Key Points
Ferroportin reduces intracellular iron, inhibits CDK2 and suppresses SAMHD1 phosphorylation thus inhibiting HIV-1 RT.
Ferroportin expression leads to overexpression of IKBα and inhibition of HIV-1 transcription.
Abstract
The low incidence of HIV-1 infection in patients with sickle cell disease (SCD) and inhibition of HIV-1 replication in vitro under the conditions of low intracellular iron or heme treatment suggests a potential restriction of HIV-1 infection in SCD. We investigated HIV-1 ex vivo infection of SCD peripheral blood mononuclear cells (PBMCs) and found that HIV-1 replication was inhibited at the level of reverse transcription (RT) and transcription. We observed increased expression of heme and iron-regulated genes, previously shown to inhibit HIV-1, including ferroportin, IKBα, HO-1, p21, and SAM domain and HD domain-containing protein 1 (SAMHD1). HIV-1 inhibition was less pronounced in hepcidin-treated SCD PBMCs and more pronounced in the iron or iron chelators treated, suggesting a key role of iron metabolism. In SCD PBMCs, labile iron levels were reduced and protein levels of ferroportin, HIF-1α, IKBα, and HO-1 were increased. Hemin treatment induced ferroportin expression and inhibited HIV-1 in THP-1 cells, mimicking the HIV-1 inhibition in SCD PBMCs, especially as hepcidin similarly prevented HIV-1 inhibition. In THP-1 cells with knocked down ferroportin, IKBα, or HO-1 genes but not HIF-1α or p21, HIV-1 was not inhibited by hemin. Activity of SAMHD1-regulatory CDK2 was decreased, and SAMHD1 phosphorylation was reduced in SCD PBMCs and hemin-treated THP-1 cells, suggesting SAMHD1-mediated HIV-1 restriction in SCD. Our findings point to ferroportin as a trigger of HIV-1 restriction in SCD settings, linking reduced intracellular iron levels to the inhibition of CDK2 activity, reduction of SAMHD1 phosphorylation, increased IKBα expression, and inhibition of HIV-1 RT and transcription.
Introduction
Sickle cell disease (SCD) is a hereditary disorder with E6V mutation in the β-globin gene.1,2 The mutated hemoglobin polymerizes and facilitates formation of “sickled” red blood cells leading to hemolysis, vaso-occlusion, and ischemia. Several previous studies pointed to a possibility that SCD patients might be protected from HIV-1 infection.3-5 Prevalence of anti-HIV-1 but not human T-cell leukemia virus type 1 antibodies was lower (2.7% vs 7.9%) in SCD patients transfused with blood that was not screened for HIV-1.3 Low or nondetectable viral load was observed in a small cohort of HIV-1–infected SCD patients.4 Our recent analysis of >400 000 medical records showed a lower frequency of HIV diagnosis among patients who have a concurrent sickle cell diagnosis (≈1.5% vs ≈3.3%; odds ratio 0.33) compared with hepatitis C and other infections.5 Although these observations suggest that SCD patients can be potentially protected from HIV-1 infection, other studies have shown an early mortality in children with SCD and HIV-1 and negative effects of antiretroviral drugs on SCD patients.6 In Africa, the lack of hydroxyurea treatment, availability of blood products, and insufficient control of bacterial infections can additionally contribute to the poor outcome of HIV-1 infection in SCD patients. In the United States, where SCD patients have access to hydroxyurea and blood transfusion, the risk of HIV-1 infection among SCD patients is significantly lower.5
Several molecular mechanisms can explain the potential protection of SCD from HIV-1 infection. Hypoxia,7 chronic inflammation producing higher levels of HIV-1 inhibitory cytokines like interleukin-10 (IL-10),8 changes in macrophage polarization,9 and induction of heme and iron regulatory pathways10 have been previously shown to inhibit HIV-1 replication. In particular, HIV-1 replication is inhibited in macrophages and T cells treated with hemin.11,12 Suppression of HIV-1 by hemin involves the induction of heme oxygenase-1 (HO-1).11 Remarkably, HIV-1 viral load dropped dramatically in a hemochromatosis patient who underwent venesection,13 suggesting an iron-mediated control of HIV-1 replication. Previously, gene expression analysis showed increased expression of HO-1, billiverdin reductase, and p21 in peripheral blood mononuclear cells (PMBCs) obtained from SCD patients in steady-state conditions.14 Along with HO-1, other iron-regulated genes like GAPDH, FTL1, ALDH1A1 and SAT2 were found to be upregulated in SCD patients.15 Thus, induction of heme and iron-regulatory pathways in SCD may contribute to the restriction of HIV-1 infection, although the mechanism remains to be clarified.
The expression of p21 among HIV-1 elite controllers16 was recently linked to a decrease in phosphorylation of the SAM domain and HD domain-containing protein 1 (SAMHD1).17 SAMHD1 restricts HIV-1 infection by controlling the intracellular deoxyribonucleotide pool, inhibiting HIV-1 reverse transcription (RT), and preventing HIV-1 infection of monocytes and dendritic cells.18,19 The transcription of p21 is activated by Egr-1,20 which is activated by HIF-1α.21 Hypoxia and alterations of iron metabolism typically found in SCD can lead to a chronic upregulation of HIF-1α.22 CDK2 positively regulates HIV-1 transcription by phosphorylating HIV-1 Tat protein23 and Ser90 residue of CDK9.24 Depletion of intracellular iron inhibits CDK2 activity and blocks HIV-1 transcription.25-27 Iron chelators have been shown to induce the expression of p21,28,29 which can inhibit CDK2.30 Physiologically, cellular iron is exported by an iron export protein, ferroportin, which is negatively regulated by hepcidin.31 In SCD patients, hepcidin levels can be either decreased32 or increased.33 We previously showed that expression of ferroportin inhibits HIV-1 replication and that hepcidin treatment increases intracellular iron and induces HIV-1 replication.34 In this study, we have defined a mechanism where HIV-1 replication is inhibited among SCD patients via the expression of ferroportin and induced expression of HIV-1 RT and transcription inhibitory factors.
Materials and methods
Study design and human subjects
This study was approved by the Institutional Review Board of Howard University (13-MED-03). Twenty-nine SCD and 16 control subjects were recruited. Clinical Laboratory Improvement Amendments–certified HPLC using the ultra2 variant system (Trinity Biotech USA, Jamestown, NY) was used to determine hemoglobin A and S levels. Number of independent experiments and control donors are defined in each figure legend. Further details are supplied in the supplemental Materials and methods.
Cells and media.
THP-1 cells were purchased from the American Type Culture Collection (Manassas, VA). PBMCs were purchased from Precision for Medicine (Flemington, NJ) or isolated from blood of SCD patients or normal subjects. PBMCs were activated with phytohemagglutinin (PHA) (0.5 μg/mL) and IL-2 (10 U/mL) for 24 hours prior to the infection. All cells were cultured at 37°C in 5% CO2 atmosphere. PBMCs and THP-1 cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum, 1% antibiotic solution (penicillin and streptomycin; Invitrogen), and supplemented with 0.1% 2-mercaptoethanol.
Plasmids.
HIV-1 proviral vector pNL4-3.Luc.R−E− (courtesy of Nathaniel Landau, NYU School of Medicine, New York, NY) was obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program.
Antibodies.
Antibodies for HIF-1α (cat. no. 610959) and transferrin receptor (TFR; cat. no. 612124) were purchased from BD Biosciences (San Jose, CA). Antibodies for ferroportin (cat. no. PAS-22993) and IKBα (cat. no. 9242) were purchased from Thermo Fisher Scientific (Waltham, MA). Antibodies for SAMHD1 (cat. no. A311-354A) were from Bethyl Laboratories (Montgomery, TX). Antibodies for SAMHD1 phosphorylated on Thr-592 (cat. no. 10-301) were from ProSci (Poway, CA). Antibodies for CDK2 (cat. no. SC-163) and β-actin (cat. no. SC47778) were purchased from Santa Cruz Biotechnology (Dallas, TX). Antibodies for HO-1 (cat. no. ADI-OSA-110) were from Enzo (Farmingdale, NY).
Ferritin and p24 ELISA.
Ferritin and p24 enzyme-linked immunosorbent assays (ELISA) were performed using Spectro Ferritin MT kit (Ramco Laboratories, Stafford, TX) and Retrotek HIV-1 p24 antigen kit (ZeptoMetrix, Buffalo, NY) using manufacturer’s recommendations.
Western blot analysis.
THP-1 cells or PBMCs were lysed in whole cell lysis buffer (50 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS]) supplemented with protease cocktail. Proteins were resolved on gradient 4% to 12% Bis-Tris gels (Invitrogen), transferred to polyvinylidene fluoride membranes and detected with appropriate antibodies.
Fluorescence-activated cell sorting.
