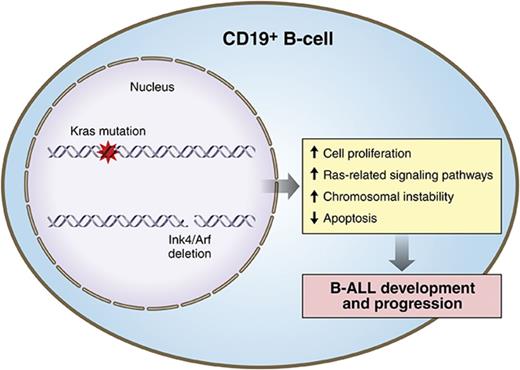
Key Points
Ras pathway activation cooperates with Ink4a/Arf locus deletion in B cells to induce a fully penetrant lymphoma/leukemia phenotype in mice.
These tumors resemble high-risk subtypes of human B-ALL, providing a convenient and highly reproducible model of refractory B-ALL.
Visual Abstract
Professional illustration by Patrick Lane, ScEYEnce Studios.
Abstract
Despite recent advances in treatment, human precursor B-cell acute lymphoblastic leukemia (B-ALL) remains a challenging clinical entity. Recent genome-wide studies have uncovered frequent genetic alterations involving RAS pathway mutations and loss of the INK4A/ARF locus, suggesting their important role in the pathogenesis, relapse, and chemotherapy resistance of B-ALL. To better understand the oncogenic mechanisms by which these alterations might promote B-ALL and to develop an in vivo preclinical model of relapsed B-ALL, we engineered mouse strains with induced somatic KrasG12D pathway activation and/or loss of Ink4a/Arf during early stages of B-cell development. Although constitutive activation of KrasG12D in B cells induced prominent transcriptional changes that resulted in enhanced proliferation, it was not sufficient by itself to induce development of a high-grade leukemia/lymphoma. Instead, in 40% of mice, these engineered mutations promoted development of a clonal low-grade lymphoproliferative disorder resembling human extranodal marginal-zone lymphoma of mucosa-associated lymphoid tissue or lymphoplasmacytic lymphoma. Interestingly, loss of the Ink4a/Arf locus, apart from reducing the number of apoptotic B cells broadly attenuated KrasG12D-induced transcriptional signatures. However, combined Kras activation and Ink4a/Arf inactivation cooperated functionally to induce a fully penetrant, highly aggressive B-ALL phenotype resembling high-risk subtypes of human B-ALL such as BCR-ABL and CRFL2-rearranged. Ninety percent of examined murine B-ALL tumors showed loss of the wild-type Ink4a/Arf locus without acquisition of highly recurrent cooperating events, underscoring the role of Ink4a/Arf in restraining Kras-driven oncogenesis in the lymphoid compartment. These data highlight the importance of functional cooperation between mutated Kras and Ink4a/Arf loss on B-ALL.
Introduction
B-cell acute lymphoblastic leukemia (B-ALL) is the most common neoplasm among children.1 By using current therapy regimens, complete remission is attainable in more than 85% of young patients. However, the prognosis after relapse is poor, and B-ALL remains a significant cause of cancer mortality among this age group.2 Furthermore, as a result of a worse prognosis and higher relapse rate, B-ALL remains a significant clinical problem in adult patients.1,3,4 B-ALL is a heterogeneous malignancy of B-cell–committed precursor cells that affects bone marrow (BM) and peripheral blood (PB) and is most commonly characterized by a precursor B-ALL immunophenotype (CD10+CD19+TdT+cyt-IgM+).1 Over the years, recurrent cytogenetic abnormalities have been used to classify disease subtypes and study their pathogenesis and clinical features.5
On a more positive note, recent advances in molecular biology methodology have uncovered a complex genetic landscape, helped to identify new subtypes of B-ALL, and demonstrated alterations in cellular pathways independent of cytogenetic subclasses.5-8 Among the numerous recently recognized pathogenetic processes in B-ALL, such as lymphoid development, tumor suppression, cytokine receptor signaling, kinase signaling, and chromatin remodeling,5 Ras pathway mutations (eg, KRAS, NRAS, FLT3, PTPN11, and NF1) seem particularly important, inasmuch as they are present in up to 40% of relapsed cases and probably originate in small subclones, already present at diagnosis, that for various reasons are unresponsive to chemotherapy.9-13 Moreover, the true incidence of activated RAS/MAPK signaling may be underestimated, because it can be induced by chromosomal rearrangements such as BCR-ABL,14,15 CRLF2,16-18 and/or other genomic alterations, including those acquired during relapse. Indeed, genomic analysis of presentation/relapsed B-ALL samples provides strong indication of convergence in WNT and RAS-related MAPK pathways.19
Altered expression of tumor suppression and cell-cycle regulation genes such as TP53 and CDKN2A/B are also frequently acquired during relapse of B-ALL.19-21 CDKN2A encodes p16INK4A and p14ARF tumor suppressors in humans (p16Ink4a and p19Arf in mice; hereafter referred to as Ink4a and Arf) that play non-overlapping crucial roles in the regulation of cell-cycle progression.22,23 Both proteins counteract excessive mitogenic signals to promote cellular senescence. The INK4A/ARF locus is most frequently deleted in unfavorable hypodiploid, BCR-ABL, and CRLF2 subtypes of B-ALL.24,25 Therefore, to comprehensively characterize the functional roles of somatic RAS pathway activation and INK4A/ARF alterations in B cells, we generated conditional mutant mice overexpressing constitutively active mutant KrasG12D and/or harboring a knockout of a single Ink4a/Arf allele. These animals show that Ink4a/Arf restrains the oncogenic potential of KrasG12D mutation and that concurrent Kras activation and loss of Ink4a/Arf in CD19+ B cells results in spontaneous development of an aggressive form of B-ALL.
Materials and methods
Engineered mice
Mouse strains bearing either a mutant LoxP-STOP-LoxP-KrasG12D knockin allele (KrasG12D), Ink4a/Arf with floxed exons 2 and 3 (Ink4a/ArfL), or a CD19-driven Cre recombinase knockin allele (CD19Cre) were crossed to generate experimental cohorts.26-29 CD19Cre/+ animals were used as controls. Genotyping was performed as previously described.30 Cre-mediated excision of the Ink4a/Arf conditional allele was tested by Southern blot analysis of genomic DNA isolated from whole testes or CD19+ splenocytes isolated by using magnetic beads (Miltenyi Biotech). Reverse transcription polymerase chain reaction/restriction fragment length polymorphism analyses were performed with RNA isolated from whole testes or CD19+ splenocytes.30 Survival analysis was performed by using the Kaplan-Meier method and compared by using the log-rank test. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Dana-Farber Cancer Institute.
