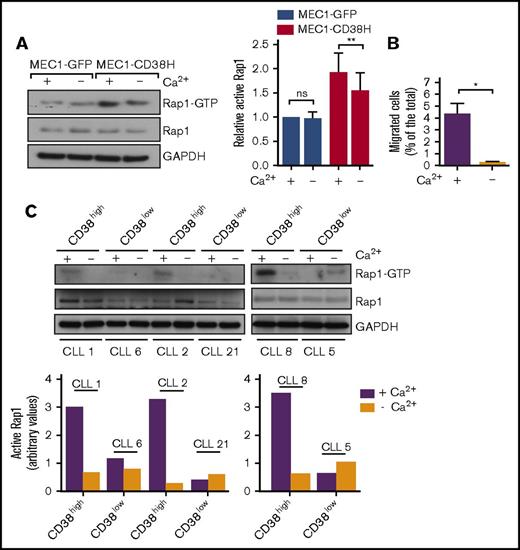
Previous studies have shown a role of agonist-mediated increases in intracellular Ca2+ in Rap1 activation.36 However, little is known about how basal Ca2+ levels affect Rap1 activity. We therefore asked whether the elevated basal Ca2+ observed in CD38-expressing cells was linked to their levels of active Rap1. We depleted intracellular Ca2+ in MEC1 cells by treatment with the Ca2+ ionophore ionomycin in a Ca2+-free solution. This lowered intracellular Ca2+ concentration in MEC1-GFP and MEC1-CD38H cells to the same level (supplemental Figure 3C). Rap1 activity was significantly reduced in MEC1-CD38H cells but not in MEC1-GFP cells after Ca2+ depletion (Figure 4A). This indicates that the higher Rap1 activity in MEC1-CD38H cells, compared with MEC1-GFP cells, is linked to their higher basal intracellular Ca2+. Consistent with our observations that Rap1 is important for CLL migration, Ca2+ depletion strongly decreased basal MEC1-CD38H cell migration (Figure 4B) without affecting cell viability (supplemental Figure 3D).
Elevated Rap1 activity and migration in CD38-expressing CLL cells is linked to intracellular basal Ca2+. Cells were depleted of intracellular Ca2+, as indicated (Methods). (A) Rap1 activity assays. Representative western blot (left) and quantification (right) of basal active Rap1 in MEC1-GFP and MEC1-CD38H cells. Relative active Rap1 levels were obtained by normalizing each value to the MEC1-GFP control cells. Graph shows the mean of 3 independent experiments ± SEM. *P < .05; **P < .01 determined by 2-tailed Student t test. (B) Migration of MEC1-CD38H cells through transwell filters after calcium depletion. Values represent the percentage of migrated cells divided by the total number of cells added to the filter. Data shown are the mean of 3 independent experiments ± SEM. *P < .05 determined by 2-tailed Student t test. (C) Western blot (top) and quantification (bottom) of basal active Rap1 in primary CLL cells with varying CD38 expression levels (n = 6 CLL patient samples). ns, not significant.
Elevated Rap1 activity and migration in CD38-expressing CLL cells is linked to intracellular basal Ca2+. Cells were depleted of intracellular Ca2+, as indicated (Methods). (A) Rap1 activity assays. Representative western blot (left) and quantification (right) of basal active Rap1 in MEC1-GFP and MEC1-CD38H cells. Relative active Rap1 levels were obtained by normalizing each value to the MEC1-GFP control cells. Graph shows the mean of 3 independent experiments ± SEM. *P < .05; **P < .01 determined by 2-tailed Student t test. (B) Migration of MEC1-CD38H cells through transwell filters after calcium depletion. Values represent the percentage of migrated cells divided by the total number of cells added to the filter. Data shown are the mean of 3 independent experiments ± SEM. *P < .05 determined by 2-tailed Student t test. (C) Western blot (top) and quantification (bottom) of basal active Rap1 in primary CLL cells with varying CD38 expression levels (n = 6 CLL patient samples). ns, not significant.