Activated PBMCs (1 × 106 cells) were fixed for 5 minutes at room temperature in 4% paraformaldehyde and then permeabilized 15 minutes in cytofix/cytoperm buffer (cat. no. 554714; BD Pharmingen). Cells were stained with CD4-PE antibodies (cat no. 555347; BD Pharmingen) for 1 hour at 4°C in the dark. After staining, the cells were washed and analyzed in BD FACSCalibur (BD Biosciences) using FlowJo software. Unpaired Student t test was used to test statistical significance.
Knockdown of iron and hemin regulated genes.
Lentiviruses expressing small hairpin RNA (shRNA)-targeting human CDK2, ferroportin, HIF-1α, HO-1, IKBα, p21, SAMHD1, and control shRNA were purchased from Santa Cruz Biotechnology or Sigma-Aldrich (St. Louis, MO). THP-1 cells were infected with lentivirus with 1.1 multiplicity of infection per 1000 cells. Spinoculation was carried out at 800g for 30 minutes. Cells were then incubated for 24 hours prior to adding puromycin (0.75 μg/mL) for selection of the shRNA-expressing clones. Efficiency of knockdown was assessed by quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis.
HIV-1 infection assays.
Vesicular stomatitis virus G protein–pseudotyped pNL4-3.Luc.R-E-virus was prepared as previously described.26 The cells were infected with 1 ng of p24 per 5 × 106 cells, and cells were incubated for 48 hours, collected, washed with phosphate-buffered saline (PBS), and resuspended in 100 μL PBS. Then, 100 μL reconstituted luciferase buffer (Luclite Kit, Perkin Elmer) was added to each well, and after 10 minutes of incubation, the lysates were transferred into white plates (Perkin Elmer) and luminescence was measured using GloMax luminometer (Promega). Further details are supplied in the supplemental Materials and methods. HIV-1 BaL isolate was purchased from Advanced Biotechnologies (Columbia, MD). Monocytes were isolated from PBMCs (see details in the supplemental Materials and methods) and converted to monocyte-derived macrophages (MDMs) by culturing in Dulbecco’s modified Eagle’s media supplemented with 10% fetal bovine serum, 20 μg/mL gentamicin, and 1000 U/mL macrophage colony-stimulating factor (all from Invitrogen). After 5 days in culture, MDMs were infected with HIV-1 (BaL) isolate for 2 hours at 37°C. Then the cells were washed and cultured for 2 more days. At day 7, the cells were lyzed in Trizol for RNA isolation.
LC-MS analysis of hepcidin in plasma.
Synthetic human wild-type and isotope-labeled hepcidin peptides were purchased from Peptide Institute (Ozaka, Japan). Rabbit plasma was obtained from Sigma-Aldrich. Preparation of Hepcidin stock solution, working solutions, and calibration solution are described in the supplemental materials and methods. Plasma samples were collected from 13 SCD patients and 12 healthy subjects. Sample preparation is described in the supplemental materials and methods. Liquid chromatography-mass spectrometry (LC-MS) analysis was performed on an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) coupled to a Prominence Nano LC (Shimadzu, Columbia, MD) using the Xcalibur version 2.7.0 (Thermo Scientific). Ten microliters of samples were loaded on a C18-packed analytical column (25 cm × 150 μm, 5 μm, 200 Å; Michrom Bioresources, Auburn, CA) and separated with a linear gradient of 0 to 15 minutes, 2% to 80% B, 15–20 minutes, 80% B (vol/vol, B was 0.1% FA in acetonitrile) at the flow rate of 600 nL/min. The mass spectra acquisition was performed in high-resolution/selective ion monitoring (HR/SIM) scan of the most abundant isotope ion peaks of hepcidin m/z 698.27 and isotopically labeled hepcidin peptides m/z 703.28. The spray voltage, capillary temperature, and capillary voltage were set to 1.5 kV, 200°C, and 35 V, respectively. Extracted ion chromatograms (EICs) were based on a ±0.01 Da mass extraction window (MEW) centered on the m/z theoretical.
Measurement of labile iron pool (LIP).
LIP in PBMCs was examined as described previously.35 Cells were loaded with Calcein-AM, a nonfluorescent, hydrophobic compound that easily permeates intact, viable cells. Treatment of the cells with iron chelators removes the calcein-bound iron and produces Calcein, a hydrophilic, strongly fluorescent compound that is well retained in the cell cytoplasm. The fluorescence intensity is related to the amount of cellular iron that had been effectively removed by the chelators. The chelating efficiency was quantified using the formula: (F0 − F)/F, where F0 = the fluorescence intensity in the presence of chelator at time 0 and F = the fluorescence intensity at a given time after the addition of the chelator. This formula was derived from the equation (F0 − F)/F = Ks*Q, which was obtained from the equation Ks = F·Q/F*Q and the assumption of F0 = F + F·Q, where F is concentration of fluorophore, Q is the concentration of the quencher (iron), F·Q is the concentration of the complex of a quencher with the fluorophore, and Ks is dissociation constant of fluorophore and quencher. The values derived from (F0 − F)/F are proportional to the concentrations of the chelated iron present when reaction equilibrium is reached.
Ferritin and p24 ELISA.
Ferritin and p24 ELISA assays were performed using Spectro Ferritin MT kit (Ramco Laboratories) and Retrotek HIV-1 p24 antigen kit (ZeptoMetrix) using manufacturer’s recommendations.
CDK2 activity assay.
PBMCs or THP-1 cells were lysed in whole cell lysis buffer (50 mM Tris-HCl, pH 7.5, 0.3 M NaCl, 1% NP-40, 0.1% SDS) supplemented with protease cocktail (Sigma-Aldrich). CDK2 was immunoprecipitated using anti-CDK2 antibodies. Kinase assay was performed at 30°C for 20 minutes in the kinase assay buffer containing 2 µg Histone H1 as a substrate, 200 μM adenosine triphosphate, and 5 μCi of (γ-32P) adenosine triphosphate. At the end of the incubation, SDS-containing electrophoresis sample buffer was added to stop the reaction, and protein bands were resolved on 10% SDS-polyacrylamide gel electrophoresis. A gel was stained with Coomassie blue to visualize histone H1 and then exposed to a Phosphor Imager screen to measure histone H1 (32P) incorporation using PhosphoImager (Packard).
Analysis of mRNA expression.
Total RNA was extracted using Trizol reagent according to the manufacturer’s protocol (Invitrogen, Grand Island, NY). THP-1 cells or activated PBMCs were pretreated where indicated with 100 μM hemin for 36 hours. Total RNA (100 ng) was reverse-transcribed to complementary DNA using Superscript RT-PCR kit (Invitrogen), and hexamers and oligo-dT were used as primers. For RT-PCR analysis, complementary DNA was amplified using Roche LightCycler 480 and SYBR Green1 Master mix (Roche Diagnostics, Indianapolis, IN). PCR was carried with denaturation at 95°C for 10 seconds, annealing at 60°C for 10 seconds, and extension at 72°C for 10 seconds for 45 cycles. The 18S ribosomal RNA (rRNA) was used as a housekeeping normalization standard for quantification of messenger RNA (mRNA) levels of BACH1, CDK2, Cyclin A, Cyclin E, EGR1, FPN1, Hepcidin, HIF-1α, HO-1 IKBα, p21, p27, PP1α and SAMHD1. Primer sequences are listed in supplemental Materials and methods.
Determination of HIV-1 RNA and DNA levels.
Quantitative analysis of HIV-1 RNA and DNA was conducted as recently described.36 For quantitative analysis of HIV-1 RNA, total RNA was isolated from PBMCs or THP-1 cells, untreated or treated with hemin. RNA was purified using Trizol Reagent (Invitrogen) according to the manufacturer's protocol. Mean Cp values for target genes and 18S rRNA were determined, and ΔΔCt method was used to calculate relative expression levels.
For quantification of HIV-1 DNA, SCD and normal PBMCs were infected with HIV-1-LUC-G. Total DNA was extracted from 1 × 106 cells using total DNA isolation kit from Thermo Fisher. For the RT-PCR analysis, 100 ng DNA was amplified using Roche Light Cycler 480 (Roche Diagnostics) and SYBR Green1 Master mix (Roche Diagnostics). PCR was carried with initial preincubation for 5 minutes at 45°C and then for 3 minutes at 95°C followed by 45 cycles of denaturation at 95°C for 15 seconds, annealing and extension at 60°C for 45 seconds, and final extension at 72°C for 10 seconds. Quantification of Early-LTR and Late-LTR transcripts was carried using β-globin DNA as a normalization standard. Mean Cp values for Early-LTR, Late-LTR transcripts, and β-globin DNA were determined, and ΔΔCt method was used to calculate relative expression levels.
Statistical analysis.
All graphs were prepared using GraphPad prism 6 software. Data are presented as mean ± standard deviation (SD) or standard error of the mean (SEM) as indicated in the figure legends. Unpaired Student t test was used to determine statistical significance.
Meta-analysis.