Histopathology, immunohistochemistry, TUNEL assay, proliferation assay, and flow cytometry
Mice were euthanized at specified time points or when clinical signs of disease were evident; a full histologic examination was performed on each animal to assess for tumor formation. Histology, immunohistochemistry (IHC), and terminal deoxynucleotidyl transferase (TdT) deoxyuridine triphosphate (dUTP) nick-end labeling (TUNEL) assay were performed as in previous studies.31 Immunofluorescence was performed on paraffin-embedded tissue with anti-mouse CD19 antibody and 4′,6-diamidino-2-phenylindole (DAPI) nuclear counterstain. Apoptotic cells were detected by using the APO-BrdU TUNEL kit (Invitrogen). For quantification of IHC analysis, 5 representative high-power fields were quantified by using ImageJ software (Fiji distribution).32,33 Flow cytometry using antibody against B220/CD45R and proliferation assay using tritiated thymidine ([3H]dThd) incorporation into DNA were performed as described.31 For a detailed list of antibodies, see supplemental Methods. Figures display means from 3 independent replicates; P values were calculated by using Welch’s t test.34
Western and Southern blotting
Whole-cell protein extracts prepared from CD19+ splenocytes of 8-week-old mice were immunoblotted by using indicated antibodies as described.31 For a detailed list of antibodies, see supplemental Methods. Genomic DNA collected via the Puregene kit (Gentra Systems) from lymph nodes (LNs) or lungs was used to analyze the immunoglobulin heavy-chain (IgH) gene and the T-cell receptor (TCR) gene by Southern blotting as described.31
Gene expression analysis, array comparative genomic hybridization, and whole-exome sequencing
Duplicate samples of RNA from CD19+ cells from premalignant 8-week-old mice (GSE69367) and from 10 mouse B-ALL tumors (GSE69368) were hybridized to Affymetrix Gene Chip Mouse Genome 430 2.0 arrays (supplemental Methods).
Array comparative genomic hybridization (aCGH) and whole-exome sequencing (WES) were performed by using genomic DNA isolated as for Southern blotting. For aCGH, DNA was hybridized and analyzed as described previously.35 For WES, libraries were prepared from 100 ng of DNA by using the SureSelectXT2 kit (Agilent Genomics) according to the manufacturer’s protocol; thereafter, concentration and quality were checked using Qubit (Life Technologies), Bioanalyzer (Agilent), and quantitative polymerase chain reaction using KAPA Library Quantification kits (Kapa Biosystems) with a 7900HT thermocycler (Applied Biosystems). Samples were loaded onto paired-end 75-bp high-throughput flow cells (Illumina) at a final concentration of 2 pM, and then were sequenced with NextSeq500 (Illumina). FastQ files (PRJNA397401) were aligned, processed with Mutect2,36 and filtered to remove remaining germ line variants (supplemental Methods).
Results
KrasG12D mutation and Ink4a/Arf loss result in expansion of B cells in young engineered mice
To model the cooperative interactions of 2 concomitant genetic lesions found frequently in relapsed human B-ALL, mouse strains bearing either a mutant KrasG12D knockin allele, a conditional Ink4a/Arf knockout allele, or a CD19-driven Cre recombinase knockin allele were crossed to generate cohorts of single-mutant CD19Cre/+KrasG12D/+ and CD19Cre/+Ink4a/ArfL/+ as well as CD19Cre/+KrasG12D/+Ink4a/ArfL/+ compound-mutant mice.26-28 The CD19Cre allele drives Cre recombinase expression during most stages of B-cell development starting with pre-B cells.26 We confirmed that Cre-mediated deletion of the Ink4a/Arf allele and activation of the mutated KrasG12D/+ allele were specific and efficient in CD19+ splenic B cells from engineered mice (supplemental Figure 1).
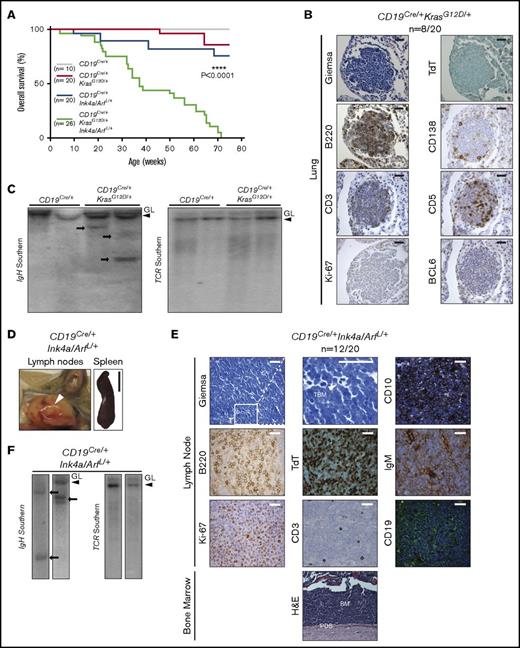
To initially assess the role of KrasG12D and/or loss of Ink4a/Arf during B-cell development, we performed physical examination of young CD19Cre/+KrasG12D/+, CD19Cre/+Ink4a/ArfL/+, and CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice. Significant differences in weight, posture, and overall appearance between young control CD19Cre/+ and engineered mice were not observed. Therefore, groups of 3 single- and double-mutant animals were euthanized at 8 weeks of age (ie, preceding disease development) and rigorously assessed for alterations of lymphoid organs and B cells. Pathologic examination revealed significantly enlarged spleens with increased numbers of mononuclear cells in KrasG12D-mutated animals with or without Ink4a/Arf inactivation (Figure 1A; Table 1), but obvious histologic differences in spleens and LNs between the mutant genotypes and CD19Cre/+ controls were not found (Figure 1B; supplemental Figure 2A). IHC analysis for the CD3 and B220 markers revealed normal T- and B-cell ratios (Figure 1B; supplemental Figure 2A). Of note, there was no obvious increase in number of progenitor or blast cells as measured by TdT staining in the BM, spleen, and LNs from young mutant mice compared with CD19Cre/+ controls (Figure 1B-C; supplemental Figure 2A-B).
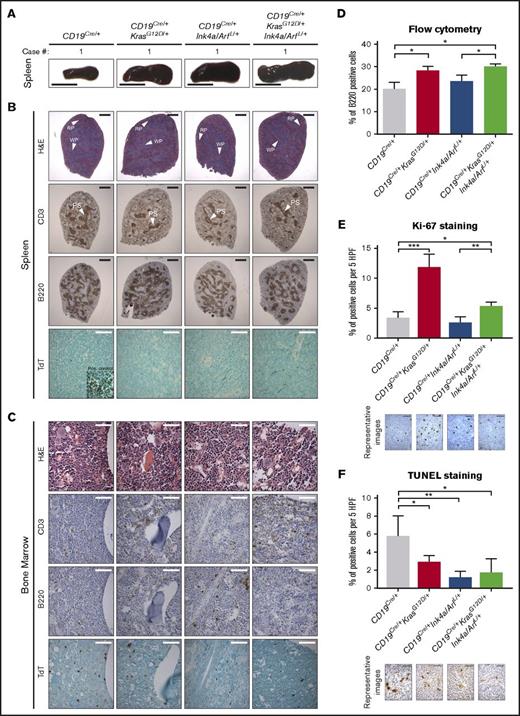
Analysis of 8-week-old mice with conditional expression of KrasG12Dand loss of Ink4a/Arf. (A) Representative gross pictures of dissected spleens from mice of the indicated genotypes. Scale bar, 1 cm. (B-C) Representative histologic and IHC analyses of CD3, B220, and TdT expression in (B) spleens and (C) BM from mice of the indicated genotypes. Inset, positive control: TdT staining of thymus from the same mouse. Scale bars: black, 1 mm; white, 50 µm (see supplemental Figure 2). (D) Mean percentage of B220+ cells assessed by flow cytometry in splenocytes from mice of the indicated genotype; error bars represent ± standard deviation (SD) of 3 different mice per genotype. P values were calculated by using Welch’s t test. (E-F) Differences in the number of proliferating and apoptotic cells assessed by Ki-67 (E) IHC analysis of B-cell areas and (F) TUNEL assay of germinal center areas, respectively, in spleens from mice of the indicated genotypes, calculated using ImageJ. Bar plots represent mean number of cells per 5 high-power fields (HPFs); error bars ± SD. P values were calculated by using Welch’s t test. Representative images are shown below the graphs. Scale bar, 25 µm. Two independent assays were performed for all experiments for which statistics were calculated. *P ≤ .05, **P ≤ .01, ***P ≤ .001. H&E, hematoxylin and eosin; PS, periarterial sheath; RP, red pulp; WP, white pulp.