We then investigated Rap1 activity and the effect of Ca2+ depletion in CD38high and CD38low primary CLL samples. In line with the results in MEC1 cells, the CD38high samples had a markedly higher basal Rap1 activity compared with the CD38low samples (Figure 4C), whereas there was only a small variation in the total Rap1 expression between samples (supplemental Figure 3E). Rap1 activates integrins, and surface α4-integrin levels positively correlated with CD38 expression (supplemental Figure 3F), in agreement with previous reports.10,37 Moreover, Ca2+ depletion reduced Rap1 activity in all the CD38high samples. Because of the very low Rap1-GTP levels in the CD38low samples, it was not possible to observe the effect of Ca2+ depletion.
These results indicate that high CD38 expression in CLL cells is associated with elevated basal intracellular Ca2+ levels, which leads to increased Rap1 activity and, in turn, affects integrin-mediated adhesion and intrinsic cell migration.
Cell migration in CD38-expressing cells is dependent on the Rap1 GEF RasGRP2
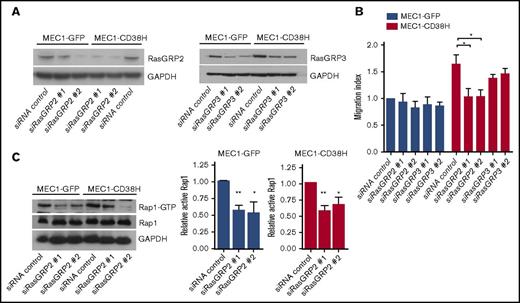
Rap1 is activated by GEFs, which stimulate exchange of GDP for GTP. The Rap-GEFs RasGRP2 and RasGRP3 have 2 Ca2+-binding domains as well as the GEF domain,38 suggesting that RasGRPs could link Rap1 to Ca2+ signaling.39-41 We therefore investigated the contribution of these Rap-GEFs to MEC1 cell migration. Depletion of RasGRP2 (Figure 5A; supplemental Figure 4A) reduced basal migration in MEC1-C38H but not MEC1-GFP cells (Figure 5B; supplemental Figure 4B). In contrast, RasGRP3 depletion did not affect the migration of either cell population (Figure 5B; supplemental Figure 4B). RasGRP2 expression was similar in the 2 MEC1 cell populations (supplemental Figure 4A). Surprisingly, RasGRP2 knockdown in MEC1-GFP cells resulted in a reduction of basal Rap1 activity to a similar extent as in MEC1-CD38H cells (Figure 5C), suggesting that the RasGRP2-Rap1 signaling axis is active in both cell populations, but its role in cell migration is limited to MEC1-CD38H cells.
The Rap1 GEF RasGRP2 is required for the enhanced migration of MEC1-CD38H cells. MEC1-GFP and MEC1-CD38H cells were transfected with siRNAs targeting RasGRP2 or RasGRP3. (A) Representative blots of RasGRP2 and RasGRP3 expression levels. (B) Migration of MEC1-GFP and MEC1-CD38H cells through transwell filters upon RasGRP2 and RasGRP3 depletion. The migration index was obtained by normalizing the values to the MEC1-GFP control cells. Data shown are the mean of 3 independent experiments ± SEM. *P < .05, determined by 2-tailed Student t test. (C) Representative blot (left) and quantification (right) of active Rap1-GTP levels in MEC1-GFP and MEC1-CD38H cells after RasGRP2 depletion. Graph shows the mean of 3 independent experiments ± SEM. *P < .05; **P < .01 determined by 2-tailed Student t test.
The Rap1 GEF RasGRP2 is required for the enhanced migration of MEC1-CD38H cells. MEC1-GFP and MEC1-CD38H cells were transfected with siRNAs targeting RasGRP2 or RasGRP3. (A) Representative blots of RasGRP2 and RasGRP3 expression levels. (B) Migration of MEC1-GFP and MEC1-CD38H cells through transwell filters upon RasGRP2 and RasGRP3 depletion. The migration index was obtained by normalizing the values to the MEC1-GFP control cells. Data shown are the mean of 3 independent experiments ± SEM. *P < .05, determined by 2-tailed Student t test. (C) Representative blot (left) and quantification (right) of active Rap1-GTP levels in MEC1-GFP and MEC1-CD38H cells after RasGRP2 depletion. Graph shows the mean of 3 independent experiments ± SEM. *P < .05; **P < .01 determined by 2-tailed Student t test.