Differentially expressed genes from PBMCs of 24 SCD patients and 10 controls (Geoset accession number GSE53441) were analyzed by Geo2R software. The top 250 upregulated and downregulated genes were selected and further analyzed by Ingenuity Pathway Analysis (IPA) software.
Results
Inhibition of 1 round of HIV-1 infection in SCD-derived PBMCs
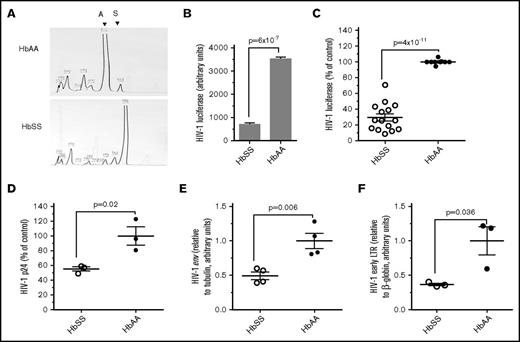
We tested ex vivo whether SCD patients are refractory for HIV-1 infection by analyzing 1 round of HIV-1 infection. PBMCs obtained from SCD (HbSS) and Control (HbAA) individuals were infected with vesicular stomatitis virus G protein–pseudotyped HIV-1 pNL4-3 virus expressing luciferase in place of nef (HIV-1-LUC-G).37 Blood from HbSS patients and HbAA controls was obtained using an Institutional Review Board–approved protocol (see supplemental Table 1 for Patient Data). HbSS or HbAA status was confirmed using a high-resolution HPLC system (Figure 1A). PBMCs were activated with PHA and IL-2 and then infected with HIV-1-LUC-G and cultured for 3 days. Significantly lower levels of luciferase activity were observed in HIV-1–infected PBMCs obtained from an SCD patient (Figure 1B). Expanded analysis for PBMCs obtained from 15 HbSS patients and 9 HbAA control individuals showed threefold reduction of 1 round of HIV-1 infection in PBMCs from HbSS individuals (Figure 1C). We did not detect any significant changes in CD4 expression levels (supplemental Figure 1). Production of HIV-1 p24 was significantly reduced in SCD-derived PBMCs infected with HIV-1-LUC-G (Figure 1D). Also, expression of HIV-1 env gene was reduced in HIV-1–infected SCD PBMCs (Figure 1E). Similar reduction in the expression of HIV-1 gag gene was also observed (not shown). To elucidate whether HIV-1 infection in SCD PBMCs was inhibited at an earlier postentry step of HIV-1 replication, we analyzed early RT by quantifying HIV-1 DNA for early LTR.38 We observed a marked 2.5-fold inhibition of early long terminal repeat (LTR) sequence production in HIV-1–infected SCD PBMCs compared with that of normal controls (Figure 1F). The same samples showed a reduction of luciferase expression (not shown). Thus, the inhibition from 1 round of HIV-1 infection was significantly inhibited at the transcription level as well as at the level of HIV-1 RT, suggesting the involvement of an HIV-1 transcription restriction factor and an RT restriction factor, such as SAMHD1.
Inhibition of HIV-1 in SCD-derived PBMCs. (A) HPLC analysis of SCD hemoglobin. HPLC of representative blood samples obtained from an SCD subject with HbSS hemoglobin and normal control with HbAA hemoglobin. (B-C) Inhibition of 1 round of HIV-1 infection in SCD-derived PBMCs. PBMCs were purified from whole blood obtained from SCD patients and healthy controls. The cells were activated with PHA and IL-2 and infected with HIV-1-LUC-G virus expressing luciferase. Luciferase activity was measured at 24 hours postinfection (PI) and normalized to the cell numbers. (B) A representative sample that corresponds to panel A. (C) Comparison of 1 round of HIV-1 replication in a cohort of 15 SCD patients. The results are expressed relative to infection of PBMCs from 9 control subjects, which were set to 100%. The means ± standard error (SE) and P value calculated with Student t test are shown. (D) Inhibition of p24 production in SCD-derived PBMCs. PBMCs obtained from 3 SCD patients and 3 controls were activated and infected as described above. Supernatants were collected 72 hours PI, and p24 was measured by ELISA. The means ± SE and P value calculated with Student t test are shown. (E) Inhibition of HIV-1 env expression in SCD-derived PBMCs. PBMCs obtained from 4 SCD patients and 4 controls were activated and infected as described above for 48 hours. RNA was extracted, reverse transcribed, and analyzed with primers for HIV-1 env gene by RT-PCR using 18S RNA as a reference. The means ± SE and P value calculated with Student t test are shown. (F) Inhibition HIV-1 RT in SCD PBMCs. PBMCs obtained from 3 SCD patients and 3 controls were activated with PHA and IL-2 and infected with HIV-1-LUC-G. At 6 hours PI, DNA was extracted and analyzed by RT-PCR on Roche 480 using primers for early LTR and β-globin gene as a reference. The means ± SE are shown (n = 3 for each sample). The means ± SE and P value calculated with Student t test are shown.
Inhibition of HIV-1 in SCD-derived PBMCs. (A) HPLC analysis of SCD hemoglobin. HPLC of representative blood samples obtained from an SCD subject with HbSS hemoglobin and normal control with HbAA hemoglobin. (B-C) Inhibition of 1 round of HIV-1 infection in SCD-derived PBMCs. PBMCs were purified from whole blood obtained from SCD patients and healthy controls. The cells were activated with PHA and IL-2 and infected with HIV-1-LUC-G virus expressing luciferase. Luciferase activity was measured at 24 hours postinfection (PI) and normalized to the cell numbers. (B) A representative sample that corresponds to panel A. (C) Comparison of 1 round of HIV-1 replication in a cohort of 15 SCD patients. The results are expressed relative to infection of PBMCs from 9 control subjects, which were set to 100%. The means ± standard error (SE) and P value calculated with Student t test are shown. (D) Inhibition of p24 production in SCD-derived PBMCs. PBMCs obtained from 3 SCD patients and 3 controls were activated and infected as described above. Supernatants were collected 72 hours PI, and p24 was measured by ELISA. The means ± SE and P value calculated with Student t test are shown. (E) Inhibition of HIV-1 env expression in SCD-derived PBMCs. PBMCs obtained from 4 SCD patients and 4 controls were activated and infected as described above for 48 hours. RNA was extracted, reverse transcribed, and analyzed with primers for HIV-1 env gene by RT-PCR using 18S RNA as a reference. The means ± SE and P value calculated with Student t test are shown. (F) Inhibition HIV-1 RT in SCD PBMCs. PBMCs obtained from 3 SCD patients and 3 controls were activated with PHA and IL-2 and infected with HIV-1-LUC-G. At 6 hours PI, DNA was extracted and analyzed by RT-PCR on Roche 480 using primers for early LTR and β-globin gene as a reference. The means ± SE are shown (n = 3 for each sample). The means ± SE and P value calculated with Student t test are shown.
Upregulation of heme and iron regulatory and antiviral genes in SCD PMBC
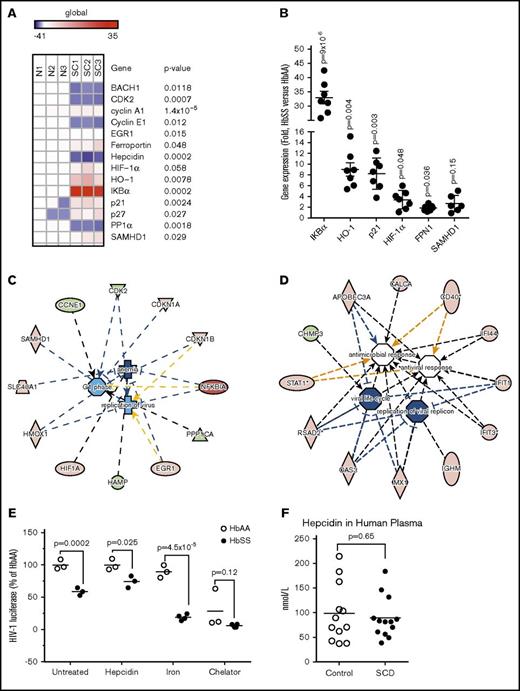
To identify a potential host HIV-1 restriction factor or factors, we tested expression of selected heme and iron regulatory genes, including BACH1, EGR1, ferroportin, hepcidin, HIF-1α, HO-1, p21, and p27. In addition, we analyzed cellular HIV-1 regulatory factors that may inhibit HIV-1 RT and transcription, SAMHD1, CDK2, and PP1.10 We observed a strong downregulation of BACH1, CDK2, cyclin E, hepcidin, and PP1, and an upregulation of ferroportin, HIF-1α, HO-1, IKBα, p21, p27, and SAMHD1 (Figure 2A). No appreciable changes were detected in the expression of cyclin A and EGR1 (Figure 2A). Overexpression of IKBα, HO-1, p21, HIF-1α, and ferroportin were confirmed in the analysis of additional SCD patient samples, whereas SAMHD1 did not shown a statistical significance in upregulation in the SCD samples (Figure 2B). IPA was used to link the analyzed genes in a network that showed a relation to the control of viral replication, degree of anemia, and cell cycle progression (Figure 2C). SCD-derived PBMC host cell proteins, which modulate iron metabolism and regulate HIV-1 replication, were also linked in the IPA.