Analysis of 8-week-old mice with conditional expression of KrasG12Dand loss of Ink4a/Arf. (A) Representative gross pictures of dissected spleens from mice of the indicated genotypes. Scale bar, 1 cm. (B-C) Representative histologic and IHC analyses of CD3, B220, and TdT expression in (B) spleens and (C) BM from mice of the indicated genotypes. Inset, positive control: TdT staining of thymus from the same mouse. Scale bars: black, 1 mm; white, 50 µm (see supplemental Figure 2). (D) Mean percentage of B220+ cells assessed by flow cytometry in splenocytes from mice of the indicated genotype; error bars represent ± standard deviation (SD) of 3 different mice per genotype. P values were calculated by using Welch’s t test. (E-F) Differences in the number of proliferating and apoptotic cells assessed by Ki-67 (E) IHC analysis of B-cell areas and (F) TUNEL assay of germinal center areas, respectively, in spleens from mice of the indicated genotypes, calculated using ImageJ. Bar plots represent mean number of cells per 5 high-power fields (HPFs); error bars ± SD. P values were calculated by using Welch’s t test. Representative images are shown below the graphs. Scale bar, 25 µm. Two independent assays were performed for all experiments for which statistics were calculated. *P ≤ .05, **P ≤ .01, ***P ≤ .001. H&E, hematoxylin and eosin; PS, periarterial sheath; RP, red pulp; WP, white pulp.
Summary of mouse phenotypes
| . | CD19Cre/+ . | CD19Cre/+KrasG12D/+ . | CD19Cre/+Ink4a/Arf L/+ . | CD19Cre/+KrasG12D/+Ink4a/Arf L/+ . |
|---|---|---|---|---|
| Premalignant | ||||
| Number of mice | 3 | 3 | 3 | 3 |
| Spleen weight, mg | 127 ± 12 | 194 ± 6** | 173 ± 12** | 213 ± 6**†* |
| Spleen cells × 107 | 10.6 ± 0.6 | 14.0 ± 0.8** | 12.6 ± 1.2 (ns) | 16.5 ± 1.0**†* |
| Malignant | ||||
| No. of mice | 10 | 20 | 20 | 26 |
| Lymphoma type | None(10 of 10) | Low-grade B lymphoma(8 of 20) | Pre-B-ALL(12 of 20) | Pre-B-ALL(26 of 26) |
| . | CD19Cre/+ . | CD19Cre/+KrasG12D/+ . | CD19Cre/+Ink4a/Arf L/+ . | CD19Cre/+KrasG12D/+Ink4a/Arf L/+ . |
|---|---|---|---|---|
| Premalignant | ||||
| Number of mice | 3 | 3 | 3 | 3 |
| Spleen weight, mg | 127 ± 12 | 194 ± 6** | 173 ± 12** | 213 ± 6**†* |
| Spleen cells × 107 | 10.6 ± 0.6 | 14.0 ± 0.8** | 12.6 ± 1.2 (ns) | 16.5 ± 1.0**†* |
| Malignant | ||||
| No. of mice | 10 | 20 | 20 | 26 |
| Lymphoma type | None(10 of 10) | Low-grade B lymphoma(8 of 20) | Pre-B-ALL(12 of 20) | Pre-B-ALL(26 of 26) |
Engineered mice of the indicated genotypes were analyzed before overt malignancy (premalignant; 8 weeks of age) or following development of leukemia/lymphoma (malignant). For spleen weight and cell number, the standard deviation is provided.
ns, not significant.
P values were calculated relative to CD19Cre/+ or CD19Cre/+Ink4a/Arf L/+ (†) using Welch’s t test. *P ≤ .05; **P ≤ .01.
However, in agreement with the gross pathologic examination, an increase was noted by flow cytometry in splenic B220+ B cells of mice with the KrasG12D mutation compared with CD19Cre/+ and CD19Cre/+Ink4a/Arf L/+ mice (Figure 1D). Likewise, a significant increase in percentage of proliferating (Ki-67+) cells within spleen B-cell areas of CD19Cre/+KrasG12D/+, and CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice was observed in comparison with CD19Cre/+ controls and CD19Cre/+Ink4a/ArfL/+ mice (Figure 1E). TUNEL staining revealed fewer apoptotic cells within germinal center B-cell areas in CD19Cre/+KrasG12D/+, CD19Cre/+Ink4a/ArfL/+, and CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice compared with CD19Cre/+ controls (Figure 1F). Collectively, these results suggest that the KrasG12D mutation strongly enhances proliferation and reduces apoptosis of B cells, whereas Ink4a/Arf loss inhibits B-cell apoptosis and, to a certain degree, reduces KrasG12D-induced proliferation in vivo.
KrasG12D mutation induces transcriptional changes that prime cells for hypersensitivity to proliferative stimuli
To elucidate molecular mechanisms responsible for KrasG12D mutation and/or Ink4a/Arf inactivation–driven expansion of B cells, we performed microarray gene expression analysis in CD19+ B cells from 8-week-old engineered mice. The most differentially expressed genes (fold change >2; P < .01) between control, single-, and double-mutant mice are shown in the heat map (Figure 2A; supplemental Table 1). Next, to identify the biological processes underlying observed phenotypical changes, we performed gene-set enrichment analysis (GSEA) of expression data from mutant compared with control mice. Marked upregulation of gene signatures related to proliferation, Ras-related signaling pathways, apoptosis, rewired cellular metabolism, DNA repair, and pre-B stage of B-cell development were observed in both CD19Cre/+KrasG12D/+ and CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice (Figure 2B; supplemental Figure 3A-B). Consistent with our previous observations, the proliferative signature in CD19+ B cells from CD19Cre/+Ink4a/ArfL/+ mice was less pronounced; instead, upregulation of several signaling pathways related to independence from external growth signals and retention of stemness was noted (Figure 2B; supplemental Figure 3A,C). Interestingly, Ink4a/Arf inactivation suppressed, at least in part, a signature upregulated by Kras signaling.
