Key Points
Terminally differentiated CD56neg NK cells expand in children after chronic malaria exposure and in those diagnosed with eBL.
NK cells in eBL patients express high levels of MIP-1β in lieu of TNF-α, and normal NK cell profiles appear to be restored in eBL survivors.
Abstract
Natural killer (NK) cells are critical for restricting viral infections and mediating tumor immunosurveillance. Epstein-Barr virus (EBV) and Plasmodium falciparum malaria are known risk factors for endemic Burkitt lymphoma (eBL), the most common childhood cancer in equatorial Africa. To date, the composition and function of NK cells have not been evaluated in eBL etiology or pathogenesis. Therefore, using multiparameter flow cytometry and in vitro killing assays, we compared NK cells from healthy children and children diagnosed with eBL in Kenya. We defined 5 subsets based on CD56 and CD16 expression, including CD56negCD16pos. We found that licensed and terminally differentiated perforin-expressing CD56negCD16pos NK cells accumulated in eBL children, particularly in those with high EBV loads (45.2%) compared with healthy children without (6.07%) or with (13.5%) malaria exposure (P = .0007 and .002, respectively). This progressive shift in NK cell proportions was concomitant with fewer CD56dimCD16pos cells. Despite high MIP-1β expression, CD56negCD16pos NK cells had diminished cytotoxicity, with lower expression of activation markers NKp46, NKp30, and CD160 and the absence of TNF-α. Of note, the accumulation of poorly cytotoxic CD56negCD16pos NK cells resolved in long-term eBL survivors. Our study demonstrates impaired NK cell–mediated immunosurveillance in eBL patients but with the potential to restore a protective NK cell repertoire after cancer treatment. Characterizing NK cell dysfunction during coinfections with malaria and EBV has important implications for designing immunotherapies to improve outcomes for children diagnosed with eBL.
Introduction
Endemic Burkitt lymphoma (eBL) is a pediatric Epstein-Barr virus (EBV)–associated B-cell malignancy that occurs in malarious regions of equatorial Africa.1 Children at risk for eBL experience their primary EBV infection before 2 years of age, accompanied by repeated Plasmodium falciparum (Pf) coinfections and episodes of viral reactivation resulting in higher EBV loads.2-4 Malaria is postulated to diminish EBV-specific T-cell immune surveillance in eBL etiology.5-12 However, the complementary role of natural killer (NK) cells in viral and tumor immunosurveillance has not been studied within the context of eBL pathogenesis.
NK cell immunosurveillance involves orchestrating activation and inhibitory signals to identify virally infected or tumorigenic cells.13-16 Killer cell immunoglobulin-like receptors (KIRs; reviewed in Lanier17 and Saunders et al18 ) interact with human leukocyte antigen (HLA) class I molecules for target cell recognition,15,19,20 distinguishing “self” from “nonself.” NK cells also receive coordinated signals from other cell surface receptors that determine their cytolytic function or lack thereof: the NKG2 family induces inhibitory signals (via NKG2A) or activation signals (via NKG2C and NKG2D),21 and natural cytotoxicity receptors (NCRs) are crucial for triggering cytotoxic activity and targeted cell death (reviewed in Kruse et al22 ).
In addition to the well-characterized CD56brightCD16neg, CD56dimCD16pos NK cells, an increase in the CD56dimCD16neg NK cell subset with a concomitant decrease in CD56dimCD16pos proportions has been observed after coculture with the K562 tumor cell line or cytokine cocktails in healthy American donors.23,24 The presence of surface metalloproteases appears to regulate CD16 expression after degranulation.23,24 Another NK cell subset, CD56negCD16pos, has been described in HIV-viremic and hepatitis C virus–infected adults, particularly those who failed hepatitis C virus treatment.25-28 Compared with CD56pos NK cells, this subset has been characterized as “dysfunctional” because of its lower expression of activating receptors and cytokine production.
EBV, like most herpesviruses, has evolved to evade immune clearance and, therefore, is able to persist as a life-long asymptomatic infection in immunocompetent individuals.29 A recent study described a role for viral microRNAs in mediating immune evasion from CD8 T cells30 ; however, little is known about NK cell immune escape mechanisms indulgent of EBV-infected B cells. Clues from studying EBV infection during adolescence that results in acute infectious mononucleosis suggest that a distinct CD56dimCD16posNKG2AposKIRneg NK cell subset expands and accompanies disease resolution.31 Studies of human cytomegalovirus (CMV) revealed an accumulation of a CD56dimCD16posCD57posNKG2Chigh NK cell subset during CMV/EBV coinfection that was not found in children who were infected only with EBV.32,33 “Memory-like” NKG2Cpos NK cells found in patients receiving bone marrow or peripheral blood stem cell transplants from CMVpos donors displayed more durable IFN-γ production compared with NK cells from CMVneg donors.34 NK cell subset phenotypes and cytotoxic capacity during asymptomatic pediatric EBV infections are not known.
Studies of malaria infections in African children have reported high IL-12 and IL-18 plasma levels compared with children without malaria.35,36 These cytokines are essential to induce IFN-γ secretion by NK cells in response to Pf-infected red blood cells (Pf-iRBCs) in vitro.37 This model identified IFN-γ secretion predominantly from CD56pos cells during peripheral blood mononuclear cell (PBMC)/Pf-iRBC coculture experiments.37 Cytotoxic CD56dimCD16pos NK/NK T cells and CD8pos T cells are protective against malaria38 ; however, EBV or CMV coinfections were not evaluated in their study populations.
To improve our understanding of NK cell composition and function during malaria and EBV coinfections in children, as well as how NK cell alterations may contribute to eBL pathology, we compared healthy children with divergent malaria exposure histories and children diagnosed with eBL in Kenya. We characterized NK cell subset frequencies and function by flow cytometry and in vitro killing assays, with further stratification of children by peripheral blood EBV load. In addition, we conducted evaluations of NK cell subset phenotypes and functions in eBL survivors.