Altered iron metabolism in SCD-derived PBMCs is critical for HIV-1 inhibition. (A) Heat map showing expression of selected iron and HIV-1 regulatory genes in SCD PBMCs. RNA was extracted from PBMCs obtained from 3 SCD patients and 3 controls. RNA was reverse transcribed and analyzed by RT-PCR using primers for BACH1, CDK2, Cyclin A, Cyclin E, EGR1, ferroportin, hepcidin, HIF-1α, HO-1, IKBα, p21, p27, PP1α, and SAMHD1. The 18S RNA was used as a housekeeping control gene. Heat map was constructed using Morpheus software. P values were determined using paired Student t test. (B) Validation of candidate genes expression. Expression of IKBα, HO-1, p21, HIF-1α, and ferroportin mRNA was analyzed from additional 3 controls and 7 SCD PBMCs. SAMHD1 expression was analyzed in 3 controls and 6 SCD PBMCs. RNA was isolated, reverse transcribed, and analyzed by real time. 18S rRNA was used as a reference for ΔΔCt analysis. The means ± SE and P value calculated with Student t test are shown (C) IPA of the iron and HIV-1 regulatory genes. Ingenuity software analysis of genes shown in panel A identified a protein network that connected iron and HIV-1 regulatory genes through virus replication, anemia, and cell cycle progression networks (D). Upregulated genes are colored in red and downregulated genes are colored in green. (D) IPA of viral and pathogen restricting genes in SCD. Ingenuity software analysis of 250 upregulated and downregulated genes determined by meta-analysis of Geoset GSE53441 (data from PBMCs of 24 SCD patients and 10 controls) identified a protein network of 12 genes involved in antiviral response and viral replication. (E) Hepcidin restores HIV-1 replication in SCD PBMCs. Activated PBMCs from 3 SCD patients and 3 control individuals were infected with HIV-1-LUC-G virus and treated with 0.9 μM hepcidin, 20 μM ferric ammonium citrate (iron), or 10 μM PPYeT iron chelator. At 48 hours PI, luciferase activity was measured. The means and P values determined by Student t test are shown. (F) Hepcidin in plasma from SCD patients. Hepcidin was measured in plasma obtained from 13 SCD and 12 healthy subjects by HR/SIM method. The means and P values determined by Student t test are shown.
Altered iron metabolism in SCD-derived PBMCs is critical for HIV-1 inhibition. (A) Heat map showing expression of selected iron and HIV-1 regulatory genes in SCD PBMCs. RNA was extracted from PBMCs obtained from 3 SCD patients and 3 controls. RNA was reverse transcribed and analyzed by RT-PCR using primers for BACH1, CDK2, Cyclin A, Cyclin E, EGR1, ferroportin, hepcidin, HIF-1α, HO-1, IKBα, p21, p27, PP1α, and SAMHD1. The 18S RNA was used as a housekeeping control gene. Heat map was constructed using Morpheus software. P values were determined using paired Student t test. (B) Validation of candidate genes expression. Expression of IKBα, HO-1, p21, HIF-1α, and ferroportin mRNA was analyzed from additional 3 controls and 7 SCD PBMCs. SAMHD1 expression was analyzed in 3 controls and 6 SCD PBMCs. RNA was isolated, reverse transcribed, and analyzed by real time. 18S rRNA was used as a reference for ΔΔCt analysis. The means ± SE and P value calculated with Student t test are shown (C) IPA of the iron and HIV-1 regulatory genes. Ingenuity software analysis of genes shown in panel A identified a protein network that connected iron and HIV-1 regulatory genes through virus replication, anemia, and cell cycle progression networks (D). Upregulated genes are colored in red and downregulated genes are colored in green. (D) IPA of viral and pathogen restricting genes in SCD. Ingenuity software analysis of 250 upregulated and downregulated genes determined by meta-analysis of Geoset GSE53441 (data from PBMCs of 24 SCD patients and 10 controls) identified a protein network of 12 genes involved in antiviral response and viral replication. (E) Hepcidin restores HIV-1 replication in SCD PBMCs. Activated PBMCs from 3 SCD patients and 3 control individuals were infected with HIV-1-LUC-G virus and treated with 0.9 μM hepcidin, 20 μM ferric ammonium citrate (iron), or 10 μM PPYeT iron chelator. At 48 hours PI, luciferase activity was measured. The means and P values determined by Student t test are shown. (F) Hepcidin in plasma from SCD patients. Hepcidin was measured in plasma obtained from 13 SCD and 12 healthy subjects by HR/SIM method. The means and P values determined by Student t test are shown.
To test for additional HIV-1 restriction factors, we conducted a meta-analysis of gene expression data from PBMCs of 24 SCD patients and 10 controls (Geoset accession number GSE53441) using Geo2R software, which identified 250 upregulated and downregulated genes. In addition, IPA analysis of the selected genes further identified 12 candidate genes (supplemental Table 2) that were linked to a network that modulated viral replication, viral life cycle, antiviral response, and antimicrobial response (Figure 2D). The top 3 candidates, radical S-adenosyl methionine domain 2 protein (RSAD2), interferon-induced protein with teratricopeptide repeats 1 (IFIT1) and IFIT2, were previously shown to have an effect on HIV-1 (see Discussion).39,40
We tested the importance of ferroportin expression in SCD PBMCs as we previously showed it to inhibit HIV-1 replication.34 The HIV-1–infected SCD PBMCs were treated with hepcidin, which promotes ferroportin internalization and degradation. The addition of hepcidin reduced HIV-1 inhibition in the SCD PBMCs (Figure 2E, hepcidin). Treatment with ferric ammonium citrate further increased HIV-1 inhibition in SCD PBMCs but had no effect on HIV-1 infection in control PBMCs (Figure 2E, iron). Treatment with iron chelator, PPYeT,25 inhibited HIV-1 replication in control PBMCs in accord with our previous study,25 and further inhibited HIV-1 in SCD PBMCs (Figure 2E, chelator). These observations suggested that iron metabolism was altered in SCD PBMCs, and that iron depletion by an iron chelator or ferroportin inhibited HIV-1.
Hepcidin levels were tested in our cohort of SCD patients, using HR/SIM MS. Synthetic human hepcidin (HEP) and isotopically labeled hepcidin peptides were used for the analysis (supplemental Figure 2A). Both synthetic HEP added externally to rabbit plasma (RP) and native HEP in human plasma were efficiently detected by HR/SIM method (supplemental Figure 2B). The RP was used as a blank plasma control lacking any residual human HEP to build a calibration curve (supplemental Figure 2C). Quality control samples were obtained using hepcidin added at 150 nM, 15 nM, and 1.5 nM in RP (not shown). Analysis of 13 SCD serum samples vs 12 healthy controls showed no significant difference in hepcidin levels between SCD and control subjects (Figure 2F). Ferroportin expression was sustained in SCD patients, as hepcidin levels were not elevated in these patients.
Intracellular iron levels are lower in SCD PBMCs
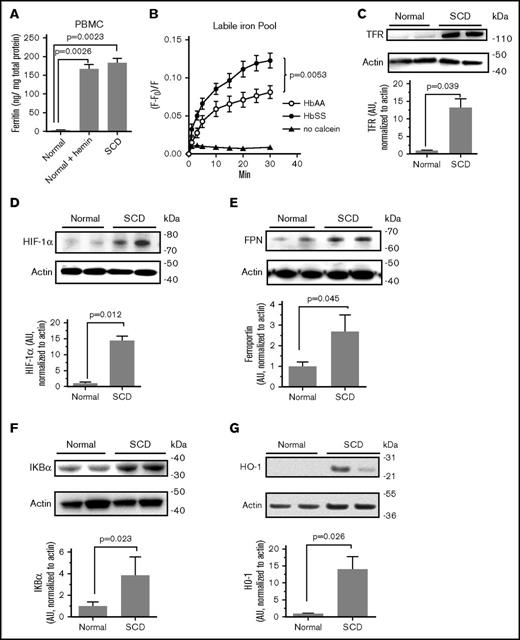
The expression of iron regulatory proteins and intracellular iron levels in SCD PBMCs were analyzed. Cellular ferritin levels were significantly increased in SCD PBMCs compared with normal controls (Figure 3A). Treatment of the control PBMCs with hemin led to elevation of ferritin level comparable to that in SCD PBMCs (Figure 3A), suggesting that the increase in ferritin level can be due to the hemolysis. Intracellular iron levels were then analyzed using calcein fluorescent sensor.25 PBMCs were treated with nonfluorescent cell-permeable calcein AM, which is converted to calcein in the cells whose fluorescence is quenched upon binding to iron. A fractional fluorescence (F − F0)/F, which is inversely proportional to the amount of labile chelatable iron, was markedly higher in SCD PBMCs (Figure 3B), suggesting a reduced LIP in SCD PBMCs. To further confirm that intracellular iron levels were low in SCD PBMCs, we analyzed protein expression of TFR mRNA, which is stabilized when its iron responsive elements located in the 3′-untranslated region are bound to iron responsive protein.41 We also analyzed HIF-1α, which is stabilized when intracellular iron levels are low.42 TFR expression was increased by 13-fold and HIF-1α was increased by >15-fold in SCD PBMCs (Figure 3C-D), further suggesting low intracellular iron levels in SCD PBMCs. Protein expression of ferroportin was found 2.7-fold higher in SCD PBMCs (Figure 3E). The expression of IKBα was fourfold higher in SCD PBMCs (Figure 3F). At the protein level, HO-1 expression was 12-fold higher in SCD PBMCs (Figure 3G). These results suggest that the altered metabolism in SCD PBMCs of iron causes upregulation of proteins involved in iron export and storage. Also, proteins that respond to low levels of intracellular iron were upregulated in SCD PBMCs.