Gene expression analysis of CD19+B cells from 8-week-old engineered mice. (A) Heat map of the most distinctive sets of differentially expressed genes across indicated genotypes. Values are row-normalized. Cyclin D2 (CCND2) is highlighted in red and validated in panels C and D (see supplemental Table 1). (B) Summary of changes in biological processes and pathways measured by GSEA in CD19+ B cells from mice of indicated genotypes compared with CD19Cre/+ control mice. Color scale represents –log10 false discovery rate (FDR) from GSEA; positive sign reflects correlation to mutant genotype, negative sign reflects correlation to CD19Cre/+ control (see supplemental Figure 3). (C-D) Cyclin D2 expression evaluated by (C) immunoblotting in CD19+ B cells and (D) IHC in the B-cell areas of spleens from mice of the indicated genotypes, calculated by using ImageJ. Bar plots present mean percentage of cells per 5 high-power fields; error bars ± SD. P values were calculated by using Welch’s t test. Representative images are shown below the graphs. Scale bar, 25 µm. (E) Impact of KrasG12D mutation and Ink4a/Arf loss on proliferative potential of CD19+ B cells from mice of the indicated genotype, evaluated by tritiated thymidine ([3H]dThd) incorporation. CD19+ B cells were isolated, stimulated with lipopolysaccharide (20 µg/mL) for 48 hours, and pulsed with [3H]dThd for the final 16 hours of growth. Incorporated [3H]dThd was measured by scintillation counting. Bars represent mean and error bars ± SD of triplicate cultures. P values were calculated by using Welch’s t test. Two independent assays were performed for all experiments for which statistics were calculated. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
Gene expression analysis of CD19+B cells from 8-week-old engineered mice. (A) Heat map of the most distinctive sets of differentially expressed genes across indicated genotypes. Values are row-normalized. Cyclin D2 (CCND2) is highlighted in red and validated in panels C and D (see supplemental Table 1). (B) Summary of changes in biological processes and pathways measured by GSEA in CD19+ B cells from mice of indicated genotypes compared with CD19Cre/+ control mice. Color scale represents –log10 false discovery rate (FDR) from GSEA; positive sign reflects correlation to mutant genotype, negative sign reflects correlation to CD19Cre/+ control (see supplemental Figure 3). (C-D) Cyclin D2 expression evaluated by (C) immunoblotting in CD19+ B cells and (D) IHC in the B-cell areas of spleens from mice of the indicated genotypes, calculated by using ImageJ. Bar plots present mean percentage of cells per 5 high-power fields; error bars ± SD. P values were calculated by using Welch’s t test. Representative images are shown below the graphs. Scale bar, 25 µm. (E) Impact of KrasG12D mutation and Ink4a/Arf loss on proliferative potential of CD19+ B cells from mice of the indicated genotype, evaluated by tritiated thymidine ([3H]dThd) incorporation. CD19+ B cells were isolated, stimulated with lipopolysaccharide (20 µg/mL) for 48 hours, and pulsed with [3H]dThd for the final 16 hours of growth. Incorporated [3H]dThd was measured by scintillation counting. Bars represent mean and error bars ± SD of triplicate cultures. P values were calculated by using Welch’s t test. Two independent assays were performed for all experiments for which statistics were calculated. *P ≤ .05, **P ≤ .01, ***P ≤ .001.
To validate one of the most compelling transcriptomic findings at the protein level, we examined both cyclin D2 (CCND2) expression and the proliferative potential of CD19+ B cells. Higher cyclin D2 expression in B cells from CD19Cre/+KrasG12D/+ and CD19Cre/+KrasG12D/+Ink4a/ArfL/+ compared with CD19Cre/+ and CD19Cre/+Ink4a/ArfL/+ mice was observed by immunoblotting and IHC (Figure 2C-D). Next, isolated CD19+ B cells from the animals’ spleens were treated with lipopolysaccharide and assessed for in vitro proliferative potential. Consistently, proliferation was highest in CD19+ B cells with KrasG12D mutation with or without loss of Ink4a/Arf (Figure 2E). These results indicate that KrasG12D and Ink4a/Arf mutations drive perturbations in B-cell signaling and proliferation and that Ink4a/Arf deletion restrains KrasG12D-induced changes.
CD19Cre/+KrasG12D/+ and CD19Cre/+Ink4a/Arf L/+ mice develop distinct subtypes of B-cell leukemia/lymphoma
Given the molecular and functional changes induced by mutant KrasG12D and loss of Ink4a/Arf, we evaluated cohorts of CD19Cre/+KrasG12D/+ and CD19Cre/+Ink4a/ArfL/+ mice over a span of 75 weeks, after which we euthanized any remaining animals and subjected them to thorough pathologic and histologic examination. Notably, during this aging period, 2 of a total of 20 CD19Cre/+KrasG12D/+ mice developed weight loss and hunched posture, requiring euthanasia (Figure 3A; Table 1). Upon histologic examination, only minimal distortion of LNs and spleen architecture was noted in both animals. In the lungs, however, a nodular lymphoid infiltrate composed predominantly of small atypical cells was observed (Figure 3B). All other tissues, including the BM and PB, seemed to be unaffected. The atypical lymphoid cells were predominantly composed of B220+BCL6–TdT– B cells with a low proliferative rate as evaluated by Ki-67 immunostaining and were varyingly infiltrated by CD3+CD5+ T cells and CD138+ plasma cells (Figure 3B). Southern blot analysis of the IgH gene locus showed a minor clonal band in relation to the corresponding band in the germ line (no clonal bands were observed for the TCR locus; Figure 3C), a finding consistent with a clonal low-grade B-cell lymphoproliferative disorder such as extranodal marginal zone lymphoma of mucosa-associated lymphoid tissue or lymphoplasmacytic lymphoma in humans.1 This phenotype was likewise noted in 6 of the 18 CD19Cre/+KrasG12D/+ mice that seemed to be disease free at 75 weeks (supplemental Figure 4). Moreover, WES analysis (supplemental Figure 6D) revealed the lack of any highly recurrent secondary loss-of-function or missense mutations in 6 analyzed lymphomas, underscoring the important role of Kras mutation in initiating this low-grade B-cell malignancy. All 10 of the CD19Cre/+ controls lacked this phenotype and were histologically unremarkable (Figure 3A; supplemental Figure 4).
Development of a clonal low-grade B-cell lymphoma and pre-B-ALL in engineered mice. (A) Kaplan-Meier survival plots illustrating overall survival of mouse cohorts according to genotype. P values were calculated by using the log-rank test. (B) Histologic and IHC stains of indicated markers in a representative example of low-grade B-cell lymphoma involving the lungs of a CD19Cre/+KrasG12D/+ mouse. Scale bar, 25 µm (see supplemental Figure 4). (C) Southern blot analysis for the immunoglobulin heavy chain (IgH) (left) and T-cell receptor (TCR) (right) genes in the same genomic DNA isolated from CD19Cre/+ LNs and CD19Cre/+KrasG12D/+ lymphomas. Clonal bands are present for the IgH gene (arrows) but not for TCR (see supplemental Figure 6D). (D) Representative gross pictures of enlarged LNs (white arrow) and spleen from CD19Cre/+Ink4a/ArfL/+ mouse with pre-B-ALL. Scale bar, 1 cm. (E) Histologic, IHC, and immunofluorescent stains of indicated markers in LNs and BM from representative CD19Cre/+Ink4a/ArfL/+ mouse with B-ALL. CD19 staining shown in green with blue 4′,6-diamidino-2-phenylindole (DAPI) nuclear counterstain. Note cytoplasmic expression of IgM in B-ALL cells. TBG, tingible body macrophage. Scale bars: white, 25 µm; black, 50 µm (see supplemental Figure 5). (F) Southern blot analysis for the IgH (left) and TCR (right) genes in the same genomic DNA isolated from CD19Cre/+Ink4a/ArfL/+ leukemias. Clonal bands are present for the IgH gene (arrows) but not for TCR. GL, germ line band.
