Key Points
Buccal epithelial cells harbor an MPN-associated CALR mutation in a patient with CALR-mutant essential thrombocytosis, Ph+ CML, and no germ line CALR mutation.
Introduction
CALR mutations are present in 70% to 84% of JAK2 wild-type myeloproliferative neoplasms (MPNs).1,2 With rare exceptions, CALR mutations are mutually exclusive with JAK2 or MPL mutations and have rarely been reported in conjunction with a BCR-ABL1 translocation.3-6 Most cases of MPNs are sporadic, but 7% to 11% of cases have evidence of familial predisposition.7-9 Of these MPN families, approximately 60% have a single MPN phenotype in all affected kindreds, but the remaining 40% present with a variety of phenotypes. In such families, the propensity to develop MPNs, not the MPN-associated mutation, is hereditary. Accordingly, several MPN-predisposition alleles have been described.10-16 Different MPN-associated alterations can be found within the same family, including BCR-ABL1 and mutations in JAK2 and CALR.10,17 In familial MPNs, all CALR mutations identified have been somatic.18 Germ line mutations in CALR have not been described in MPNs but have been described in 0.6% of healthy patients without MPNs.19
The appropriate tissue to use for germ line studies in hematological malignancies is controversial because leukocytes infiltrate many tissues.20,21 Cultured fibroblasts, derived from skin biopsies, are the gold-standard germ line material10,20 but are costly and invasive to obtain. Hair follicles and nails clippings are readily available and generally free of leukocytes, but adequate DNA is difficult to obtain from these tissues. Many investigators use buccal swabs and washes for germ line tissue because these are easy to obtain and provide adequate DNA.19,22,23
Case description
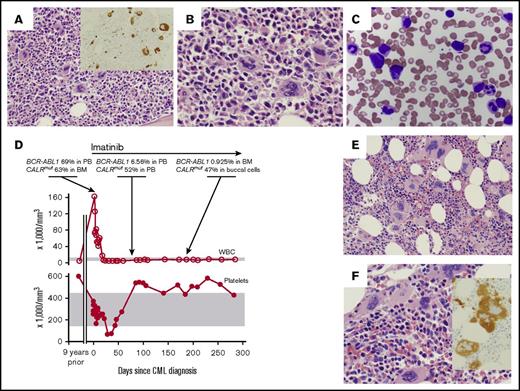
A 67-year-old woman with no significant medical history presented with abdominal pain. Peripheral blood (PB) revealed a marked leukocytosis, with a white blood cell (WBC) count of 160 × 109/L: 66% neutrophils, 16% myelocytes, 6.5% monocytes, 3.5% basophils, 2.5% promyelocytes, 2.5% metamyelocytes, 1.5% lymphocytes, and 1.5% blasts. She was nonanemic (hemoglobin [Hb], 12.7 g/dL) and had a normal platelet count (340 × 109/L). Bone marrow (BM) biopsy revealed a hypercellular marrow (>90%) with myeloid-predominant trilineage hematopoiesis and 1% to 2% blasts. (Figure 1A-C). A subset of the megakaryocytes had increased nuclear-to-cytoplasmic ratios with hyperchromatic nuclei and irregularly shaped nuclear contours, which are not typically seen in CML but are common in essential thrombocythemia and primary myelofibrosis. Reticulin fibrosis was mildly increased (MF-1 of 3) by reticulin stain. Megakaryocytes appeared slightly increased in number, as can be seen in CML,24 and were rarely clustered. Fluorescence in situ hybridization of PB identified a BCR-ABL1 fusion in 98.5% of interphase cells, confirming a diagnosis of CML.
Chronic myeloid leukemia (CML) therapy unmasks effects of coexistent CALR mutation. (A-B) BM biopsy obtained at the time of CML diagnosis. Representative hematoxylin and eosin–stained sections from the core biopsy are shown at ×20 (A) and ×40 magnification (B). (A) Inset shows CD61 immunohistochemical stain highlighting megakaryocytes at ×20 magnification. (C) Wright Giemsa–stained PB smear obtained at time of CML diagnosis. Original magnification ×50. (D) BCR-ABL1 fusion transcript and CALR-mutant allele frequency are quantitated in relationship to blood parameters as a function of time. Normal range of blood parameters is indicated by gray bars. (E-F) BM biopsy obtained at 6 months after initiation of imatinib therapy. Representative hematoxylin and eosin–stained sections from the core biopsy are shown at ×20 (E) and ×40 magnification (F). (F) Inset shows CD61 immunohistochemical stain highlighting megakaryocytes at ×20 magnification.
Chronic myeloid leukemia (CML) therapy unmasks effects of coexistent CALR mutation. (A-B) BM biopsy obtained at the time of CML diagnosis. Representative hematoxylin and eosin–stained sections from the core biopsy are shown at ×20 (A) and ×40 magnification (B). (A) Inset shows CD61 immunohistochemical stain highlighting megakaryocytes at ×20 magnification. (C) Wright Giemsa–stained PB smear obtained at time of CML diagnosis. Original magnification ×50. (D) BCR-ABL1 fusion transcript and CALR-mutant allele frequency are quantitated in relationship to blood parameters as a function of time. Normal range of blood parameters is indicated by gray bars. (E-F) BM biopsy obtained at 6 months after initiation of imatinib therapy. Representative hematoxylin and eosin–stained sections from the core biopsy are shown at ×20 (E) and ×40 magnification (F). (F) Inset shows CD61 immunohistochemical stain highlighting megakaryocytes at ×20 magnification.