Deregulation of intracellular iron in SCD PBMCs. (A) Increased ferritin expression in SCD PBMCs. PBMCs obtained from 3 SCD patients and 3 controls were cultured in media supplemented with PHA for 36 hours and then activated with IL-2 for 36 hours. Where indicated, control PBMCs were further treated with 100 μM hemin for 24 hours. Ferritin was measured in cell lysates by ELISA. The means ± SD are shown. (B) Reduced intracellular iron levels in SCD PBMCs. Labile intracellular iron pool was measured in PBMCs obtained from 3 SCD patients or 3 normal controls. Cells were treated with 0.1 μM calcein AM for 10 minutes at 37°C. After washing with PBS, cells were incubated at 37°C, and calcein fluorescence was measured in a Glomax Multidetection system at different time points. Fractional fluorescence (F − F0)/F, which is inversely proportional to the amount of chelatable iron plotted on the y-axis. Paired Student t test was used to determine P value of the iron values at 30 minutes. (C-G) Elevated expression of TFR, HIF-1α, ferroportin, IKBα, and HO-1 in SCD PBMCs. Cell lysates from activated PBMCs obtained from SCD patients and normal controls were resolved on 4% to 12% Bis-Tris gel and probed with antibodies against TFR, HIF-1α, ferroportin (FPN), IKBα, and HO-1. The β-actin was used as loading control. Results were quantified using Image Quant Software. Bars represent independent experiments on 4 patients and 4 controls for HIF-1α and IKBα; 6 SCD patients and 6 controls for ferroportin; and 3 SCD patients and 3 controls for HO-1. The blots show 2 representative SCD and control samples. The means ± SD are shown. P values were calculated using Student t test.
Deregulation of intracellular iron in SCD PBMCs. (A) Increased ferritin expression in SCD PBMCs. PBMCs obtained from 3 SCD patients and 3 controls were cultured in media supplemented with PHA for 36 hours and then activated with IL-2 for 36 hours. Where indicated, control PBMCs were further treated with 100 μM hemin for 24 hours. Ferritin was measured in cell lysates by ELISA. The means ± SD are shown. (B) Reduced intracellular iron levels in SCD PBMCs. Labile intracellular iron pool was measured in PBMCs obtained from 3 SCD patients or 3 normal controls. Cells were treated with 0.1 μM calcein AM for 10 minutes at 37°C. After washing with PBS, cells were incubated at 37°C, and calcein fluorescence was measured in a Glomax Multidetection system at different time points. Fractional fluorescence (F − F0)/F, which is inversely proportional to the amount of chelatable iron plotted on the y-axis. Paired Student t test was used to determine P value of the iron values at 30 minutes. (C-G) Elevated expression of TFR, HIF-1α, ferroportin, IKBα, and HO-1 in SCD PBMCs. Cell lysates from activated PBMCs obtained from SCD patients and normal controls were resolved on 4% to 12% Bis-Tris gel and probed with antibodies against TFR, HIF-1α, ferroportin (FPN), IKBα, and HO-1. The β-actin was used as loading control. Results were quantified using Image Quant Software. Bars represent independent experiments on 4 patients and 4 controls for HIF-1α and IKBα; 6 SCD patients and 6 controls for ferroportin; and 3 SCD patients and 3 controls for HO-1. The blots show 2 representative SCD and control samples. The means ± SD are shown. P values were calculated using Student t test.
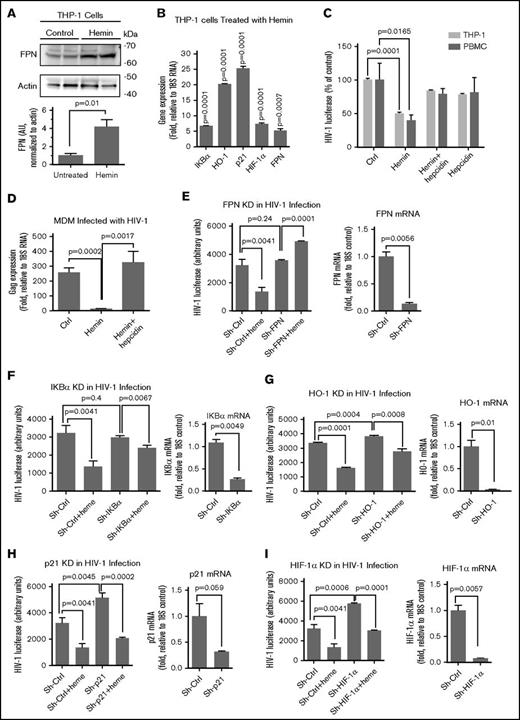
Ferroportin mediates the inhibition of HIV-1 by hemin
Next, we analyzed whether treatment with hemin replicated gene expression pattern and inhibition of HIV-1 infection observed in SCD PBMCs. Hemin treatment led to a fourfold increase of ferroportin expression in THP-1 cells (Figure 4A). Gene expression analysis showed upregulation of IKBα, HO-1, p21, HIF-1α, and ferroportin mRNA in hemin-treated THP-1 cells (Figure 4B) and PBMCs (supplemental Figure 3). The gene expression patterns resembled the one observed among the SCD PBMCs except that IKBα levels were significantly elevated in SCD PBMCs. Hemin treatment suppressed HIV-1 infection in both PBMCs and THP-1 cells, whereas subsequent treatment with hepcidin restored HIV-1 replication (Figure 4C), suggesting a critical role of ferroportin in HIV-1 inhibition. Primary MDMs infected with M-tropic (BaL) HIV-1 showed inhibition of HIV-1 infection when treated with hemin (Figure 4D). Meanwhile, treatment with hepcidin restored HIV-1 replication (Figure 4D), confirming the critical role of ferroportin.
FPN, hepcidin, and iron regulatory genes play a role in heme-mediated HIV-1 inhibition. (A) Hemin treatment induced ferroportin expression. THP-1 cells were treated with hemin for 24 hours. Cell lysates were resolved on 4% to 12% Bis-Tris gel and probed with antibodies against FPN. The β-actin was used as loading control. Results were quantified using Image Quant Software. Bars represent quantification from 4 SCD and control samples. The means ± SD are shown. P values were calculated using Student t test. (B) Expression of candidate genes in hemin-treated THP-1 cells. THP-1 cells were treated with hemin for 24 hours. RNA was extracted, reverse transcribed, and analyzed by RT-PCR using primers for IKBα, HO-1, p21, HIF-1α, and FPN. The18S rRNA was used as a reference for ΔΔCt analysis. The means ± SD are shown (n = 3 for each sample). P values were calculated using Student t test. (C) Restoration of hemin-inhibited HIV-1 replication in THP-1 cells and PBMCs. THP-1 cells and PBMCs were treated with hemin or with hemin and hepcidin and infected with HIV-1-LUC-G. Luciferase activity was measured 2 days PI. The means ± SD are shown (n = 3 for each sample). P values were calculated using Student t test. (D) Restoration of hemin-inhibited HIV-1 replication in MDMs. MDMs were treated with hemin or with hemin and hepcidin and infected with M-tropic HIV-1 (BAL) isolate. Expression of gag was measured by RT-PCR after 6 days PI. The means ± SD are shown (n = 3 for each sample). P values were calculated using Student t test. (E-I) Restoration of hemin-mediated HIV-1 replication by knockdowns of FPN, IKBα, HO-1, p21, and HIF-1α. THP-1 cells were stably transduced with lentiviruses expressing corresponding shRNA and infected HIV-1-LUC-G virus. Where indicated, the cells were also treated with 75 μM hemin. Luciferase activity was measured 2 days PI (left panels). The expression of the corresponding mRNAs was measured by RT-PCR with 18S RNA primers for internal control (right panels). Quantification from 3 independent experiments show the means ± SD and P values calculated using Student t test.