Development of a clonal low-grade B-cell lymphoma and pre-B-ALL in engineered mice. (A) Kaplan-Meier survival plots illustrating overall survival of mouse cohorts according to genotype. P values were calculated by using the log-rank test. (B) Histologic and IHC stains of indicated markers in a representative example of low-grade B-cell lymphoma involving the lungs of a CD19Cre/+KrasG12D/+ mouse. Scale bar, 25 µm (see supplemental Figure 4). (C) Southern blot analysis for the immunoglobulin heavy chain (IgH) (left) and T-cell receptor (TCR) (right) genes in the same genomic DNA isolated from CD19Cre/+ LNs and CD19Cre/+KrasG12D/+ lymphomas. Clonal bands are present for the IgH gene (arrows) but not for TCR (see supplemental Figure 6D). (D) Representative gross pictures of enlarged LNs (white arrow) and spleen from CD19Cre/+Ink4a/ArfL/+ mouse with pre-B-ALL. Scale bar, 1 cm. (E) Histologic, IHC, and immunofluorescent stains of indicated markers in LNs and BM from representative CD19Cre/+Ink4a/ArfL/+ mouse with B-ALL. CD19 staining shown in green with blue 4′,6-diamidino-2-phenylindole (DAPI) nuclear counterstain. Note cytoplasmic expression of IgM in B-ALL cells. TBG, tingible body macrophage. Scale bars: white, 25 µm; black, 50 µm (see supplemental Figure 5). (F) Southern blot analysis for the IgH (left) and TCR (right) genes in the same genomic DNA isolated from CD19Cre/+Ink4a/ArfL/+ leukemias. Clonal bands are present for the IgH gene (arrows) but not for TCR. GL, germ line band.
During the same 75-week period, 4 of 20 CD19Cre/+Ink4a/ArfL/+ mice developed more aggressive disease characterized by weight loss, hunched posture, and lymphadenopathy (Figure 3A; Table 1). Enlarged LNs and splenomegaly were observed at necropsy in all affected mice (Figure 3D). Histologic examination revealed massive distortion of LNs and spleen evidenced by a diffuse monotonous population of blasts (Figure 3E; supplemental Figure 5). A starry-sky pattern with frequent tingible body macrophages and mitotic figures was observed (Figure 3E). Atypical cells were predominantly of the B220+CD19+TdT+CD3–CD5–CD10+cyt-IgM+ phenotype and had a very high proliferation rate according to Ki-67 immunostaining (Figure 3E), all of which were consistent with a high-grade B-cell lymphoproliferative disorder such as precursor B-ALL.1 The atypical lymphoid cells diffusely infiltrated other organs, including the BM, PB, liver, skin, lungs, intestine, and peridural space (Figure 3E; supplemental Figure 5; data not shown). Clonal bands for the IgH gene were detected (Figure 3F). Precursor B-ALL was also observed in 8 of the remaining 16 CD19Cre/+Ink4a/ArfL/+ mice that seemed to be disease free according to visual examination at 75 weeks of age, but whose disease was evident at necropsy (supplemental Figure 5). In sum, although KrasG12D mutation induced significant transcriptional and functional changes in B cells, it gave rise to only a low-grade mature B-cell malignancy in our model, in contrast with Ink4a/Arf loss, which drives the high-grade immature B-cell malignant phenotype.
Kras activation and Ink4a/Arf loss cooperate in B-ALL development
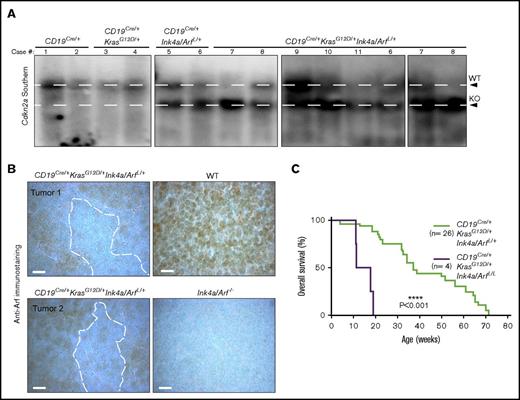
In contrast to the relatively low penetrance and long latency observed in single-mutant mice, significantly faster tumor development and higher penetrance were seen in CD19Cre/+KrasG12D/+Ink4a/ArfL/+ animals, with each of 26 mice developing cancer within 75 weeks (Figure 3A; Table 1). Animals seemed ill with hunched posture, weight loss, anemia, hind-limb paralysis, and adenopathy (Figure 4A). Massively enlarged LNs and spleens, in addition to pale long bones of the legs and white nodules in the livers and spleens, were observed at necropsy (Figure 4A; data not shown). Histologic examination revealed that most tissues were infiltrated with B220+CD19+TdT+CD3–CD5–CD10+ lymphoblasts with a proliferative index similar to that seen in CD19Cre/+Ink4a/ArfL/+ mice (Figure 4B; supplemental Figure 5). One or 2 major bands for the IgH gene, but not for the TCR gene, were detected in most samples by Southern blot analysis, indicating that they were monoclonal or biclonal B-cell neoplasms with immunophenotypes of precursor B-ALL (Figure 4C).
Development of a fully penetrant, highly aggressive B-ALL in KrasG12D/+and Ink4a/Arf double-mutant mice. (A) Representative images of enlarged LNs (middle, white arrows) and spleen (right) from a CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mouse with pre-B-ALL. Representative whole-body photograph of CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mouse with pre-B-ALL (left). White arrow denotes cervical lymphadenopathy; black arrow denotes lagging hind-limb paralysis due to peridural space involvement. Scale bar, 1 cm. (B) Histologic, IHC, and immunofluorescent stains of indicated markers in LN and BM from a representative CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mouse with B-ALL. CD19 staining is shown in green with blue DAPI nuclear counterstain. Note cytoplasmic expression of IgM in B-ALL cells and infiltration of B-ALL cells in the BM and peridural space (PDS) contributing to paralysis (bottom H&E). Scale bars: white, 25 µm; black, 50 µm (see supplemental Figure 5). (C) Southern blot analysis for IgH (left) and TCR (right) genes in the same genomic DNA isolated from CD19Cre/+KrasG12D/+Ink4a/ArfL/+ leukemias. Clonal bands are present for the IgH gene (arrows) but not for TCR. Absence of GL band in some tumor DNA samples indicates almost complete replacement with lymphoma cells.
Development of a fully penetrant, highly aggressive B-ALL in KrasG12D/+and Ink4a/Arf double-mutant mice. (A) Representative images of enlarged LNs (middle, white arrows) and spleen (right) from a CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mouse with pre-B-ALL. Representative whole-body photograph of CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mouse with pre-B-ALL (left). White arrow denotes cervical lymphadenopathy; black arrow denotes lagging hind-limb paralysis due to peridural space involvement. Scale bar, 1 cm. (B) Histologic, IHC, and immunofluorescent stains of indicated markers in LN and BM from a representative CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mouse with B-ALL. CD19 staining is shown in green with blue DAPI nuclear counterstain. Note cytoplasmic expression of IgM in B-ALL cells and infiltration of B-ALL cells in the BM and peridural space (PDS) contributing to paralysis (bottom H&E). Scale bars: white, 25 µm; black, 50 µm (see supplemental Figure 5). (C) Southern blot analysis for IgH (left) and TCR (right) genes in the same genomic DNA isolated from CD19Cre/+KrasG12D/+Ink4a/ArfL/+ leukemias. Clonal bands are present for the IgH gene (arrows) but not for TCR. Absence of GL band in some tumor DNA samples indicates almost complete replacement with lymphoma cells.