She started treatment with imatinib and achieved a hematological remission within 2.5 weeks. Ninety days later, polymerase chain reaction showed 6.56% BCR-ABL1/ABL1 in the PB (a reduction from 69.7% at diagnosis; Figure 1D). However, at 90 days, her platelet count was elevated at 539 ×109/L, with normal WBC, Hb, and differential values. She had had a single complete blood count performed 9 years before CML presentation, with a platelet count of 600 ×109/L and normal Hb, WBC, and differential values. No additional historic complete blood count was available. Thrombocytosis persisted for the subsequent follow-up period of >2 years, with a maximal platelet count of 584 × 109/L. A BM biopsy obtained 6 months after starting imatinib treatment showed resolution of the typical CML morphology in the myeloid lineage but persistent megakaryocytosis (Figure 1E-F). In addition, this biopsy showed more pronounced hyperchromatic and irregularly shaped megakaryocytes, as seen in non-CML MPNs.
Methods
Next-generation (NGS) and Sanger sequencing were performed. These studies were performed in accordance with the Declaration of Helsinki and with approval of the University of Minnesota institutional review board.
Results and discussion
Given her thrombocytosis in the setting of appropriate molecular response to imatinib, a PB sample obtained after 90 days of imatinib underwent NGS of JAK2, MPL, and CALR genes. A 52-bp out-of-frame deletion in exon 9 of the CALR gene (NM_004343.3, p.Leu367fs*46) was detected, with a variant allele fraction (VAF) of 52% (Figures 1D and 2A). Material from her BM biopsy obtained at the time of CML diagnosis revealed the same 52-bp CALR deletion at a VAF of 63% (Figures 1D and 2A-B). A buccal specimen obtained 6 months after starting imatinib (day 174), when the BCR-ABL1 transcript was 1% in the PB, harbored the CALR mutation at an allele frequency of 52% (Figures 1D and 2A,C). Cytospin and cell pellet preparations of her buccal material showed large cells with small, condensed nuclei and abundant cytoplasm, consistent with mature cells of epithelial origin (Figure 2D-G), without significant numbers of lymphocytes or macrophages. Accordingly, immunostaining was diffuse and strongly positive for cytokeratin AE1/AE3 and negative for CD45 (a panleukocyte marker) and OCT4 (an undifferentiated cell marker; Figure 2F-G; supplemental Figure 1A). SSE-A (CD15; found on multiple cell types including squamous epithelium and granulocytes) was weak to moderate in approximately half of the cells (supplemental Figure 1B). These findings indicate that the buccal cells were mature and epithelial without significant leukocyte contamination. A second buccal specimen, collected at day 194, showed the same CALR mutation at a VAF of 29%. NGS analysis of her day-194 BM specimen detected the CALR mutation at a VAF of 56%.
A somatic 52-bp deletion in the CALR gene is detected in buccal epithelial cells. (A) Table summarizing the detection of CALR mutation by NGS and Sanger sequencing in various tissues. (B-C) NGS of diagnostic BM (B) and buccal cells (C) visualized through Integrated Genome Viewer. Sequence reads are aligned to the CALR locus. Gray bars represent read alignments. White gaps in alignment represent areas of deletion. (D) Cytospin preparation of buccal cells stained with Diff-Quick (Romanowski) stain. (E-G) Paraffin-embedded buccal cell block stained with hematoxylin and eosin (E), cytokeratin AE1/AE3 (F), and CD45 (G). Original magnification ×40 in panels D-G. (H) The CALR locus was amplified by polymerase chain reaction in PB, buccal mucosa, BM, CD3-selected PB lymphocytes, CD15-selected PB myeloid cells, and hair follicles from the patient and in a normal tissue control obtained from an unrelated donor. The amplified product was separated by electrophoresis on an agarose gel and stained with ethidium bromide. The wild-type and 52-bp–deleted CALR bands are indicated. (I) Sanger sequencing traces of the wild-type (upper panel) and mutant (lower panel) buccal sample, mapped to the reference CALR sequence. Bu, buccal mucosa; H, hair follicles; Ly, CD3-selected PB lymphocytes; My, CD15-selected PB myeloid cells; NC, normal tissue control.