FPN, hepcidin, and iron regulatory genes play a role in heme-mediated HIV-1 inhibition. (A) Hemin treatment induced ferroportin expression. THP-1 cells were treated with hemin for 24 hours. Cell lysates were resolved on 4% to 12% Bis-Tris gel and probed with antibodies against FPN. The β-actin was used as loading control. Results were quantified using Image Quant Software. Bars represent quantification from 4 SCD and control samples. The means ± SD are shown. P values were calculated using Student t test. (B) Expression of candidate genes in hemin-treated THP-1 cells. THP-1 cells were treated with hemin for 24 hours. RNA was extracted, reverse transcribed, and analyzed by RT-PCR using primers for IKBα, HO-1, p21, HIF-1α, and FPN. The18S rRNA was used as a reference for ΔΔCt analysis. The means ± SD are shown (n = 3 for each sample). P values were calculated using Student t test. (C) Restoration of hemin-inhibited HIV-1 replication in THP-1 cells and PBMCs. THP-1 cells and PBMCs were treated with hemin or with hemin and hepcidin and infected with HIV-1-LUC-G. Luciferase activity was measured 2 days PI. The means ± SD are shown (n = 3 for each sample). P values were calculated using Student t test. (D) Restoration of hemin-inhibited HIV-1 replication in MDMs. MDMs were treated with hemin or with hemin and hepcidin and infected with M-tropic HIV-1 (BAL) isolate. Expression of gag was measured by RT-PCR after 6 days PI. The means ± SD are shown (n = 3 for each sample). P values were calculated using Student t test. (E-I) Restoration of hemin-mediated HIV-1 replication by knockdowns of FPN, IKBα, HO-1, p21, and HIF-1α. THP-1 cells were stably transduced with lentiviruses expressing corresponding shRNA and infected HIV-1-LUC-G virus. Where indicated, the cells were also treated with 75 μM hemin. Luciferase activity was measured 2 days PI (left panels). The expression of the corresponding mRNAs was measured by RT-PCR with 18S RNA primers for internal control (right panels). Quantification from 3 independent experiments show the means ± SD and P values calculated using Student t test.
The HIV-1 inhibitory role of each individual gene overexpressed in the hemin treated cells was assessed by shRNA-mediated RNA interference knockdowns. Our results showed that ferroportin knockdown in THP-1 cells led to the complete prevention of HIV-1 inhibition by hemin (Figure 4E), further pointing to the key role of ferroportin. Knockdowns of IKBα and HO-1 also significantly reduced hemin-mediated HIV-1 inhibition (Figure 4F-G). In contrast, knockdowns of p21 or HIF-1α, while rescuing HIV-1 replication in hemin-treated cells and inducing HIV-1 infection in untreated cells, did not have a significant effect on hemin-mediated HIV-1 inhibition (Figure 4H-I). Taken together, these results conclude that ferroportin and IKBα are novel molecular mediators of HIV-1 inhibition in hemin-treated cells.
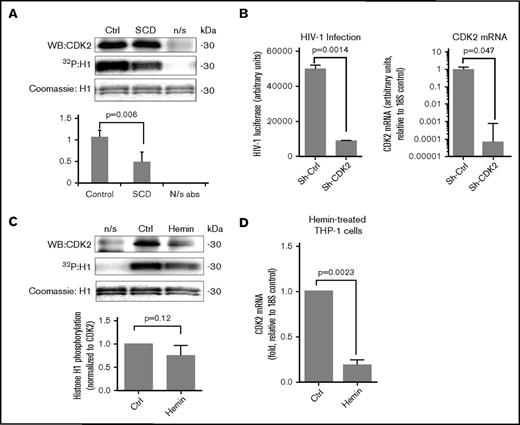
CDK2 activity is decreased in SCD PBMCs
Our previous work revealed that CDK2 activity in iron chelators–treated cells was reduced.26,27 Thus, we tested whether CDK2 activity would be decreased in SCD PBMCs. CDK2 was immunoprecipitated from cell lysates using anti-CDK2 antibodies, and its activity was assayed using histone H1 as a substrate. There was more than twofold reduction of CDK2 activity (Figure 5A). To confirm that CDK2 is critical for HIV-1 replication, we tested stable shRNA-mediated CDK2 knockdowns in THP-1 cells that showed significant reduction in virus replication (Figure 5B). To determine whether hemin treatment affected CDK2 activity, we analyzed CDK2 expression and activity in hemin-treated THP-1 cells (Figure 5C). Hemin treatment of THP-1 cells showed a decrease in both CDK2 expression and activity (Figure 5C), thus reducing the overall CDK2 activity. CDK2 mRNA expression was significantly reduced in hemin-treated THP-1 cells (Figure 5D), suggesting that hemin treatment can also decrease CDK2 mRNA expression.
Reduction of CDK2 activity in SCD PBMCs. (A) CDK2 activity is reduced in SCD PBMCs. PBMCs were obtained from 3 SCD patients and 3 control subjects and activated with PHA and IL-2. Cells were lysed, and CDK2 was immunoprecipitated using anti-CDK2 antibodies from lysates that were equalized by protein. Kinase assays were performed using histone H1 as a substrate. Upper panels show a representative immunoblot of CDK2, a radioactive image of phosphorylated histone H1, and a Coomassie-stained image of histone H1. Lower panel shows quantification from 3 independent experiments. The means ± SD and P values calculated using Student t test are shown. (B) CDK2 knockdown inhibits HIV-1 infection in THP-1 cells. THP-1 cells were stably transduced with lentiviruses expressing CDK2-targeting shRNA or control shRNA and then infected with HIV-1-LUC-G virus. Left panel shows luciferase activity measured at 48 hours PI. Right panel shows expression of CDK2 mRNA determined by RT-PCR with 18S RNA as an internal reference for ΔΔCt analysis. The means ± SD are shown for 3 CDK2 KD and 3 control shRNA clones. (C) CDK2 activity is reduced in hemin-treated THP-1 cells. THP-1 cells treated with 75 μM hemin and control cells were lysed, and CDK2 was immunoprecipitated using anti-CDK2 antibodies. Kinase assays were performed using histone H1 as a substrate. Lower panel shows quantification from 3 independent experiments. P value was calculated using Student t test. (D) Hemin treatment reduces CDK2 mRNA expression. THP-1 cells were treated with 75 μM hemin for 24 hours. RNA was extracted, reverse transcribed, and analyzed by RT-PCR using primers for CDK2 and 18S rRNA as an internal reference for ΔΔCt analysis. The means ± SD and P values calculated using Student t test are shown for 3 independent samples. N/s abs, nonspecific antibodies.
Reduction of CDK2 activity in SCD PBMCs. (A) CDK2 activity is reduced in SCD PBMCs. PBMCs were obtained from 3 SCD patients and 3 control subjects and activated with PHA and IL-2. Cells were lysed, and CDK2 was immunoprecipitated using anti-CDK2 antibodies from lysates that were equalized by protein. Kinase assays were performed using histone H1 as a substrate. Upper panels show a representative immunoblot of CDK2, a radioactive image of phosphorylated histone H1, and a Coomassie-stained image of histone H1. Lower panel shows quantification from 3 independent experiments. The means ± SD and P values calculated using Student t test are shown. (B) CDK2 knockdown inhibits HIV-1 infection in THP-1 cells. THP-1 cells were stably transduced with lentiviruses expressing CDK2-targeting shRNA or control shRNA and then infected with HIV-1-LUC-G virus. Left panel shows luciferase activity measured at 48 hours PI. Right panel shows expression of CDK2 mRNA determined by RT-PCR with 18S RNA as an internal reference for ΔΔCt analysis. The means ± SD are shown for 3 CDK2 KD and 3 control shRNA clones. (C) CDK2 activity is reduced in hemin-treated THP-1 cells. THP-1 cells treated with 75 μM hemin and control cells were lysed, and CDK2 was immunoprecipitated using anti-CDK2 antibodies. Kinase assays were performed using histone H1 as a substrate. Lower panel shows quantification from 3 independent experiments. P value was calculated using Student t test. (D) Hemin treatment reduces CDK2 mRNA expression. THP-1 cells were treated with 75 μM hemin for 24 hours. RNA was extracted, reverse transcribed, and analyzed by RT-PCR using primers for CDK2 and 18S rRNA as an internal reference for ΔΔCt analysis. The means ± SD and P values calculated using Student t test are shown for 3 independent samples. N/s abs, nonspecific antibodies.