Oncogenic Ras signaling is constrained in some tissue types by Ink4a/Arf tumor suppressors.23,30,37 Therefore, we suspected that selective pressure might cause loss of heterozygosity (LOH) at the wild-type (WT) Ink4a/Arf locus during B-ALL development. Southern blot analysis of this locus revealed preservation of the WT allele in every low-grade B-cell lymphoma in the CD19Cre/+KrasG12D mice; however, 4 of 8 precursor B-ALL tumors in CD19Cre/+Ink4a/ArfL/+ mice showed only a faint WT band indicative of LOH (Figure 5A). The residual faint band is most likely the result of contaminating WT hematopoietic and/or stromal cells as evidenced by positive anti-Arf immunostaining (Figure 5B). Moreover, 9 of 10 CD19Cre/+KrasG12D/+Ink4a/ArfL/+ precursor B-ALL tumors had Ink4a/Arf LOH (Figure 5A). Given these intriguing results, we generated a small (n = 4) cohort of CD19Cre/+KrasG12D/+Ink4a/ArfL/L mice with homozygous inactivation of Ink4a/Arf and found that these animals succumbed within the first 20 weeks of life with a rapidly progressive form of pre-B-ALL (Figure 5C). Collectively, these data indicate that activated Ras signaling promotes the pathogenesis of the pre-B-ALL, but is restricted by Ink4a/Arf tumor suppressor control of the cell cycle, and that simultaneous Kras activation and Ink4a/Arf loss cooperate during precursor B-ALL development.
Loss of heterozygosity of the Ink4a/Arf (Cdkn2a) locus in pre-B-ALL. (A) Southern blot analysis of the Ink4a/Arf locus. WT, wild-type fragment of the Ink4a/Arf locus; KO, knockout configuration of the Ink4a/Arf locus. Loss of the upper WT band is indicative of loss of heterozygosity (LOH) of the Ink4a/Arf locus (see supplemental Figure 6). (B) Anti-Arf immunostains in the LNs of mice of the indicated genotypes. The specificity of Anti-Arf antibody was confirmed in LNs from Ink4a/Arf−/− mice. Scale bar, 10 µm. (C) Kaplan-Meier survival plot illustrating the impact of homozygous Ink4a/Arf deletion on survival of mice with KrasG12D mutation in the B-cell compartment. P values were calculated by using the log-rank test.
Loss of heterozygosity of the Ink4a/Arf (Cdkn2a) locus in pre-B-ALL. (A) Southern blot analysis of the Ink4a/Arf locus. WT, wild-type fragment of the Ink4a/Arf locus; KO, knockout configuration of the Ink4a/Arf locus. Loss of the upper WT band is indicative of loss of heterozygosity (LOH) of the Ink4a/Arf locus (see supplemental Figure 6). (B) Anti-Arf immunostains in the LNs of mice of the indicated genotypes. The specificity of Anti-Arf antibody was confirmed in LNs from Ink4a/Arf−/− mice. Scale bar, 10 µm. (C) Kaplan-Meier survival plot illustrating the impact of homozygous Ink4a/Arf deletion on survival of mice with KrasG12D mutation in the B-cell compartment. P values were calculated by using the log-rank test.
The next issue that had to be addressed was whether KrasG12D mutation and Ink4a/Arf inactivation are sufficient to promote leukemogenesis, or whether other cooperating genetic lesions are required. Accordingly, aCGH and WES were performed on tumors from 10 CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice to analyze them for secondary chromosomal copy number variations and mutations (supplemental Figure 6A-D). No highly recurrent secondary loss-of-function or missense mutations were identified; however, several common copy number variations were observed among 10 tumors in our engineered mice. One of the most recurrent alterations (60%) was gain of chromosome 15 (harboring the Myc oncogene)38 ; therefore, it cannot be excluded that other cooperative lesions in the genome might accelerate the onset of leukemia. Nevertheless, it was found that lymphoblasts from CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice lacked morphologic features typically associated with Myc gene translocation1 and did not exhibit increased overexpression of Myc protein (supplemental Figure 6C). Moreover, the transcriptional signatures of our murine B-ALL tumors strongly resembled those of B cells from young CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice (supplemental Figure 7; supplemental Table 1), suggesting that KrasG12D mutation and Ink4a/Arf loss profoundly influenced transcriptional profiles in our pre-B-ALL model. Taken together, the data supported the hypothesis that loss of the functional allele at the Ink4a/Arf locus is rate-limiting in precursor B-ALL tumorigenesis and is accelerated by oncogenic Ras signaling in this model.
B-ALL tumors arising in CD19Cre/+KrasG12D/+Ink4a/Arf L/+ mice most closely resemble human BCR-ABL subtype
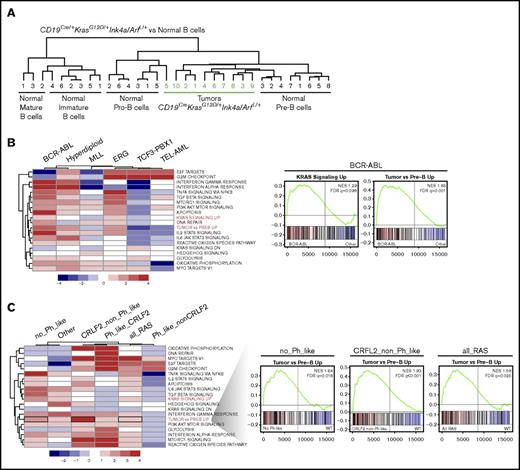
Immunohistochemical characterization of B-ALL tumors in CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice indicated that the cells were arrested at early stages of development. To further evaluate this finding, gene expression profiling was performed in 10 randomly selected CD19Cre/+KrasG12D/+Ink4a/ArfL/+ B-ALL tumors. The resulting transcriptional profiles were then compared with a reference data set from normal mouse lymphocyte populations that included purified pro-B, pre-B, immature B, and mature B cells.39 Unsupervised hierarchical clustering analysis demonstrated that 9 of 10 CD19Cre/+KrasG12D/+Ink4a/ArfL/+ B-ALL tumors were most similar to normal pre-B cells, with the tenth one more closely resembling normal pro-B cells than immature or mature B cells (Figure 6A). The findings were consistent with the pattern of Cre expression in CD19Cre/+ mice26 as well as with the gene expression profile of premalignant CD19+ B cells from CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice (supplemental Figure 3B) and suggest that KrasG12D mutation together with Ink4a/Arf deletion arrests B-cell development at the pre-B-cell stage during neoplastic transformation.
Pre-B-ALL arising in CD19Cre/+KrasG12D/+Ink4a/ArfL/+mice most closely resembles human BCR-ABL B-ALL. (A) Unsupervised hierarchical clustering of gene expression microarray profiles of B-ALL cells from CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice and purified populations of normal mouse B cells at different stages of differentiation. (B) GSEA summary in indicated human B-ALL subtypes, each compared with all other samples. Color scale represents –log10 FDR from GSEA; positive sign reflects correlation to indicated subtype, negative to all other samples (left). Mountain plots illustrating upregulation of KRAS and our murine tumor signature in human B-ALL cells harboring BCR-ABL translocation (right). (C) GSEA summary in indicated human B-ALL subtypes with RAS mutations as indicated compared with samples with WT RAS. Samples from all RAS mutations combined (all_Ras), excluding patients classified as Ph-like (no_Phlike), only patients harboring CRFL2 rearrangements with (Ph_like_CRLF2) or without Ph-like signature (CRLF2_non_Ph_like), or only patients classified as Ph-like without CRFL2 rearrangements (Ph_like_nonCRLF2). Color scale represents –log10 FDR from GSEA; positive sign reflects correlation to samples with the indicated RAS mutations, negative to those with WT RAS (left). Mountain plots illustrating upregulation of murine tumor signature in human B-ALL cells within indicated group with KRAS mutations (right).