CD38 expression did not affect RasGRP2 expression, either in MEC1 cells (Figure 5A) or primary CLL cells (supplemental Figure 5A). We therefore hypothesized that CD38 could alter the subcellular localization of RasGRP2, which could in turn explain the specific role of RasGRP2 in cell migration. Indeed, RasGRP2 localization was strongly polarized to the lamellipodial regions of MEC1-CD38H cells, whereas it was mostly diffusely localized in the cytoplasm in MEC1-GFP cells (Figure 6A-C). We next investigated RasGRP2 localization in primary CLL samples. We took advantage of the morphological roundness of CLL cells to analyze and score the variability in RasGRP2 distribution around each cell (supplemental Figure 5B-C), termed RasGRP2 polarization index. Overall, CD38high samples exhibited a stronger RasGRP2 polarization (Figure 6D-E). CD38 partially colocalized with RasGRP2 at the cell periphery, although CD38 also localized to intracellular compartments (supplemental Figure 5D-E). CD38 expression therefore induces polarized RasGRP2 localization, which likely accounts for the differential Rap1 activity associated with CLL cell migration.
CD38 increases polarized RasGRP2 localization. Cells were stained with anti-RasGRP2 antibody followed by Alexa Fluor 546–conjugated anti-rabbit antibody and imaged by confocal microscopy. F-actin and nuclei were stained with Alexa Fluor 488–conjugated phalloidin and DAPI, respectively. (A) Representative images of RasGRP2 localization in MEC1-GFP and MEC1-CD38H cells. Examples of cells with polarized RasGRP2 localization are indicated with an asterisk. Scale bar, 20 µm. (B) Fluorescence intensity profile of RasGRP2 obtained along the longitudinal axis of 2 representative MEC1-GFP and MEC1-CD38H cells. Scale bar, 5 µm. (C) The percentage of cells with polarized RasGRP2 localization was quantified by scoring n ≥ 100 cells in each of the 3 independent experiments. **P < .01, determined by 2-tailed Student t test. (D-F) RasGRP2 localization in CD38high and CD38low CLL patient samples. (D) Representative images of one CD38high (CLL 2) and one CD38low (CLL 5) CLL sample are shown. Scale bar, 10 μm. (E) The RasGRP2 polarization index, indicating distribution of the fluorescence intensity, was calculated using ImageJ Oval Profile plug-in (supplemental Data). Graph shows the mean values obtained for each CLL patient sample (n ≥ 30 cells analyzed per sample), horizontal bars indicate mean values of the RasGRP2 polarization index obtained for the 2 groups by grouping CD38low (n = 7, green) and CD38high (n = 7 red) samples. **P < .01, determined by 2-tailed Student t test.
CD38 increases polarized RasGRP2 localization. Cells were stained with anti-RasGRP2 antibody followed by Alexa Fluor 546–conjugated anti-rabbit antibody and imaged by confocal microscopy. F-actin and nuclei were stained with Alexa Fluor 488–conjugated phalloidin and DAPI, respectively. (A) Representative images of RasGRP2 localization in MEC1-GFP and MEC1-CD38H cells. Examples of cells with polarized RasGRP2 localization are indicated with an asterisk. Scale bar, 20 µm. (B) Fluorescence intensity profile of RasGRP2 obtained along the longitudinal axis of 2 representative MEC1-GFP and MEC1-CD38H cells. Scale bar, 5 µm. (C) The percentage of cells with polarized RasGRP2 localization was quantified by scoring n ≥ 100 cells in each of the 3 independent experiments. **P < .01, determined by 2-tailed Student t test. (D-F) RasGRP2 localization in CD38high and CD38low CLL patient samples. (D) Representative images of one CD38high (CLL 2) and one CD38low (CLL 5) CLL sample are shown. Scale bar, 10 μm. (E) The RasGRP2 polarization index, indicating distribution of the fluorescence intensity, was calculated using ImageJ Oval Profile plug-in (supplemental Data). Graph shows the mean values obtained for each CLL patient sample (n ≥ 30 cells analyzed per sample), horizontal bars indicate mean values of the RasGRP2 polarization index obtained for the 2 groups by grouping CD38low (n = 7, green) and CD38high (n = 7 red) samples. **P < .01, determined by 2-tailed Student t test.