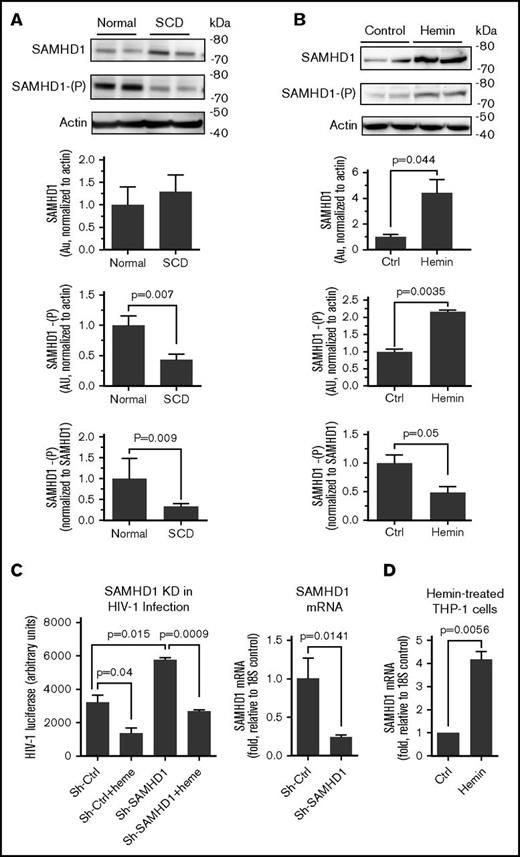
Phosphorylation of SAMHD1 is reduced in SCD-derived and heme-treated cells
HIV-1 is restricted by nonphosphorylated SAMHD1, whereas phosphorylation of SAMHD1 on Thr-592 inhibits its activity and prevents HIV-1 restriction.43 Because CDK2 was shown to phosphorylate SAMHD1 and inhibit its activity,44 we analyzed SAMHD1 expression and SAMHD1 Thr-592 phosphorylation in SCD PBMCs. Among SCD PBMCs, SAMHD1 protein expression was slightly increased but it was not statistically significant (Figure 6A). However, SAMHD1 Thr-592 phosphorylation was significantly reduced in SCD PBMCs (Figure 6A). In hemin-treated THP-1 cells, SAMHD1 expression was significantly elevated (Figure 6B). SAMHD1 phosphorylation was reduced when normalized to nonphosphorylated SAMHD1 protein level (Figure 6B). These results suggest that HIV-1 inhibition in SCD PBMCs can be mediated by a decreased SAMHD1 phosphorylation. Also, heme can induce production of nonphosphorylated SAMHD1 contributing to HIV-1 inhibition.
Reduction of SAMHD1 phosphorylation in SCD PBMCs and hemin-treated THP-1 cells. (A) Reduced SAMHD1 Thr-529 phosphorylation in SCD PBMCs. PBMCs obtained from 4 SCD patients and 4 controls were activated with PHA and IL-2 and treated with 75 μM hemin for 24 hours. Cell lysates were resolved on 4% to 12% Bis-Tris gel and probed with antibodies against SAMHD1 and Thr-592 phosphorylated SAMHD1 (SAMHD1-(P)) and β-actin as loading control. Lower panels show quantification for 4 individual samples. Shown are SAMHD1 normalized to β-actin (upper panel), SAMHD1-(P) normalized to β-actin (middle panel), and SAMHD1-(P) normalized to SAMHD1 expression (bottom panel). The means ± SD and P values calculated using Student t test are shown. (B) Increased SAMHD1 expression and reduced Thr-592 phosphorylation in hemin-treated THP-1 cells. THP-1 cells were treated with 75 μM hemin for 24 hours. Cell lysates were analyzed and quantified as in panel A for 4 independent samples. (C) Restoration of hemin-mediated HIV-1 replication by knockdowns of SAMHD1. THP-1 cells were stably transduced with lentiviruses expressing SAMHD1-targeting shRNA and infected with HIV-1-LUC-G virus. Where indicated, the cells were treated with 75 μM hemin. Luciferase activity was measured 2 days PI. The mRNAs were measured by RT-PCR with 18S rRNA as an internal reference for ΔΔCt analysis. The means ± SD and P values calculated using Student t test are shown for 3 independent samples. (D) Hemin treatment increased SAMHD1 mRNA expression. THP-1 cells were treated with hemin for 24 hours. RNA was extracted, reverse transcribed, and analyzed by RT-PCR using primers for SAMHD1 and 18S rRNA as an internal reference for ΔΔCt analysis. The means ± SD and P values calculated using Student t test are shown for 3 independent samples.
Reduction of SAMHD1 phosphorylation in SCD PBMCs and hemin-treated THP-1 cells. (A) Reduced SAMHD1 Thr-529 phosphorylation in SCD PBMCs. PBMCs obtained from 4 SCD patients and 4 controls were activated with PHA and IL-2 and treated with 75 μM hemin for 24 hours. Cell lysates were resolved on 4% to 12% Bis-Tris gel and probed with antibodies against SAMHD1 and Thr-592 phosphorylated SAMHD1 (SAMHD1-(P)) and β-actin as loading control. Lower panels show quantification for 4 individual samples. Shown are SAMHD1 normalized to β-actin (upper panel), SAMHD1-(P) normalized to β-actin (middle panel), and SAMHD1-(P) normalized to SAMHD1 expression (bottom panel). The means ± SD and P values calculated using Student t test are shown. (B) Increased SAMHD1 expression and reduced Thr-592 phosphorylation in hemin-treated THP-1 cells. THP-1 cells were treated with 75 μM hemin for 24 hours. Cell lysates were analyzed and quantified as in panel A for 4 independent samples. (C) Restoration of hemin-mediated HIV-1 replication by knockdowns of SAMHD1. THP-1 cells were stably transduced with lentiviruses expressing SAMHD1-targeting shRNA and infected with HIV-1-LUC-G virus. Where indicated, the cells were treated with 75 μM hemin. Luciferase activity was measured 2 days PI. The mRNAs were measured by RT-PCR with 18S rRNA as an internal reference for ΔΔCt analysis. The means ± SD and P values calculated using Student t test are shown for 3 independent samples. (D) Hemin treatment increased SAMHD1 mRNA expression. THP-1 cells were treated with hemin for 24 hours. RNA was extracted, reverse transcribed, and analyzed by RT-PCR using primers for SAMHD1 and 18S rRNA as an internal reference for ΔΔCt analysis. The means ± SD and P values calculated using Student t test are shown for 3 independent samples.
To further elucidate the effect of SAMHD1 in heme-mediated inhibition of HIV-1 infection, we tested the effect of its shRNA-mediated knockdown in THP-1 (Figure 6C). SAMHD1 knockdown rescued HIV-1 replication in the hemin-treated cells while strongly inducing HIV-1 infection in the untreated cells (Figure 6C), which was similar to the effect of p21 and HIF-1α knockdowns seen above. Hemin treatment also significantly induced SAMHD1 mRNA expression level (Figure 6D), suggesting that hemolysis might upregulate SAMHD1 expression, thus contributing to HIV-1 restriction.
Our study indicates that increased iron export by ferroportin restricts HIV-1 replication in biologically relevant settings. Collectively reduced iron levels, increased IKBα expression, decreased CDK2 activity, and reduced SAMHD1 phosphorylation can ultimately lead to the inhibition of HIV-1 infection.
Discussion
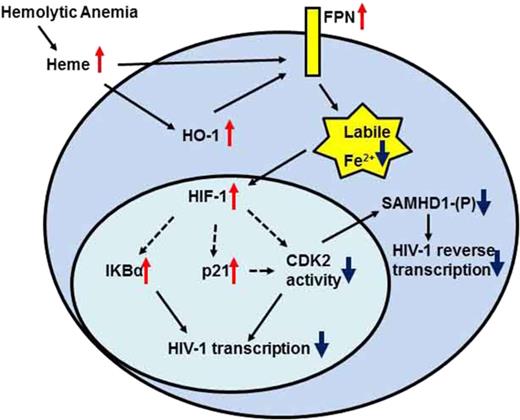
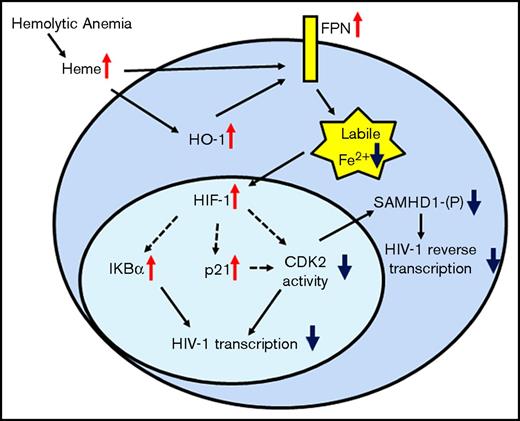
In Figure 7, we created a model of a protein network that could represent the response to hemolysis seen in SCD patients. During hemolysis, the release of heme leads to the activation of HO-1 and induction of HO-1 and ferroportin mRNA expression, facilitating cellular iron export. Meanwhile, the reduction of cellular iron stabilizes HIF-1α, increases p21 expression, and inhibits CDK2 enzymatic activity either indirectly by suppressing mRNA production or directly through p21-mediated inhibition. The reduced CDK2 activity contributes to the reduction of SAMHD1 phosphorylation and inhibition of HIV-1 RT. In addition, overexpression of nonphosphorylated SAMHD1 by heme causes accumulation of active nonphosphorylated SAMHD1. HIV-1 transcription is inhibited through the expression of IKBα, which sequesters NF-κB, critical for HIV-1 transcription activation. Finally, the lack of hepcidin upregulation in SCD allows sustained ferroportin expression to collectively block HIV-1 RT and transcription, which restricts HIV-1 replication.