Pre-B-ALL arising in CD19Cre/+KrasG12D/+Ink4a/ArfL/+mice most closely resembles human BCR-ABL B-ALL. (A) Unsupervised hierarchical clustering of gene expression microarray profiles of B-ALL cells from CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice and purified populations of normal mouse B cells at different stages of differentiation. (B) GSEA summary in indicated human B-ALL subtypes, each compared with all other samples. Color scale represents –log10 FDR from GSEA; positive sign reflects correlation to indicated subtype, negative to all other samples (left). Mountain plots illustrating upregulation of KRAS and our murine tumor signature in human B-ALL cells harboring BCR-ABL translocation (right). (C) GSEA summary in indicated human B-ALL subtypes with RAS mutations as indicated compared with samples with WT RAS. Samples from all RAS mutations combined (all_Ras), excluding patients classified as Ph-like (no_Phlike), only patients harboring CRFL2 rearrangements with (Ph_like_CRLF2) or without Ph-like signature (CRLF2_non_Ph_like), or only patients classified as Ph-like without CRFL2 rearrangements (Ph_like_nonCRLF2). Color scale represents –log10 FDR from GSEA; positive sign reflects correlation to samples with the indicated RAS mutations, negative to those with WT RAS (left). Mountain plots illustrating upregulation of murine tumor signature in human B-ALL cells within indicated group with KRAS mutations (right).
The utility of genetically engineered mouse models of cancer can sometimes be limited by clinico-pathologic incongruence with their human counterparts. It was surmised, however, that the cellular characteristics and aggressive phenotype of B-ALL in CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice would resemble a poor-prognosis form of human B-ALL. To investigate this possibility, the most differentially expressed genes in murine CD19Cre/+KrasG12D/+Ink4a/ArfL/+ B-ALL were identified and compared with normal mouse pre-B cells.39 Next, the enrichment of our tumor signature was assessed by GSEA, along with other hallmark signatures in human B-ALL, classified according to BCR-ABL, hyperdiploid, TEL-AML, TCF3-PBX1, and MLL subgroups.40,41 Genes upregulated in CD19Cre/+KrasG12D/+Ink4a/ArfL/+ B-ALL (labeled “tumor vs Pre-B up”) as well as the hallmark Kras (up- but not downregulated) and other signaling signatures were most significantly enriched in the BCR-ABL subtype of human B-ALL (Figure 6B; supplemental Figure 7), which has historically been associated with poor outcome.42 The gene-set profiles of the hyperdiploid human B-ALL also correlated with the murine tumor signature, although to a lesser degree and without any enrichment of the Kras signatures.
To minimize the possibility that the resemblance of our mouse model to human leukemia results from the convergence of Ras-activated signaling pathways with pro-proliferative activated kinase and/or cytokine receptor signals associated with BCR-ABL/Philadephia chromosome–like (Ph-like) as well as CRLF2 alterations, GSEA was next performed in human B-ALL tumors with known RAS mutational status,43,44 and in relation to the Ph-like and CRLF2 subtypes. Importantly, significant upregulation of the CD19Cre/+KrasG12D/+Ink4a/ArfL/+ B-ALL transcriptional signature was observed in leukemic blasts with RAS mutations compared with WT RAS from all B-ALL patients (Figure 6C; all_RAS, black box) regardless of Ph-like expression profile (Figure 6C; no_Ph_like, black box). Moreover, the murine tumor signature was most highly enriched in human blasts with CRLF2 rearrangements but lacking the Ph-like signature compared with other categories and with RAS mutations compared with WT RAS (Figure 6C; CRLF2_non_Ph_like, black box). It is worth mentioning that CRLF2 rearrangements are also associated with poor clinical outcome.16 The cross-species analyses consistently demonstrated that B-ALL tumors in the CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mice recapitulate gene expression programs in poor-prognosis subgroups of human B-ALL that harbor BCR-ABL and CRLF2 rearrangements. Furthermore, these observations suggest that RAS mutations have a high impact on B-ALL blast transcriptional profiles and that our engineered mouse models, at least to a certain degree, recapitulate the alterations observed in human B-ALL.
Discussion
In this study, we demonstrate that somatic activation of 1 KrasG12D/+ allele and concurrent heterozygous deletion of Ink4a/Arf during early stages of B-cell development alter transcriptional profiles of CD19+ B cells and result in enhanced proliferation and decreased apoptosis. As a consequence, 40% of CD19Cre/+KrasG12D/+ mice developed a clonal low-grade B-cell lymphoproliferative disorder, whereas 60% of CD19Cre/+Ink4a/ArfL/+ mice developed a more aggressive precursor B-ALL phenotype. Concurrent KrasG12D/+ mutation and Ink4a/Arf deletion in CD19+ B cells cooperatively induced a fully penetrant, highly aggressive B-ALL phenotype that resembled BCR-ABL and CRLF2 subtypes of human B-ALL.
The high prevalence of activating RAS pathway mutations (∼35%), either direct or through other means such as BCR-ABL and CRLF2 chromosomal translocations in newly diagnosed and relapsed cases of human B-ALL indicates its importance in the pathogenesis of this form of leukemia.9,11-19,45 However, the exact role of RAS activation in B-ALL is still debatable. Previous studies show that RAS mutations can be initiating as well as secondary events.9 Our findings presented in this article demonstrate that, although Ras activation as the initiating event may not be sufficient by itself to induce high-grade leukemia/lymphoma, it can in fact induce dramatic transcriptional changes that prime cells to become hypersensitive to proliferative stimuli, as well as leading to the development of clonal low-grade B-cell lymphoproliferative disorder. However, because Kras mutations have not been reported in human MALT and LPL,46,47 the possible role of such mutations in the development of these types of low-grade B-cell lymphoproliferative disorders needs to be further investigated.
Interestingly, RAS activation has been implicated in leukemogenesis in the myeloid compartment, promoting the development of indolent chronic myeloid leukemia (CML)48 which in early stages before blast transformation, exhibits no alterations in the INK4A/ARF locus.49 The transition of Ph+ CML to lymphoid blast crisis resembling pre-B-ALL is often accompanied by biallelic INK4A/ARF deletion.50 Similarly, our finding that concerted KrasG12D mutation and Ink4a/Arf deletion induce LOH at the WT Ink4a/Arf locus in mice without acquisition of universal secondary lesions before tumor development confirms that Ink4a/Arf is an important tumor suppressor of oncogenic Ras signaling in this in vivo model. Consistent with our findings, Arf inactivation in pre-B cells has been shown to augment BCR-ABL–induced development of B-ALL in transplantation models.51,52 Although Arf was shown to be more important than Ink4a for B-ALL development,51,52 we must presume that deletion of both proteins contributes to our phenotype because mice with Ink4a deletion were found to likewise develop high-grade B-cell malignancy.53
Acute oncogene activation is known to induce cellular stress resulting in senescence or apoptosis.54 Kras-induced oncogenic stress in low-grade leukemia/lymphoma may not suffice by itself to activate cell-cycle checkpoints or might be accompanied by additional cooperating events (as in CML48 ) whose identification would necessitate larger cohorts of animals than the one used in this study. Nevertheless, loss of Ink4a/Arf (2 powerful tumor suppressors) presumably removes the cell-cycle control mechanisms restricting KrasG12D-induced proliferation23,49 and thus enhances B-ALL development. Our results suggest that additional regulatory mechanisms may well be at play. For instance, deletion of Ink4a/Arf in CD19+ B cells with KrasG12D mutation not only suppresses oncogenic stress-related signatures such as DNA replication or oxidative stress55 but also inhibits KrasG12D-induced transcriptional profiles almost globally and reduces the number of proliferating cells. In line with these findings, it has been shown that activation of oncogenes such as BCR-ABL in pre-B cells induces apoptosis. Importantly, rare surviving cells acquired mutations in genes that negatively regulate ERK kinase activity, a downstream target of RAS/MAPK pathway,56 which could point to attenuation of RAS activity as a necessary condition for induction of leukemogenesis, and which would be important to address when designing rational therapeutic strategies. Taken together, although these genomic changes may have different and in some way opposite effects, they nonetheless cooperate in the induction of B-ALL in vivo.