Methods
Study participants
Children with suspected eBL were enrolled at Jaramogi Oginga Odinga Teaching and Referral Hospital between 2003 and 2011.39 Two independent pathologists in Kenya confirmed the diagnosis by cytopathology and May-Grünwald Giemsa staining. In this region, eBL is the most common pediatric cancer; 95% of children clinically suspected of having the disease are pathologically confirmed with eBL. Samples from our biorepository were further characterized by transcriptome and mutational profiling to confirm eBL diagnosis40 and were found to be 90% concordant with the initial diagnosis. For cancer patients, baseline samples were collected before induction of chemotherapy and, for those in remission, 3 and 9 months later. Age-matched healthy Kenyan controls were selected and stratified based on divergent malaria exposure histories and correlated EBV loads.2,39 Nandi children experience low exposure to malaria infections and have been shown previously to harbor low EBV loads.2,4 They also reside in an area with a low incidence of eBL.41 In contrast, Kisumu children reside in a high malaria–transmission region (ie, holoendemic malaria), experience higher EBV loads, and reside in an area with a higher incidence of eBL.41 After obtaining written informed consent from parents, PBMCs from children with eBL were compared with those from Kisumu children (with malaria) and Nandi children (without malaria). All children in this study were EBV seropositive, HIV-negative, and born of HIV-negative mothers. Ethical approval was obtained from the Kenya Medical Research Institute and the University of Massachusetts Medical School.
Flow cytometry phenotyping, functional assays, EBV viral load, IgG levels in plasma, and statistical analysis
See supplemental Methods for details.
Results
Study population characteristics
The 3 groups of children did not differ significantly with regard to median age (Nandi: 6 years, range 2.5-6; Kisumu: 6.5 years, range 2.5-6.5; and eBL: 6.1 years, range 2.5-7.9), sex, or absolute lymphocyte counts (>2000 cells/mm3). Serology profiles confirmed expected malaria exposure histories, with eBL patients and Kisumu children having higher antibody titers to malaria antigens compared with Nandi children (supplemental Figure 1). All of the children included in this analysis were EBV seropositive. None of the children had malaria blood-stage infections at the time of blood collection (data not shown). The survival rate for eBL children included in this study was 57%, similar to our larger study cohort.39 Our eBL group was divided into 2 subgroups based on EBV status: no/low EBV loads detected in peripheral blood by quantitative polymerase chain reaction and high EBV copy numbers (>3 log EBV copies per microgram of DNA). This threshold was determined previously in comparisons of healthy Kisumu and eBL children.42
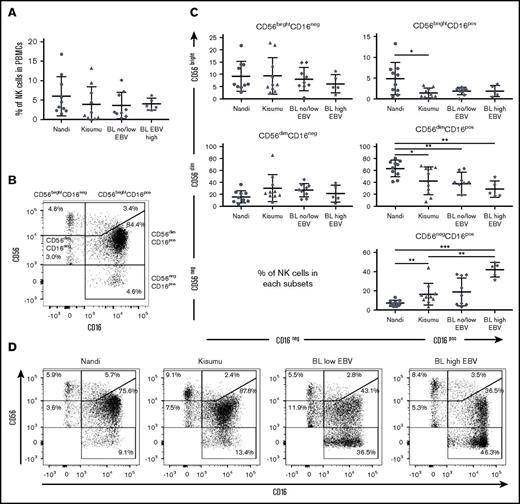
CD56negCD16pos NK cells accumulate in eBL patients
Overall, no statistical differences were observed among the 3 groups of children with regard to the frequency of total NK cells (Figure 1A), although we noted that our study populations had generally lower frequencies (median 3.06% for healthy and 2.84% for eBL) compared with healthy children from Cameroon (median 8% for 2- to 6-year-old children).43 We examined 5 subsets defined by CD56 and CD16 expression (gating strategy is shown in supplemental Figure 2): CD56brightCD16neg, CD56brightCD16pos, CD56dimCD16pos, CD56dimCD16neg, and CD56negCD16pos (Figure 1B). We observed significantly more CD56negCD16pos NK cells in eBL patients with high EBV loads (median 45.2%) compared with Kisumu (median 13.5%) and Nandi (median 6.07%) children (P = .002 and .0007, respectively; Figure 1C). This observation was concomitant with a lower median number of CD56dimCD16pos NK cells in eBL patients with high EBV loads (22.2%) and no/low EBV (39.7%) compared with Nandi children (67.25%) (P = .002 and .003, respectively). Interestingly, we observed the same NK cell subset skewing between the 2 groups of healthy children: Kisumu children had significantly lower CD56dimCD16pos (median 45.85%; P = .02) and higher CD56negCD16pos (median 13.5%; P = .006) NK cells compared with Nandi children (median 67.25% and 6.07%, respectively). This alteration in relative NK cell subset proportions is illustrated in Figure 1D using representative flow cytometry plots from children within each group.
Characterization of NK cell subsets in Kenyan children. Children were categorized by malaria and EBV exposure, as well as eBL diagnosis: Nandi (EBVlow/malarialow; n = 10), Kisumu (EBVhigh/malariahigh; n = 10), and eBL patients (n = 14) with no/low EBV and high EBV loads. (A) The percentage of NK cells within the circulating lymphocytes was defined as the number of NK cells (CD56pos and/or CD16pos)/total number of live lymphocytes. (B) Five NK cell subsets were defined by CD56 and CD16 expression levels: CD56brightCD16neg, CD56brightCD16pos, CD56dimCD16neg, CD56dimCD16pos, and CD56negCD16pos. (C) Percentages of NK cell subsets for each group of children within our study. (D) Representative proportions of NK cell subsets in different groups of children. Data in panels A and C are mean ± standard deviation (SD). *P < .05, **P < .01, ***P < .001.
Characterization of NK cell subsets in Kenyan children. Children were categorized by malaria and EBV exposure, as well as eBL diagnosis: Nandi (EBVlow/malarialow; n = 10), Kisumu (EBVhigh/malariahigh; n = 10), and eBL patients (n = 14) with no/low EBV and high EBV loads. (A) The percentage of NK cells within the circulating lymphocytes was defined as the number of NK cells (CD56pos and/or CD16pos)/total number of live lymphocytes. (B) Five NK cell subsets were defined by CD56 and CD16 expression levels: CD56brightCD16neg, CD56brightCD16pos, CD56dimCD16neg, CD56dimCD16pos, and CD56negCD16pos. (C) Percentages of NK cell subsets for each group of children within our study. (D) Representative proportions of NK cell subsets in different groups of children. Data in panels A and C are mean ± standard deviation (SD). *P < .05, **P < .01, ***P < .001.