Protein network of iron-activated HIV-1 restriction factors in hemolytic anemia. SCD condition or hemin treatment increases expression of HO-1 and FPN. FPN reduces intracellular iron levels stabilizing HIF-1α, and increasing IKBα and p21 production. It also decreases CDK2 expression and inhibits CDK2 activity. Decreased CDK2 activity reduces SAMHD1 phosphorylation and inhibits HIV-1 RT. Decreased CDK2 activity and increased IKBα expression can also inhibit HIV-1 transcription.
Protein network of iron-activated HIV-1 restriction factors in hemolytic anemia. SCD condition or hemin treatment increases expression of HO-1 and FPN. FPN reduces intracellular iron levels stabilizing HIF-1α, and increasing IKBα and p21 production. It also decreases CDK2 expression and inhibits CDK2 activity. Decreased CDK2 activity reduces SAMHD1 phosphorylation and inhibits HIV-1 RT. Decreased CDK2 activity and increased IKBα expression can also inhibit HIV-1 transcription.
HIV-1 replication can be affected by multiple host restriction factors, including APOBEC3G, TRIM5α, MxB, SAMHD1, BST-2/tetherin, SERIN5/6,45 and likely others. All mentioned factors are counteracted by HIV-1 accessory proteins except SAMHD1, which restricts HIV-1 infection in myeloid cells.18,46 Here we showed that expression of ferroportin can regulate SAMHD1 phosphorylation and also the expression of IKBα, thus expanding the inhibitory activity to RT and transcription. Ferroportin expression is likely to be sustained as we detected similar levels of hepcidin in plasma of SCD patients in comparison with controls. Interestingly, recent analysis of hepcidin production during viral infection showed a virus-specific pattern with upregulation of hepcidin production during HIV-1 but not HCV or HBV infection.47 Thus, containment of hepcidin expression in SCD can add additional control to HIV-1 infection. Iron treatment further inhibited HIV-1 in SCD PBMCs but not in control PBMCs, further pointing to the altered iron metabolism in SCD PBMCs. Iron chelators, on the other hand, inhibited HIV-1 infection in both SCD and normal controls in accord with our previous observations.25-27 Remarkably, ferroportin expression in SCD PBMCs explains the long standing paradox of the near absence of HIV-1 infection in patients with SCD. Expression of ferroportin was shown to control intracellular bacterial pathogens, including Salmonella, Chlamydia, and Legionella.48 Our current study highlights our previous observations and extends the previous findings to show that HIV-1 can also be inhibited by overexpression of ferroportin.34
Our meta-analysis identified additional potentially antiviral genes, including RSAD2, IFIT1, and IFIT2. RSAD2 was found among the 16 antiviral genes overexpressed in patients with psoriasis and HIV-1 elite controllers.39 RSAD2, IFIT1, IFIT2, APOBEC3A, and MX1 are interferon-induced genes that are also induced by HIV-1 viral gene and protein product, Vpr.40 Further studies are needed to determine if any of these genes may play a role in HIV-1 restriction in SCD.
Our previous studies have shown that HIV-1 transcription is negatively affected by low intracellular iron levels, which affects the activity of CDK2 and CDK9.25-27 We recently showed that iron chelators induce IKBα expression.25 Because the HIV-1 promoter contains 2 NF-κB binding sites,49 its activity is critically dependent on NF-κB. The NF-κB availability was shown to be increased by iron efflux that activates IκB kinase.50-52 Upregulation of IKBα expression in SCD PBMCs suggests a mechanism for reduced HIV-1 transcription in hemolytic conditions. Previously, hemoglobin-stimulated macrophages were shown to undergo a selective differentiation program toward anti-inflammatory hemoglobin macrophages, which exhibited increased ferroportin expression and reduced intracellular iron levels.53 Our current study demonstrated significantly reduced intracellular iron levels and increased expression of ferroportin, suggesting induction of anti-inflammatory type macrophages. Additional in vivo analysis is needed to test whether this is the case in SCD. Iron depletion removes iron from prolyl hydroxylases and also increases HIF-1α and HIF-2α protein levels.42 We observed a significant upregulation of HIF-1α protein level and increased expression of TFR. HIF-1α may contribute to the regulation of HIV-1 replication, as seen in HIF-1α knockdown that induced 1 round of HIV-1 infection. In addition to the inhibition of CDK2,27,54 iron depletion can also have an effect on hypusine hydroxylation of eIF5α.55 This translation factor is critical for mRNA export by HIV-1 Rev protein, and this process can also be inhibited by an iron chelator, deferiprone.55 We also observed here a reduction of PP1α expression. Our previous studies showed that CDK9 activity is regulated in part by PP1.7,56-58 Previously, PP1 has been shown to be negatively affected by hypoxia either through a decrease of mRNA expression59 or by increased association with nuclear inhibitor of PP1 (NIPP1).60 Thus, hypoxia-mediated PP1 inhibition may lead to changes in CDK9 phosphorylation and modulation of its activity. Hypoxia can also modulate the activity of CDK2 through the expression of p27,61 which may also have a downstream effect on CDK9.
Previously, hemin was shown to inhibit HIV-1 infection in vitro and in vivo,11 and the inhibition required expression of HO-1. We observed significant induction of HO-1 expression and showed that HO-1 knockdown prevented HIV-1 inhibition by hemin, further confirming that HO-1 contributed to the HIV-1 inhibition. HO-1 activation was shown to protect a person with SCD trait against cerebral malaria through the production of CO.62 Whether SCD trait may have a protective effect against HIV-1 infection remains to be determined. Our current study suggests that hemolysis and excess of heme in SCD play a key role in HIV-1 restriction in SCD. Heme induces inflammation through the activation of inflammasome and, specifically, the nucleotide-binding domain and leucine-rich repeat containing family, pyrin domain containing 3 protein.63 Heme is among the growing list of danger molecules of pathogen recognition.64 Future studies will address the role of heme-induced inflammation in the setting of SCD.
Although our study focused on CDK2 and SAMHD1, the observed induction of p21 expression and downregulation of CDK2 can also affect HIV-1 RT as CDK2 was shown to phosphorylate HIV-1 RT.65 In addition, p21 controls the expression of ribonucleotide reductase 2 (RNR2) that may lower deoxyribonucleotide pool and impair HIV-1 RT.46,66,67 Expression of p21 inhibited RNR2 transcription by repressing E2F1 transcription factor, which activates RNR2 transcription.66 In the future, we will analyze RNR2 activity to determine its contribution to the HIV-1 inhibition in SCD PBMCs.
In conclusion, we demonstrated that hemolytic conditions of SCD upregulate hypoxia and iron regulatory pathways leading to the inhibition of HIV-1 replication. Our study links HIV-1 inhibition in a biologically relevant setting to the reduction of cellular iron levels and expression of ferroportin. For the first time, we linked ferroportin expression to the inhibition of CDK2 activity and reduction of SAMHD1 phosphorylation. Our results provide the first direct evidence that HIV-1 replication is restricted in SCD-derived PBMCs. Understanding the mechanism of HIV-1 inhibition in physiologically relevant settings might open therapeutic possibilities for treatment of hidden HIV-1 reservoirs that harbor nonreplicating HIV-1 provirus or provide new approaches for permanent HIV-1 inhibition.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Christian Parry for careful reading and insightful comments.
This work was supported by Research grants from the NIH National Heart, Lung, and Blood Institute (1P50HL118006 and 1R01HL125005), National Institute on Minority Health and Health Disparities (5G12MD007597), and a District of Columbia Developmental Center for AIDS Research grant (P30AI087714). The content is solely the responsibility of the authors and does not necessarily represent the official view of National Heart, Lung, and Blood Institute, National Institute of Allergy and Infectious Diseases, National Institute on Minority Health and Health Disparities, NIH, or US Food and Drug Administration.
Authorship
Contribution: N.K. designed research, performed experiments, and wrote the manuscript; T.A., X.L, X.N., and A.I. designed research, performed experiments, and analyzed the data; S. Diaz and P.O. enrolled patients and edited the manuscript; M.J. analyzed the data and edited the manuscript; S. Dhawan designed research, performed experiments, analyzed the data, and edited the manuscript; and S.N. designed the project, supervised research, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sergei Nekhai, Center for Sickle Cell Disease, Howard University, 1840 7th St NW, Suite 202, Washington DC 20001; e-mail: snekhai@howard.edu.
References
Author notes
The data reanalyzed in this article have been deposited in the Gene Expression Omnibus database (accession number GSE53441).