Discussion
High CD38 expression is known to correlate with increased CLL cell motility, but the molecular basis for this is unclear. Here we show that RasGRP2/Rap1 signaling is functionally linked to the CD38-associated increased CLL cell migration. We find that CD38 expression levels, either ectopically expressed in the CLL-derived cell line MEC1 or endogenously expressed in primary CLL cells, correlate with higher basal migration. This is associated with increased cell spreading and higher Rap1 activity. Basal Rap1-GTP levels are higher in CD38high compared with CD38low primary CLL samples. Rap1 is known to stimulate integrin activation, cytoskeleton rearrangements, and cell polarization both in the presence and absence of a chemokine gradient.42,43 We observed that Rap1a and Rap1b depletion abolished the migratory advantage of MEC1-CD38H cells. This indicates that Rap1 activity contributes to the increased migration and adhesion linked to CD38 expression.20,22
Intriguingly, some previous studies indicate a potential role for Rap1 in CLL pathogenesis, which may have been underestimated. First, a fraction of mice deficient for the Rap1 GTPase-activating protein SIPA1 (also known as SPA-1), hence with higher Rap1 activity, showed splenomegaly and lymphadenopathy with an increased number of self-reactive CD5+ B cells resembling human CLL.44 In addition, SIPA1 mRNA expression was lower in CLL cells compared with normal peripheral blood CD5+ B cells.45 Moreover, Rap1 activation by cAMP through the Rap-GEF EPAC reduced basal apoptosis in CLL cells,46 suggesting Rap1 regulates both CLL cell motility and survival. Furthermore, it was reported that defective chemokine-induced Rap1 activation may be linked to impaired motility in a subset of primary CLL cells.47 Therefore, the fact that we have functionally linked CD38 expression and Rap1 activity is particularly relevant in CLL pathology.
CD38 contributes to intracellular Ca2+ levels and chemotaxis in mouse neutrophils and dendritic cells, at least in part by producing NAADP, cADPR, and ADPR.48,49 In MEC1 cells, exogenous CD38 expression increases production of NAADP and cADPR.20 Here we show that ectopic CD38 expression increases Ca2+ levels in MEC1 cells. This effect was restricted to unstimulated cells; no significant difference was observed in CCL21-induced Ca2+ mobilization between CD38-expressing and control MEC1 cells. In contrast, another group reported that CD38-expressing MEC1 cells had a higher increase in intracellular Ca2+ after chemokine stimulation compared with control cells.20 Despite the apparent discrepancy, those data are consistent with our findings, as cells were stimulated with chemokines for 24 hours, suggesting that CD38 expression in CLL plays a role mainly in long-term Ca2+ homeostasis rather than in the rapid Ca2+ responses induced immediately after stimulation. Remarkably, we also observed elevated basal Ca2+ levels in primary CLL cells with high CD38 expression. Of note, we investigated CLL cases irrespective of their mutational status. Elevated Ca2+ levels have been reported in CLL samples when compared with other B-cell malignancies (eg, follicular lymphoma, multiple myeloma) or normal B cells.50,51 Moreover, higher Ca2+ levels were reported in mutated compared with unmutated CLL samples, but a correlation with CD38 was not tested.51
One intriguing finding is that, in CD38-expressing cells, basal Ca2+ affects Rap1 activity in unstimulated cells. In eosinophils, basal Rap1 activity was reported to be independent of Ca2+ concentration.52 Similarly, Ca2+ depletion in control MEC1-GFP cells did not affect the basal level of active Rap1. However, Ca2+ depletion induced a decrease of Rap1-GTP in MEC1-CD38H and primary CD38high CLL cells, suggesting that the additional intracellular Ca2+ pool in CD38-expressing cells is specifically involved in regulating Rap1 activity.