The significantly higher proportion of CD56negCD16pos NK cells in eBL and Kisumu children compared with healthy Nandi children suggests that their enrichment might be due to higher infectious disease burdens. We found that the percentage of CD56negCD16pos cells in eBL children correlated with higher antimalarial (AMA1, MSP1, and Pf SEA-1A antigens) antibody titers (P = .006, .007, and .01, respectively; supplemental Figure 3) but not with antibodies from other infectious diseases, such as schistosomiasis, measles, and CMV. It was not surprising to find no correlation with EBV antibody titers using a single time point, because our past studies have demonstrated longitudinal variability in EBV antibody titers.3,44 However, malaria antibody profiles have been used as an indicator of cumulative and recent past exposure in the absence of an active infection.45
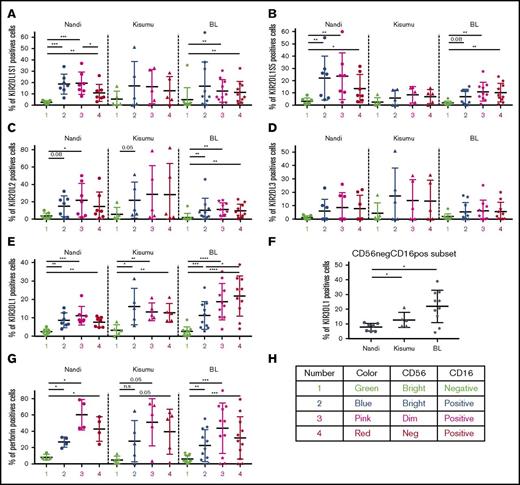
KIR licensing in malaria-exposed and eBL children favors inhibitory signals
As expected, we observed lower expression of activating and inhibitory KIRs (KIR2DL1/S1, KIR2DL1/S5, KIR2DL2, KIR2DL3, and KIR3DL1) on CD56brightCD16neg NK cells compared with other NK cell subsets across all of our study populations (Figure 2), confirming their immature nonlicensed status. Coinciding with NK cell maturation, KIR expression was elevated on CD56brightCD16pos and CD56dimCD16pos subsets in all groups of children. However, KIR2DL1/S1 levels did not decrease in CD56negCD16pos NK cells in malaria-exposed and eBL children in contrast to Nandi children, who had significantly lower KIR2DL1/S1 expression levels for this subset (Figure 2A). With regard to KIR2DL1/S5 expression (Figure 2B), Nandi children expressed this inhibition/activation marker on CD56brightCD16pos and CD56dimCD16pos NK cell subsets, whereas Kisumu and eBL children had lower expression of this KIR for all NK cells. The expression pattern of inhibitory receptors KIR2DL2 and KIR2DL3 (Figure 2C-D, respectively) was significantly lower on all NK cells from eBL patients compared with the highest levels in Kisumu children and intermediate levels in Nandi children. In contrast, expression of the KIR3DL1 inhibitory receptor (Figure 2E) increased across NK cell subsets for eBL patients, with the highest levels on CD56dimCD16pos and CD56negCD16pos NK cell subsets. Kisumu children had higher KIR3DL1 expression on mature CD16pos NK cell subsets compared with Nandi children. Comparing KIR3DL1 on the CD56negCD16pos NK cell subset across groups of children (Figure 2F), we observed significantly higher expression for Kisumu and eBL children compared with Nandi children (P = .04 and .01, respectively).
Comparison of KIR and perforin expression by different NK cell subsets for Nandi, Kisumu, and eBL children. Comparison of KIR2DL1S1 (A), KIR2DL1S5 (B), KIR2DL2 (C), KIR2DL3 (D), and KIR3DL1 (E) among the different NK cell subsets: CD56brightCD16neg (1), CD56brightCD16pos (2), CD56dimCD16pos (3), and CD56negCD16pos (4). (F) Expression of KIR3DL1 on CD56negCD16pos NK cells among Nandi, Kisumu, and eBL children. For all KIRs, Nandi, n = 7; Kisumu, n = 5; and BL, n = 10. (G) Perforin expression by NK cell subsets: CD56brightCD16neg (1), CD56brightCD16pos (2), CD56dimCD16pos (3), and CD56negCD16pos (4) (Nandi, n = 4; Kisumu, n = 5; eBL, n = 9). (H) Color code used for data. Data are mean ± SD. *P < .05, **P < .01, ***P < .001, ****P < .0001. n.s., not significant.
Comparison of KIR and perforin expression by different NK cell subsets for Nandi, Kisumu, and eBL children. Comparison of KIR2DL1S1 (A), KIR2DL1S5 (B), KIR2DL2 (C), KIR2DL3 (D), and KIR3DL1 (E) among the different NK cell subsets: CD56brightCD16neg (1), CD56brightCD16pos (2), CD56dimCD16pos (3), and CD56negCD16pos (4). (F) Expression of KIR3DL1 on CD56negCD16pos NK cells among Nandi, Kisumu, and eBL children. For all KIRs, Nandi, n = 7; Kisumu, n = 5; and BL, n = 10. (G) Perforin expression by NK cell subsets: CD56brightCD16neg (1), CD56brightCD16pos (2), CD56dimCD16pos (3), and CD56negCD16pos (4) (Nandi, n = 4; Kisumu, n = 5; eBL, n = 9). (H) Color code used for data. Data are mean ± SD. *P < .05, **P < .01, ***P < .001, ****P < .0001. n.s., not significant.
Finally, basal levels of perforin expression (Figure 2G) were higher for CD56dimCD16pos and CD56negCD16pos NK cell subsets compared with CD56bright NK cell populations for all children in our study, with no statistically significant difference between CD56dimCD16pos and CD56negCD16pos NK cell subsets. This suggests that CD56negCD16pos NK cells, which are highly enriched in eBL children, are licensed for cytotoxic functions, yet their KIR profiles differ dramatically from those of healthy Nandi children.