Although murine models of B-ALL already exist in the literature,52,57-61 including transplantation models using BCR-ABL and mutated CRLF2 proteins as well as Ink4a/Arf knockouts, we believe that additional models of this disease are needed to more comprehensively encompass the molecular heterogeneity of B-ALL and to facilitate the study of oncogenic pathways potentially amenable to therapeutic intervention. Previous transgenic experimental models have probed the effects of Ras and/or Ink4a/Arf germ line alterations during hematopoiesis and have evaluated their potential for lymphomagenesis.28,62 By using CD19-driven Cre recombinase, genomic alterations may be kept to the B-cell compartment, akin to somatic mutations in the human condition. This does not affect the cells within the BM microenvironment and other hematopoietic elements which likely play an important role in leukemogenesis.28,62 Few studies have yet created a murine model with which to predictively test new therapies specifically geared toward poor-prognosis and relapsed B-ALL.52 Genome-wide expression analysis of human B-ALL at diagnosis/relapse has demonstrated the presence of significant alterations in multiple processes, such as cell-cycle regulation, proliferation, metabolism, DNA repair, signaling, and apoptosis63,64 that are recapitulated in our CD19Cre/+KrasG12D/+Ink4a/ArfL/+ mouse model. The short latency, full penetrance, activation of signaling pathways, and CD19 expression of our CD19Cre/+KrasG12D/+Ink4a/ArfL/+ tumors in mice mimic the human setting and should facilitate future studies of kinase inhibitors as well as anti-CD19 chimeric antigen receptor T-cell therapies for pre-B-ALL.65 We believe that our model represents an attractive alternative to existing syngeneic and xenograft models in terms of reflecting the human setting.
The clinical relevance of RAS and INK4A/ARF alterations at presentation are not conclusive and could be dependent on the B-ALL subtype.41,66,67 However, the high incidence of their acquisition by leukemic cells during relapse suggests important roles in chemotherapy resistance and disease progression in humans.9-13,19,68-71 Furthermore, the functional cooperation and the negative impact on survival of concurrent Ras mutation and Ink/Arf deletion in mice suggest that these changes may also have a functional impact in human B-ALL and should be considered in the course of disease classification, as well as for prognosis and choice of therapy in human subjects.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank Ronald DePinho for providing KrasG12D/+ and Ink4a/ArfL/+ mice and Klaus Rajewsky for providing the CD19Cre mice.
This work was supported by National Institutes of Health (NIH), National Cancer Institute National Research Service Award F30 CA165857 (K.S.), by the Case Western Reserve Medical Scientist Training Program, by an NIH National Institute of General Medical Sciences grant (T32 GM007250), by a senior award from the Multiple Myeloma Research Foundation (R.D.C.), by Doctor’s Cancer Foundation, and by NIH National Cancer Institute grants 1R01CA151391-01 and 1R01CA196783-01.
Authorship
Contribution: K.S., T.S., J.N.R., D.M.W., N.E.S., and R.D.C. designed the research; K.S., M.J., T.S., Y.K., A.A., R.L., Z.S., P.S.D., J.X., and M.E.L. performed the research; J.N.R., G.S.P., K.R., and C.G.M. contributed vital new agents or analytical tools; T.S., K.S., M.J., S.S.P., B.L., Y.Z., M.E.L., N.E.S., and R.D.C. analyzed data; and T.S., K.S., S.S.P., M.E.L., D.M.W., N.E.S., and R.D.C. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruben D. Carrasco, Dana-Farber Cancer Institute, 450 Brookline Ave, JF215H, Boston, MA 02215; e-mail: ruben_carrasco@dfci.harvard.edu.

![Figure 2. Gene expression analysis of CD19+ B cells from 8-week-old engineered mice. (A) Heat map of the most distinctive sets of differentially expressed genes across indicated genotypes. Values are row-normalized. Cyclin D2 (CCND2) is highlighted in red and validated in panels C and D (see supplemental Table 1). (B) Summary of changes in biological processes and pathways measured by GSEA in CD19+ B cells from mice of indicated genotypes compared with CD19Cre/+ control mice. Color scale represents –log10 false discovery rate (FDR) from GSEA; positive sign reflects correlation to mutant genotype, negative sign reflects correlation to CD19Cre/+ control (see supplemental Figure 3). (C-D) Cyclin D2 expression evaluated by (C) immunoblotting in CD19+ B cells and (D) IHC in the B-cell areas of spleens from mice of the indicated genotypes, calculated by using ImageJ. Bar plots present mean percentage of cells per 5 high-power fields; error bars ± SD. P values were calculated by using Welch’s t test. Representative images are shown below the graphs. Scale bar, 25 µm. (E) Impact of KrasG12D mutation and Ink4a/Arf loss on proliferative potential of CD19+ B cells from mice of the indicated genotype, evaluated by tritiated thymidine ([3H]dThd) incorporation. CD19+ B cells were isolated, stimulated with lipopolysaccharide (20 µg/mL) for 48 hours, and pulsed with [3H]dThd for the final 16 hours of growth. Incorporated [3H]dThd was measured by scintillation counting. Bars represent mean and error bars ± SD of triplicate cultures. P values were calculated by using Welch’s t test. Two independent assays were performed for all experiments for which statistics were calculated. *P ≤ .05, **P ≤ .01, ***P ≤ .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/1/25/10.1182_bloodadvances.2017012211/3/m_advances012211f2.jpeg?Expires=1769091575&Signature=4VYD32-kBQzOieT72dXi6Opx5kFs~sH-epLj42ISmAdFzjbd8b6ZEVGc2CoM5ERDePeWPkq-DMJmcU2gKOIW3UzF2R~t3lmbnVSmFIPGc7ZskJNHzsW9CXJfa7Z65V-4wSCtE~W6iqrH4E2GadYZCs9~kSoT0DvmVP6HVgEVqp7oTTVE9ggweAVpA2qR3mknQSARjg3gMf1Y4NGI-7q2mux7K~D5XahtOvVBR7XTGJJsHb8GM6lCCkOQU53SoimEJ4MWPsGmwEoT2gbIBZAkiNqWk8kyRLFWlrzo432K38kP8AlyUhbo8J2Mvk6J3xhz9E1eiofbMarqt2IZZ7qk2g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)