Integration of Ca2+ and diacylglycerol signaling with Rap1 activation can be mediated by the Rap-GEFs RasGRP2 and RasGRP3.38 We found that RasGRP2 was specifically involved in both the basal and CCL21-induced migration of MEC1-CD38H, but not MEC1-GFP, cells. Surprisingly, RasGRP2 regulates Rap1 activity in both MEC1-CD38H and MEC1-GFP cells, indicating that Rap1 activation through RasGRP2 has different cellular effects depending on the levels of CD38. A possible explanation is that RasGRP2 targets different pools of Rap1 in the 2 cell populations through differential localization of RasGRP2 and/or Rap1. Indeed, RasGRP2 localization was polarized in MEC1-CD38H compared with MEC1-GFP cells. Similarly, in primary CD38high CLL cells, RasGRP2 was highly polarized compared with CD38low cells. This indicates that CD38 could affect RasGRP2 localization. To date, the mechanisms regulating RasGRP2 subcellular compartmentalization remain unclear. It has been reported that, when ectopically co-expressed in COS7 cells, Vav-RhoGEFs induce RasGRP2 translocation from the cytoplasm to the plasma membrane, where it would mediate integrin activation through Rap1.53 Vav-RhoGEFs are required for BCR-induced Ca2+ mobilization in mouse B cells,54 and thus it would be interesting to determine whether they also affect RasGRP2 localization in CLL cells.
In summary, our results indicate that CD38 enhances CLL cell migration by increasing Ca2+ levels, leading to Rap1 activation by the Ca2+-sensitive RasGRP2. Migration of CLL cells into lymphoid tissues is thought to play a key role in CLL pathogenesis because at these sites proliferation is induced by BCR activation and other microenvironment-derived signals. Our findings imply that Rap1 contributes to the aggressiveness of CLL associated with CD38 expression, and could thereby enhance BCR signaling. Rap1, together with its downstream signaling molecules RAPL and integrins VLA-4 and LFA-1, has a crucial role in leukocyte trafficking, homing, and retention in lymphoid tissues.55-57 Moreover, constitutive activation of Rap1 was shown to increase the in vivo dissemination of the lymphoma B-cell line A20.58 Given that interactions of CLL cells with their microenvironment in lymphoid organs are crucial for disease progression, it is likely that Rap1 is involved in CLL cell trafficking. Rap1 signaling could therefore potentially be targeted for CLL treatment or other diseases involving CD38, such as multiple myeloma.59
Preclinical studies indicate that CD38 is a potential target for antibody therapy in CD38-positive hematological malignancies.18,19 The anti-CD38 monoclonal antibody Daratumumab is approved for the treatment of multiple myeloma and is currently being assessed for CLL. Anti-CD38 monoclonal antibodies are believed to exert their antitumor activity through their Fc-dependent cytotoxic effect. However, recent evidence indicates that they can modulate CD38 enzymatic activity by decreasing cADPR production, which may in turn lead to decreased Ca2+ mobilization and signaling.18,60 Further characterization of CD38-dependent signaling, including the RasGRP2/Rap1 axis, is therefore important for the design of novel treatments for CD38+ hematological diseases.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors are grateful to Paul Brennan (Cardiff University, Cardiff, United Kingdom) for the lentiviral vectors encoding CD38 and GFP.
This work was funded by Cancer Research UK (A.J.R.) and Bloodwise (S.D.). S.M. was supported by the National Institute for Health Research Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London.
The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health.
Authorship
Contribution: S.M., S.D., and A.J.R. conceived the project; S.M. and E.I. performed experiments; A.G.P. provided information on primary CLL samples; S.D. and A.J.R. supervised the work; and S.M., S.D., and A.J.R. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anne J. Ridley, School of Cellular and Molecular Medicine, Biomedical Sciences Building, University of Bristol, University Walk, Bristol BS8 1TD, United Kingdom; e-mail: anne.ridley@bristol.ac.uk.