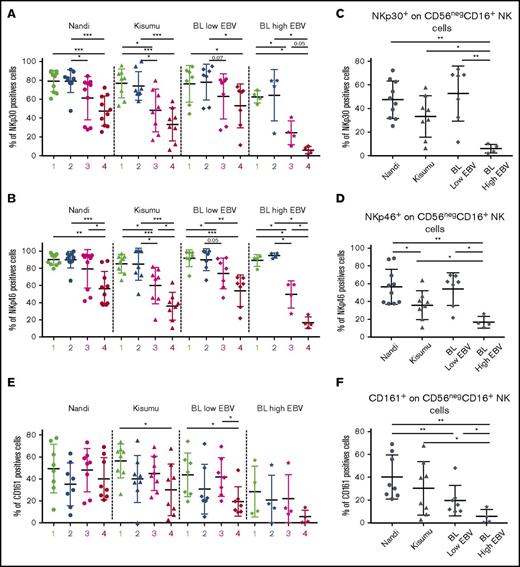
CD56negCD16pos NK cell subset found in eBL patients displays features of adaptive NK cells
To determine the phenotype (canonical vs adaptive) of the CD56negCD16pos NK cell subset, we checked the presence of activation and cytotoxic receptors. To evaluate the NCR pathways, we compared NKp30 and NKp46 expression across study populations and among the different NK cell subsets. We noted significantly lower expression of NKp30 and NKp46 for CD56negCD16pos NK cells compared with CD56brightCD16neg and CD56brightCD16pos NK cell subsets from all investigated groups of children (Figure 3A-B). Interestingly, expression of NKp30 and NKp46 was lowest on CD56negCD16pos NK cells in eBL children with the highest EBV loads compared with eBL children with low EBV titers and both healthy groups of children (Figure 3C-D). In addition, among the healthy children, Kisumu children had significantly lower expression of NKp46 on CD56negCD16pos NK cells compared with Nandi children (P = .04), suggesting an impairment in anti-infection rather than antitumor immune surveillance.
Expression of NKp30, NKp46, and CD161 consistent with an adaptive NK cell phenotype. NK cell subsets, including CD56brightCD16neg (1), CD56brightCD16pos (2), CD56dimCD16pos (3), and CD56negCD16pos (4), were evaluated for each group of children: Nandi (EBVlow/malarialow), Kisumu (EBVhigh/malariahigh), and eBL patients with low EBV and high EBV load. Comparison of NKp30 (A) and NKp46 (B) expression on the different NK cell subsets across all study groups. NKp30 (C) and NKp46 (D) expression on CD56negCD16pos NK cell subsets across study groups. Nandi, n = 10; Kisumu, n = 8; and BL, n = 11 for NCRs. (E) Comparison of CD161 on the different NK cell subsets across all study groups. (F) CD161 expression on the CD56negCD16pos NK cell subset across study groups. Nandi, n = 8; Kisumu, n = 8; and BL, n = 11 for CD161 expression. Data are mean ± SD. *P < .05, **P < .01, ***P < .001.
Expression of NKp30, NKp46, and CD161 consistent with an adaptive NK cell phenotype. NK cell subsets, including CD56brightCD16neg (1), CD56brightCD16pos (2), CD56dimCD16pos (3), and CD56negCD16pos (4), were evaluated for each group of children: Nandi (EBVlow/malarialow), Kisumu (EBVhigh/malariahigh), and eBL patients with low EBV and high EBV load. Comparison of NKp30 (A) and NKp46 (B) expression on the different NK cell subsets across all study groups. NKp30 (C) and NKp46 (D) expression on CD56negCD16pos NK cell subsets across study groups. Nandi, n = 10; Kisumu, n = 8; and BL, n = 11 for NCRs. (E) Comparison of CD161 on the different NK cell subsets across all study groups. (F) CD161 expression on the CD56negCD16pos NK cell subset across study groups. Nandi, n = 8; Kisumu, n = 8; and BL, n = 11 for CD161 expression. Data are mean ± SD. *P < .05, **P < .01, ***P < .001.
We also examined expression of CD161, which was lowest on the CD56negCD16pos NK cell subset from eBL children compared with the respective NK cells in Nandi and Kisumu children (Figure 3E-F). CD160 displayed a similarly downregulated expression pattern for eBL patients compared with healthy children (supplemental Figure 4). Interestingly, coexpression analysis revealed diminished double-positive CD160posCD161pos NK cells, as well as CD160negCD161pos NK cells, in eBL patients compared with healthy controls (supplemental Figure 5). Finally, and contrary to a previous study of HIV-infected adults that described lower CD94/NKG2A marker expression for the CD56negCD16pos NK cell subset,28 we observed no differences for this set of NK cell markers across our study populations (supplemental Figure 6).
Together, our results suggest that CD56negCD16pos NK cells display a terminally differentiated phenotype, with eBL patients unable to control EBV showing further impairments in NK cell activation markers. This would limit potent NK cell activation and is consistent with poor cytotoxic function of NK cells from eBL children with extremely high EBV loads. It is worthwhile to note that this suppressed activation profile was not observed on the CD56negCD16pos NK cell subset of eBL patients with low EBV loads and Nandi children. Based on these observations, we speculate that this NK cell phenotype may not be induced in response to tumor cells but rather by the high viral loads experienced by children residing in malaria holoendemic areas prior to tumorigenesis.