A somatic 52-bp deletion in the CALR gene is detected in buccal epithelial cells. (A) Table summarizing the detection of CALR mutation by NGS and Sanger sequencing in various tissues. (B-C) NGS of diagnostic BM (B) and buccal cells (C) visualized through Integrated Genome Viewer. Sequence reads are aligned to the CALR locus. Gray bars represent read alignments. White gaps in alignment represent areas of deletion. (D) Cytospin preparation of buccal cells stained with Diff-Quick (Romanowski) stain. (E-G) Paraffin-embedded buccal cell block stained with hematoxylin and eosin (E), cytokeratin AE1/AE3 (F), and CD45 (G). Original magnification ×40 in panels D-G. (H) The CALR locus was amplified by polymerase chain reaction in PB, buccal mucosa, BM, CD3-selected PB lymphocytes, CD15-selected PB myeloid cells, and hair follicles from the patient and in a normal tissue control obtained from an unrelated donor. The amplified product was separated by electrophoresis on an agarose gel and stained with ethidium bromide. The wild-type and 52-bp–deleted CALR bands are indicated. (I) Sanger sequencing traces of the wild-type (upper panel) and mutant (lower panel) buccal sample, mapped to the reference CALR sequence. Bu, buccal mucosa; H, hair follicles; Ly, CD3-selected PB lymphocytes; My, CD15-selected PB myeloid cells; NC, normal tissue control.
We analyzed buccal or blood samples from 2 daughters, a sister, and a brother of the case patient. None of these relatives harbored mutations in JAK2, MPL, or CALR via NGS. Next, we tested additional tissues from this patient to determine whether her CALR mutation was germ line or somatically acquired. The patient’s PB was sorted based on CD3 and CD15 status and submitted for Sanger sequencing along with a hair follicle sample and banked DNA from prior samples (day-90 PB and day-194 BM and buccal specimen). Consistent with our NGS analyses, the CALR mutation was detected in her day-90 PB, day-194 BM, and day-194 buccal specimen by Sanger sequencing. Sanger sequencing also detected the CALR deletion in the CD15+ myeloid fraction, but this mutation was not detected in her CD3+ lymphocyte fraction or her hair follicle (Figure 2A,H-I). These findings confirm that her CALR deletion was somatically acquired and not germ line.
In this patient with BCR-ABL+ CML and preexisting CALR-mutant essential thrombocythemia, buccal mucosa epithelial cells harbored the CALR mutation, but PB-derived CD3+ lymphocytes and hair follicles did not harbor this mutation, consistent with a BM origin of her buccal epithelial cells. Two independent buccal samples displayed VAFs consistent with heterozygous status in a majority of tested cells.
These findings suggest that this patient’s CALR-deleted progenitors may have engrafted her buccal mucosa and given rise to CALR-mutant buccal mucosa cells. Similarly, in a JAK2V617F-mutant case where buccal testing showed a near 50% VAF, fingernail testing confirmed absence of a germ line mutation.22 These authors concluded that lymphocyte contamination was likely the cause of their erroneous results. In our case, we used cytokeratin AE1/AE3 staining and absence of CD45 to confirm that the buccal cells were epithelial and not lymphocytic. Our data suggest that BM-derived, CALR-mutant progenitor cells engrafted within our patient’s buccal mucosa, leading to a somatically acquired CALR mutation in her buccal epithelium. Although lymphocyte contamination of buccal material is well recognized,20,21 a BM origin for buccal epithelial cells is not well known. BM-derived cells have been shown to colonize cheek epithelium after allogeneic stem-cell transplantation,25 but this has not been previously reported in a nontransplantation setting.
In some familial MPN studies, germ line status was demonstrated based on near 50% VAF in buccal tissue.19,22,23 Our case highlights the difficulty in selecting appropriate tissue for germ line testing and indicates that buccal epithelial tissue may not be a reliable source of germ line material.
Acknowledgments
Z.S. was supported by the American Cancer Society, Frederick A. DeLuca Foundation, and a Mentored Research Scholar Grant (MRSG-16-195-01-DDC); Clinical and Translational Science Institute, University of Minnesota, KL2 Career Development Awards ULI RR033183 and KL2 RR0333182 from the National Center for Advancing Translational Sciences, National Institutes of Health; the Division of Hematology, Oncology, and Transplantation, Department of Medicine, University of Minnesota; University of Minnesota Foundation donors; and a University of Minnesota Department of Medicine Women’s Early Research Career Award.
Authorship
Contribution: S.R.G. obtained samples, analyzed data, performed pathological assessment of tissues, prepared figures, designed the study, and wrote the manuscript; L.B.B. analyzed data and designed the study; M.L.S. processed samples and performed experiments; E.L.C. performed pathological assessment of tissues and prepared figures; A.C.N. analyzed data, prepared figures, designed the study, and wrote the manuscript; and Z.S. obtained samples, designed the study, prepared figures, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zohar Sachs, Division of Hematology, Oncology, and Transplantation, Department Medicine, University of Minnesota, 420 Delaware St SE, MMC 480, Minneapolis, MN 55455; e-mail: sachs038@umn.edu; and Andrew C. Nelson, Department of Laboratory Medicine and Pathology, University of Minnesota Medical Center, Fairview, 420 Delaware St SE, MMC 609, Minneapolis, MN 55455; e-mail: nels2055@umn.edu.
References
Author notes
S.R.G. and L.B.B. contributed equally to this work.
A.C.N. and Z.S. contributed equally to this work.
The full-text version of this article contains a data supplement.