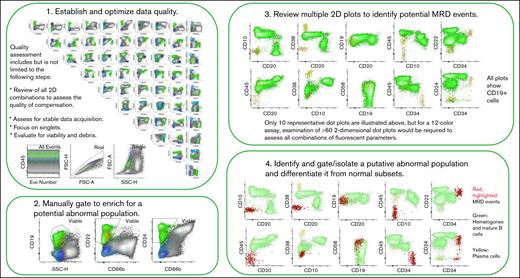
In this issue of Blood Advances, Seheult et al1 present an artificial intelligence (AI) pipeline that facilitates and expedites analysis of flow cytometry (FC) data for measurable residual disease (MRD) assessment in B-cell acute lymphoblastic leukemia (B-ALL). The presence of MRD after therapy is widely accepted as an important prognostic factor in adult and pediatric B-ALL, and has been incorporated into routine practice for prognostication and to inform therapeutic strategies.2,3 MRD assessment can be performed by FC or molecular (ie, polymerase chain reaction or next-generation sequencing) methods. Although molecular approaches offer increased sensitivity, advantages of FC include its ability to detect residual disease at clinically relevant thresholds (0.01%), applicability in nearly all cases, relatively rapid turnaround time, and ability to identify potential targets for antigen-directed therapies. However, manual analysis of FC data for MRD is labor-intensive (see figure summarizing typical steps in data analysis), requires expertise not universally available, and may even yield discordant results, although discordances can be reduced with training.4
Steps in manual FC data analysis for MRD assessment in B-ALL. 2D, 2-dimensional; FSC-A, forward scatter-area; SSC-H, side scatter-height.
Steps in manual FC data analysis for MRD assessment in B-ALL. 2D, 2-dimensional; FSC-A, forward scatter-area; SSC-H, side scatter-height.
In MRD detection by FC, small abnormal populations, often present below the limit of morphologic detection, are distinguished from normal regenerating cells by their abnormal patterns of antigen expression.5 In the marrow, these small populations of residual leukemic cells may be present in the context of background normal progenitors (hematogones) that share some overlapping immunophenotypic characteristics with the leukemic cells. Therapy may impact the appearance of normal background progenitors and/or the immunophenotype of the neoplastic population. Depending on the therapeutic regimen and time after therapy, various subsets of B cells and precursors may be proportionally increased, mimicking an abnormal population. Further, the immunophenotype of the neoplastic population can shift at relapse or with exposure to therapy, in particular targeted therapy. These factors complicate manual analysis and lead to a requirement for an experienced analyst, familiar with the impact of therapy and with the range of immunophenotypes characterizing regeneration across various treatment regimens. Assay sensitivity depends upon several factors including the number of events collected, the number and nature of antigens evaluated in the assay, and the experience of the evaluator.
The AI pipeline described by Seheult et al may circumvent some of the challenges of manual MRD analysis of FC data in B-ALL. The pipeline incorporates several previously validated machine learning (ML) algorithms to remove spurious events generated during sample acquisition, and to both dimensionally reduce and cluster unprocessed 10-color FC data. Seheult et al performed low-resolution clustering and random downsampling, with retention of clusters <5000 events to preserve rare cell populations, followed by high-resolution clustering of downsampled data to generate files of reduced size (87% cellularity reduction), while maintaining rare MRD events. This approach enriches for potential MRD, enhances computational efficiency, and enables analysis with existing software packages. In addition, the pipeline generates a quantitative measure of multidimensional phenotypic difference from normal (termed an aberrancy scale) based on overrepresentation of sample events in high-resolution clusters of merged control and sample files. This aberrancy scale simulates manual FC MRD analysis, in which differences from normal patterns of antigen expression permit identification of abnormal leukemic populations.5 A deep neural network (DNN) single-event classifier trained on expert-gated negative control samples accurately identifies normal cell populations, including various stages of B-cell maturation, while allowing separation of leukemic cells from benign mononuclear cells or B-lymphoid cells with high accuracy (area under the curve of 0.98 and 0.94, respectively). The output of the pipeline is a sample data file suitable for analysis in existing FC software packages, in which the conventional fluorescence and light scatter properties of each cell are augmented by the addition of aberrancy scale, DNN class (ie, cell type), coordinates in dimensionally-reduced space, and an upsampling factor.
Many studies have demonstrated the potential of AI to simplify, standardize, and automate analysis of FC data (reviewed in Fuda et al6), although implementation into the clinical space has been limited so far. Several groups have specifically demonstrated the potential of ML approaches to facilitate the detection of MRD in B-ALL.7-10 However, these approaches have been limited to varying extents by a requirement for manual preprocessing, inadequate sensitivity, and/or software restrictions. By contrast, the pipeline described by Seheult et al converts unprocessed FC files to smaller files annotated with ML-generated parameters that enable 83% faster manual MRD detection at clinically relevant levels with any existing clinical software platform. Whether this approach is generalizable to antibody panels used by different institutions, and for patients treated with antigen-directed targeted therapies, which may result in antigen loss or lineage switch, will require additional studies. The authors’ efforts to make the pipeline available to other laboratories as a cloud computing resource should allow these additional questions to be answered.
Advances in cytometry instrumentation will allow evaluation of larger numbers of parameters on a routine basis, which will enable more comprehensive immunophenotypic profiling. As the number of evaluable parameters increases, traditional manual methods of FC data analysis will become more cumbersome and may struggle to extract the complexity of information contained within the data. In this context, AI approaches will be integral to modern FC data analysis. The future is coming, and Seheult et al are leading the way, using ML-informed strategies to clear the path to FC MRD detection in B-ALL by bringing relevant events to the forefront.
Conflict-of-interest disclosure: The authors declare no competing financial interests.