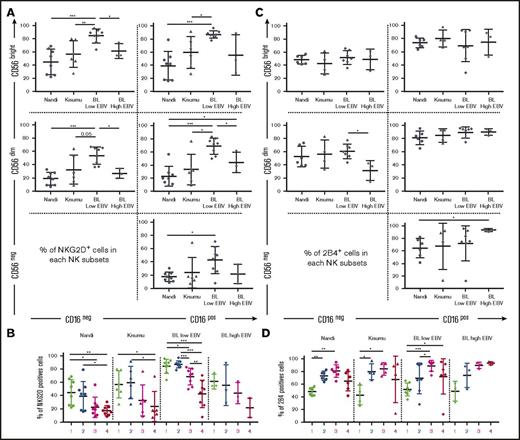
NKG2D and 2B4 coactivating receptors expressed by the CD56negCD16pos NK cell subset
We found that eBL children with low EBV loads had significantly higher NKG2D expression on most of their NK cell subsets (Figure 4A). We also noted that NKG2D expression patterns across NK cell subsets were similar among all groups of children and appear to correlate with CD56 expression (Figure 4B). Furthermore, we found no differences among groups of children for 2B4 expression on NK cell subsets, with the exception of lower expression on CD56dimCD16neg cells relative to CD56negCD16pos cells for high EBV eBL children (Figure 4C). Overall, 2B4 was expressed on >50% of cells within each NK cell subset and increased after CD16 upregulation (Figure 4D). However, it is interesting to note the inverse correlation between NKG2D and 2B4 expression for some NK cell subsets but not others (compare Figure 4B and 4D). These differences in NKG2D and 2B4 expression suggest that target cells with NKG2D ligands are recognized more efficiently by CD56bright NK cells, whereas those with 2B4 ligands preferentially stimulate CD56dim NK cells.
Expression of the NKG2D and 2B4 coactivation receptors on NK cell subsets from Nandi, Kisumu, and eBL children. NK cell subsets, including CD56brightCD16neg (1), CD56brightCD16pos (2), CD56dimCD16pos (3), and CD56negCD16pos (4), were evaluated for each group of children: Nandi (EBVlow/malarialow), Kisumu (EBVhigh/malariahigh), and BL with low EBV and BL with high EBV. NKG2D expression by (A) NK cell subset representation (A) and by linear representation (B) across study groups (Nandi, n = 8; Kisumu, n = 6; and BL, n = 11). 2B4 expression by NK cells subset representation (C) and by linear representation (D) across study groups (Nandi, n = 6; Kisumu, n = 4; and BL, n = 10). Data are mean ± SD. *P < .05, **P < .01, ***P < .001.
Expression of the NKG2D and 2B4 coactivation receptors on NK cell subsets from Nandi, Kisumu, and eBL children. NK cell subsets, including CD56brightCD16neg (1), CD56brightCD16pos (2), CD56dimCD16pos (3), and CD56negCD16pos (4), were evaluated for each group of children: Nandi (EBVlow/malarialow), Kisumu (EBVhigh/malariahigh), and BL with low EBV and BL with high EBV. NKG2D expression by (A) NK cell subset representation (A) and by linear representation (B) across study groups (Nandi, n = 8; Kisumu, n = 6; and BL, n = 11). 2B4 expression by NK cells subset representation (C) and by linear representation (D) across study groups (Nandi, n = 6; Kisumu, n = 4; and BL, n = 10). Data are mean ± SD. *P < .05, **P < .01, ***P < .001.
Absence of CD57posNKG2Cpos NK cell expansion in eBL children
CD57 was highly expressed on CD56dimCD16pos NK cells, with the overall expression pattern across NK cell subsets not differing among our study groups (Figure 5A). CD57 expression was the lowest on CD56negCD16pos NK cells for eBL children with high EBV loads; however, this trend did not achieve statistical significance (Figure 5B). Despite higher expression of NKG2C described on NK cells cocultured with Pf-iRBCs,46 we found no differences in NKG2C expression in circulating NK cells associated with malaria exposure or EBV load (comparing Kisumu and Nandi children). In contrast, eBL children with high EBV loads had the lowest expression of NKG2C on all of their NK cell subsets compared with the other groups of children (Figure 5C). NKG2C expression on the CD56negCD16pos subset was significantly lower in eBL children with high EBV loads compared with those with low EBV loads (Figure 5D; P = .03). To eliminate a potential effect modifier, we observed no significant difference in seropositivity or antibody titer to human CMV (HCMV) among our groups of children (Figure 5E). In addition, the percentage of double-positive CD57posNKG2Cpos NK cells seemed to be lower in the CD56negCD16pos NK cell subset compared with the CD56dimCD16pos NK cell population (Figure 5F). This was particularly noticeable for Nandi and eBL children with low EBV loads (Figure 5G), who had more CD57posNKG2CposCD56dimCD16pos NK cells than CD57posNKG2CposCD56negCD16pos NK cells (P = .01 and .01, respectively). These observations demonstrate that, even in healthy HCMV-infected children, CD57posNKG2Cpos NK cells were not expanded by EBV or malaria infections; however, this population of double-positive NK cells was found to be elevated in eBL children with low viral loads compared with those with high viral loads (Figure 5G). These findings suggest that high EBV loads and eBL tumor formation do not drive the formation of terminal “memory-like” NK cells, which has been associated with control of viral reactivation in HCMV.34,47
Absence of the CD57/NKG2C NK cell phenotype otherwise found in HCMV-EBV coinfection. NK cell subsets, including CD56brightCD16neg (1), CD56brightCD16pos (2), CD56dimCD16pos (3), and CD56negCD16pos (4), were evaluated for each group of children: Nandi (EBVlow/malarialow), Kisumu (EBVhigh/malariahigh), and eBL patients with no/low EBV and high EBV loads. (A) Comparison of CD57 among the different NK cell subsets across study groups. (B) CD57 expression on the CD56negCD16pos NK cell subset across study groups. (C) Comparison of NKG2C expression among different NK cell subsets across groups of children. (D) NKG2C expression on the CD56negCD16pos NK cell subset across study groups. (E) Median fluorescence intensity (MFI) of antibody titers against CMV. (F) Coexpression of CD57 and NKG2C on CD56dimCD16pos and CD56negCD16pos NK cells. (G) CD57posNKG2Cpos expression between CD56dimCD16pos and CD56negCD16pos subsets across study groups. For CD57, Nandi, n = 8; Kisumu, n = 6, and BL, n = 11. For NKG2C, Nandi, n = 7; Kisumu, n = 5; and BL, n = 11. Data are mean ± SD. *P < .05, **P < .01, ***P < .001.
Absence of the CD57/NKG2C NK cell phenotype otherwise found in HCMV-EBV coinfection. NK cell subsets, including CD56brightCD16neg (1), CD56brightCD16pos (2), CD56dimCD16pos (3), and CD56negCD16pos (4), were evaluated for each group of children: Nandi (EBVlow/malarialow), Kisumu (EBVhigh/malariahigh), and eBL patients with no/low EBV and high EBV loads. (A) Comparison of CD57 among the different NK cell subsets across study groups. (B) CD57 expression on the CD56negCD16pos NK cell subset across study groups. (C) Comparison of NKG2C expression among different NK cell subsets across groups of children. (D) NKG2C expression on the CD56negCD16pos NK cell subset across study groups. (E) Median fluorescence intensity (MFI) of antibody titers against CMV. (F) Coexpression of CD57 and NKG2C on CD56dimCD16pos and CD56negCD16pos NK cells. (G) CD57posNKG2Cpos expression between CD56dimCD16pos and CD56negCD16pos subsets across study groups. For CD57, Nandi, n = 8; Kisumu, n = 6, and BL, n = 11. For NKG2C, Nandi, n = 7; Kisumu, n = 5; and BL, n = 11. Data are mean ± SD. *P < .05, **P < .01, ***P < .001.
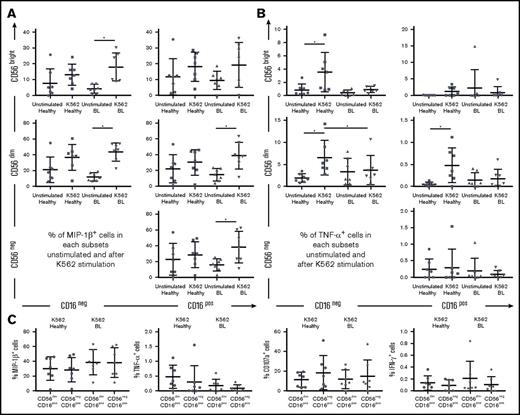
MIP-1β cytokine production by NK cells in eBL patients after K562 cell stimulation
To analyze cytokine production and degranulation as a surrogate for cytotoxicity, we combined Nandi and Kisumu children into 1 group of “healthy children” and compared them with eBL patients (irrespective of EBV load). Nearly all NK cell subsets from eBL children expressed MIP-1β upon K562 cell stimulation, including CD56negCD16pos cells (Figure 6A). MIP-1β is known to activate the synthesis and release of other proinflammatory cytokines, such as TNF-α.48 However, NK cells from eBL children were less able to produce TNF-α compared with healthy children, who had more robust responses (Figure 6B). CD56brightCD16neg, CD56dimCD16neg, and CD56dimCD16pos NK cell subsets from healthy children expressed significantly more TNF-α after stimulation, whereas all subsets from eBL children remained low and unchanged. No significant difference was observed between healthy and eBL children for IFN-γ production or CD107a expression, irrespective of NK cell subset and after K562 cell stimulation (supplemental Figure 7). Focusing on the CD56dimCD16pos and CD56negCD16pos NK cell subsets, we noted no difference in the production of the cytokines analyzed (Figure 6C). This observation supports our hypothesis that the CD56negCD16pos NK cell subset originates from the CD56dimCD16pos NK cell population. We noted that the proportion of CD56dimCD16neg NK cells seemed to expand after K562 cell stimulation compared with the unstimulated conditions for all groups of children (supplemental Figure 8). This is consistent with another study24 describing the loss of CD16 expression after degranulation of CD56dimCD16pos NK cells. Our experiments were not designed to determine the differentiation pathways giving rise to the different KIR-licensed NK cell subsets; however, this observation provides rationale for further study.
MIP-1β production and degranulation of CD56negCD16posNK cell subset after K562 cell stimulation. PBMCs from healthy children (Nandi and Kisumu; n = 7) and eBL children (n = 7) were stimulated with K562 cells in a functional in vitro killing assay. NK cell subsets were defined by CD56 and CD16 expression levels. (A) Percentage of MIP-1βpos NK cells in each subset, with and without stimulation. (B) Percentage of TNF-αpos NK cells in each subset, with and without stimulation. (C) Comparison of MIP-1β, TNF-α, CD107a, and IFN-γ positive cells after K562 cell stimulation between the CD56dimCD16pos and CD56negCD16pos NK cell subsets. The Wilcoxon test was used for related data, and Mann-Whitney U test was used for nonrelated data. Data are mean ± SD. *P < .05.
MIP-1β production and degranulation of CD56negCD16posNK cell subset after K562 cell stimulation. PBMCs from healthy children (Nandi and Kisumu; n = 7) and eBL children (n = 7) were stimulated with K562 cells in a functional in vitro killing assay. NK cell subsets were defined by CD56 and CD16 expression levels. (A) Percentage of MIP-1βpos NK cells in each subset, with and without stimulation. (B) Percentage of TNF-αpos NK cells in each subset, with and without stimulation. (C) Comparison of MIP-1β, TNF-α, CD107a, and IFN-γ positive cells after K562 cell stimulation between the CD56dimCD16pos and CD56negCD16pos NK cell subsets. The Wilcoxon test was used for related data, and Mann-Whitney U test was used for nonrelated data. Data are mean ± SD. *P < .05.
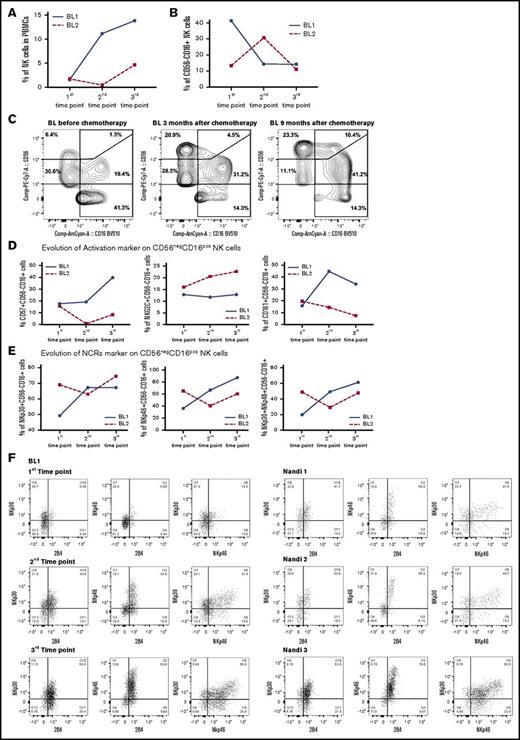
Decrease in CD56negCD16pos NK cells in eBL survivors
To determine whether skewed NK cell profiles found in eBL children would naturally re-equilibrate to resemble those found in normal Nandi children, we tracked NK cell profiles in 2 eBL survivors (with low EBV loads) over time: time point 1 was considered the time of diagnosis and prior to induction of chemotherapy, with repeated samples collected during remission at 3 and 9 months after diagnosis (time points 2 and 3, respectively). We noted markedly different rates of natural NK reconstitution for each patient (Figure 7A). However, we found markedly fewer CD56negCD16pos NK cells in both patients by 9 months postdiagnosis (Figure 7B-C), resulting in the same low levels found in healthy children described above. The proportions of immature CD56brightCD16neg and mature CD56dimCD16pos NK cell subsets also reached levels comparable to those in healthy children. Focusing on the CD56negCD16pos NK cell subset, we selected a panel of NK cell markers found to be important in the comparative studies described above and measured them longitudinally in eBL survivors (Figure 7D). We observed higher expression over time for most of the cytotoxic markers, including NKp46, NKp30, and CD161, on the surface of the CD56negCD16pos NK cell subset from 1 eBL patient. For patient “BL2” (Figure 7D), the expression of these markers is influenced by the fact that a different composition of NK cell subsets developed over time compared with “BL1.” We also checked the coexpression of cytotoxic NCR markers and found a decrease in NKp30/NKp46, as well as in NKp30/2B4 and NKp46/2B4. For both eBL patients (Figure 7E-F), these cells “reappeared” 9 months after chemotherapy and arrived at the same proportion as observed in Nandi children (Figure 7F). A limitation of this secondary analysis is the small sample size, but results from 2 randomly selected patients are provocative. This longitudinal glimpse of the proportional and expression marker changes in NK cell subsets, albeit preliminary, suggests that NK cell composition can eventually recover to normal parameters in eBL survivors. Future studies with more patients would need to be conducted to determine whether poor long-term outcomes are due to a lag in the ability to re-establish normal NK cell profiles.
Resolution of the CD56negCD16possubset from eBL children after chemotherapy. NK cell subsets were defined by CD56 and CD16 expression. The first time point was before chemotherapy, and the second and third time points were 3 and 9 months, respectively, after diagnosis (during remission). (A) Changes in the percentage of NK cells were defined as the number of NK cells (CD56pos and/or CD16pos)/total number of live lymphocytes. (B) Changes in the percentage of CD56negCD16pos NK cells at each time point for 2 eBL patients. (C) Cytograms for the 3 time points for patient “BL1.” Changes in activation (D) and NCR (E) markers over time for both eBL children (“BL1” and “BL2”) for CD56negCD16pos NK cells. (F) Plots of NKp30/NKp46, NKp30/2B4, and NKp46/2B4 coexpression for CD56negCD16pos NK cells from an eBL1 patient during the course of treatment, as well as for 3 healthy Nandi children.
Resolution of the CD56negCD16possubset from eBL children after chemotherapy. NK cell subsets were defined by CD56 and CD16 expression. The first time point was before chemotherapy, and the second and third time points were 3 and 9 months, respectively, after diagnosis (during remission). (A) Changes in the percentage of NK cells were defined as the number of NK cells (CD56pos and/or CD16pos)/total number of live lymphocytes. (B) Changes in the percentage of CD56negCD16pos NK cells at each time point for 2 eBL patients. (C) Cytograms for the 3 time points for patient “BL1.” Changes in activation (D) and NCR (E) markers over time for both eBL children (“BL1” and “BL2”) for CD56negCD16pos NK cells. (F) Plots of NKp30/NKp46, NKp30/2B4, and NKp46/2B4 coexpression for CD56negCD16pos NK cells from an eBL1 patient during the course of treatment, as well as for 3 healthy Nandi children.
Discussion
To begin to evaluate the role of NK cells in eBL etiology, our study compared healthy EBV-infected Kenyan children with those diagnosed with eBL and demonstrated that NK cell subsets were dramatically altered by Pf coinfections and even more so during this childhood cancer. We found significantly more terminally differentiated CD56negCD16pos NK cells in eBL patients with the highest EBV loads compared with healthy children. In addition, eBL children had specific defects in NK cell functions compared with healthy children, including lower TNF-α secretion, despite no overall differences in IFN-γ secretion and degranulation, as measured by CD107a after K562 cell stimulation. In contrast, NK cells from eBL children more readily secreted MIP-1β compared with healthy children, which could be a last-ditch effort by NK cells to recruit proinflammatory cells to help kill the cancer cells. Our examination of eBL survivors demonstrates that a healthy NK cell repertoire can be re-established; however, the mechanisms responsible, be they NK cell reconstitution or phenotypic restoration, have yet to be determined.
Our study confirmed that CD56negCD16pos NK cell subsets are licensed; however, KIR expression profiles varied, depending on malaria exposure and cancer diagnosis. In fact, except for higher expression of KIR3DL1 on all NK cell subsets, lower expression of other KIRs was found for eBL patients compared with healthy children. Combined with lower expression of activating receptors, this suggests that the threshold for NK cell activation is elevated in eBL patients. Interestingly, malaria-exposed Kisumu children also had higher KIR3DL1 expression within their CD56negCD16pos NK cell subset compared with Nandi children, suggesting that this may be a mechanism by which malaria subverts NK cell–mediated immunity. In other infectious disease contexts, including AIDS, variant KIR3DL1 alleles influence disease progression after activation by HLA-Bw4.49 Interestingly, higher expression of the KIR3DL1*015 allele is observed in HIV-infected adult African Americans compared with European Americans, and this particular allele has higher binding for HLA-Bw4.18,50 Future studies of eBL etiology will incorporate HLA and KIR genotypes in individual assessments of NK cell function, because higher responses to Pf-iRBCs have been reported for KIR3DL2pos NK cells.51
In addition to shifts in NK cell subset proportions and expression of licensing markers, we evaluated NKp30 and NKp46, which can be activated by the Pf erythrocyte membrane protein 1 (reviewed in Kruse et al22 ) and induce cytotoxic activity against Pf-iRBCs.36-38,52 However, our studies showed dramatically lower expression of these NCRs on NK cells from eBL children with extremely high EBV loads, as well as significantly lower NKp46 expression on NK cells from Kisumu children compared with Nandi children. This suggests that the burden of infectious diseases may be downregulating these receptors, independent of tumorigenesis. In support of this premise, albeit exploratory, we had the opportunity to evaluate NK cell subset proportions in Kenyan children enrolled in our study who were diagnosed with cancers other than eBL. Interestingly, we found that CD56negCD16pos NK cells accumulated in a pediatric patient with non-Hodgkin lymphoma who had a high EBV load commensurate with eBL patients. In contrast, patients with Hodgkin lymphoma and nephroblastoma who had low or no detectable viremia had few CD56neg NK cells, comparable to healthy children (supplemental Figure 9).
The role of CD16 is to drive antibody-dependent cellular cytotoxicity (ADCC),53 and cytokine production after CD16 activation is elevated in adaptive NK cells compared with canonical NK cells.54,55 Therefore, canonical NK cells are able to induce cytotoxicity against other immune cells56 instead of adaptive NK cells with less immunoregulatory function. Interestingly, the accumulation of CD56negCD16pos NK cells in eBL children accompanied decreased TNF-α production in target cell–killing assays. However, these children have robust IgG antibodies to EBV and malaria antigens, suggesting potential engagement of ADCC-mediated killing.57,58 Future studies using autologous lymphoblastoid cell lines and eBL tumor cell lines will allow us to directly assess which NK cell subsets are involved in ADCC and to what extent they affect innate tumor cell targeting via NKG2D and the NCRs. Another clue in favor of an adaptive NK cell profile is the decreased expression of NKp46 on the surface of CD56negCD16pos NK cells. In fact, NKp46 is thought to be necessary for immunoregulatory killing of activated T cells during canonical NK cell function.59 Thus, increased ADCC, but diminished immunoregulatory NK cell functions, could be influenced by an altered NK cell compartment in eBL children.
Our study showed that the CD56negCD16pos NK cell subset expressed proinflammatory cytokines (low IFN-γ but high MIP-1β) after K562 cell stimulation to the same degree as CD56dimCD16pos NK cells. As previously described,25 IFN-γ expression was lower in CD56neg NK cells, the predominant NK cell subset in eBL patients, compared with CD56pos NK cells (supplemental Figure 10). MIP-1β is produced by activated immune cells and has multiple biological functions, including chemotaxis, degranulation, and phagocytosis (reviewed in Maurer and von Stebut48 ), making it a key player in inflammation. A study of Ugandan children described higher MIP-1β, as well as TNF-α, in serum associated with malaria.60 However, higher MIP-1β levels were observed in children who died of severe malaria compared with survivors, in contrast to TNF-α levels, which were not differentially associated with disease outcome. High levels of MIP-1β were also described in children from Gabon with severe malaria, and an in vitro assay confirmed MIP-1β production by PBMCs after hemozoin stimulation.61 Thus, it is tempting to speculate that the overexpression of MIP-1β by NK cells in the absence of TNF-α, within the context of eBL etiology, might signify an immune adaptation that diminishes EBV immunosurveillance. Mechanistic studies examining the relative killing efficiency of MIP-1β–secreting NK cells would support this hypothesis.
Understanding the role of NK cells in fighting various cancers will help to customize NK cell–based immunotherapeutic strategies. However, within the context of infection-associated cancers, such as pediatric eBL, an immune equilibrium appears to have been established to permit asymptomatic chronic malaria and latent EBV infections in young children as a tradeoff for protection against pathologic hyperimmune responses.62,63 With their cytotoxic functions (reviewed in Morvan and Lanier64 ), NK cells are key agents to deploy against tumors cells,65 yet knowing the immunologic landscape of patients prior to introducing an immunotherapy seems essential for the success of such interventions. Results from our study are the first step in understanding the role of NK cells in eBL etiology and support the pursuit of 2 NK cell–based therapeutic approaches that could be considered to improve outcomes for children diagnosed with eBL: inhibitory KIR3DL1 blockade and ADCC boosted with tumor neoantigen–targeted antibodies. Our longitudinal results from an admittedly limited number of eBL survivors support the scientific rationale that restoring NK cell function could be of therapeutic benefit.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the children and their families for participating in this study and Peter Oluoch Owuor for correlation analysis. They also thank the clinical research priority program on human hematolymphatic diseases (KFSP HHLD) of the University of Zurich.
This research was funded by National Institutes of Health Grants R01 CA189806 and R01 CA134051 from the National Cancer Institute (A.M.M.) and U19 AI066345 from the National Institute of Allergy and Infectious Diseases (G.A.) and Thrasher Early Career Award 5926 (C.E.N.).
This manuscript was approved for publication by the Kenya Medical Research Institute.
Authorship
Contribution: C.S.F. designed research, performed research, analyzed data, and wrote the manuscript; C.P.C. designed research, performed some research and data analysis, and reviewed the manuscript; P.S.-L. contributed to experimental tool for plasma EBV viral load research; C.E.N. contributed to experimental tool for serology research; J.F. contributed to experimental tool for EBV viral load research; J.M.O. contributed to sample acquisition and research design and reviewed the manuscript; J.A.O. contributed to sample acquisition and reviewed the manuscript; G.A. contributed experimental tools and reviewed the manuscript; C.M. designed research, analyzed data, and reviewed the manuscript; and A.M.M. designed research, contributed experimental tools, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ann M. Moormann, Department of Medicine, Division of Infectious Diseases and Immunology, University of Massachusetts Medical School, 364 Plantation St, LRB 313, Worcester, MA 01605; e-mail: ann.moormann@umassmed.edu.