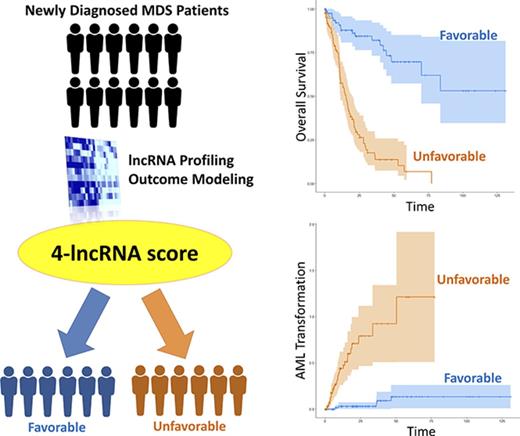
Key Points
Through lncRNA profiling, we identified an MDS patient subset with distinct clinical and mutational patterns along with inferior outcomes.
A concise yet powerful 4-lncRNA risk-scoring system was devised with the potential to improve current MDS risk stratification.
Abstract
Long noncoding RNAs (lncRNAs) not only participate in normal hematopoiesis but also contribute to the pathogenesis of acute leukemia. However, their clinical and prognostic relevance in myelodysplastic syndromes (MDSs) remains unclear to date. In this study, we profiled lncRNA expressions in 176 adult patients with primary MDS, and identified 4 lncRNAs whose expression levels were significantly associated with overall survival (OS). We then constructed a risk-scoring system with the weighted sum of these 4 lncRNAs. Higher lncRNA scores were associated with higher marrow blast percentages, higher-risk subtypes of MDSs (based on both the Revised International Prognostic Scoring System [IPSS-R] and World Health Organization classification), complex cytogenetic changes, and mutations in RUNX1, ASXL1, TP53, SRSF2, and ZRSR2, whereas they were inversely correlated with SF3B1 mutation. Patients with higher lncRNA scores had a significantly shorter OS and a higher 5-year leukemic transformation rate compared with those with lower scores. The prognostic significance of our 4-lncRNA risk score could be validated in an independent MDS cohort. In multivariate analysis, higher lncRNA scores remained an independent unfavorable risk factor for OS (relative risk, 4.783; P < .001) irrespective of age, cytogenetics, IPSS-R, and gene mutations. To our knowledge, this is the first report to provide a lncRNA platform for risk stratification of MDS patients. In conclusion, our integrated 4-lncRNA risk-scoring system is correlated with distinctive clinical and biological features in MDS patients, and serves as an independent prognostic factor for survival and leukemic transformation. This concise yet powerful lncRNA-based scoring system holds the potential to improve the current risk stratification of MDS patients.
Introduction
Myelodysplastic syndromes (MDSs) comprise a heterogeneous group of clonal hematopoietic disorders, and are the most common class of acquired bone marrow (BM) failure syndromes in adults. MDSs are clinically characterized by BM dysplasia with ineffective hematopoiesis, peripheral blood cytopenia, and an increased risk of transformation to acute myeloid leukemia (AML) in ∼30% to 40% of patients.1,2 The clinical presentations and treatment outcomes of MDS patients may vary considerably,3,4 underscoring the necessity of individualized management.5-8
Although cytogenetic abnormalities are detected in about half of MDS patients,9-11 recurrent somatic mutations, most notably those involving RNA-splicing machinery and epigenetic regulators, can be identified in over 80% of them.12 Not only have these coding gene mutations been linked to disease progression and prognosis in MDSs,13-17 but dysregulation of noncoding RNAs, such as the microRNAs, have also been implicated as important elements in the pathogenesis and disease evolution of MDSs.18-21 Recent research works have revealed that the long noncoding RNAs (lncRNAs), a novel class of versatile noncoding RNAs operationally defined as transcripts longer than 200 nucleotides, play crucial roles in a constellation of biological processes including chromatin modification, regulation of gene expression in various signaling pathways, genomic imprinting, and epigenetic control.22-25 Although a myriad of previous publications have established the association between deranged lncRNA expression and carcinogenesis, tumor progression, and metastasis in a wide variety of solid cancers,26-33 relatively limited data exist so far on how these lncRNAs are involved in normal and malignant hematopoiesis.29,34-37
In this study, we aimed to perform lncRNA expression profiling in a large cohort of primary MDS patients to investigate the link between lncRNA expression and the clinical and genetic features of MDS patients. A concise, integrated risk-scoring system was constructed by incorporating the expression levels of 4 lncRNAs that were significantly associated with survival, taking into account the statistical weight of each component lncRNA. To our knowledge, this is the first report that provides a lncRNA platform for risk stratification of MDS patients. Furthermore, by analyzing the messenger RNA (mRNA) expression profiles, we identified several pathways highly correlated with the lncRNA prognostic signature.
Materials and methods
Patients
We recruited 176 consecutive adult patients with primary MDS diagnosed between November 1992 and December 2010 at the National Taiwan University Hospital (NTUH) as the training cohort; we confirmed their diagnoses according to the 2008 World Health Organization (WHO) classification.38 Patients with therapy-related MDSs or antecedent hematological diseases were excluded.
Another independent set of 30 patients, who were diagnosed with MDSs with these criteria between January 2011 and May 2012 at NTUH, were collected as the validation cohort. This study was approved by the institutional review board of the NTUH. Informed consent was obtained from the patients in accordance with the Declaration of Helsinki. Cytogenetic analyses were performed as described previously,39 and interpreted according to the International System for Human Cytogenetic Nomenclature.40 Mutation analyses of 17 genes relevant to myeloid malignancies, such as genes related to epigenetic regulation (including ASXL1,41 IDH1,42 IDH2,43 EZH2,44 TET2,45 and DNMT3A46 ), genes related to the RNA-splicing machinery (including SF3B1,47 U2AF1,48 SRSF2,49 and ZRSR250 ), as well as FLT3/ITD,51 NRAS,52 KRAS,52 RUNX1,53 MLL/PTD,54 TP53,55 and SETBP1,52 were performed by Sanger sequencing as previously described. Abnormal sequencing results were confirmed by at least 2 repeated analyses.
Microarray analysis of lncRNA and data processing
We extracted RNA by the TRIzol method from the BM mononuclear cells obtained at diagnosis. One microgram of RNA from each sample was processed with the miRNeasy mini kit (Qiagen, Hilden, Germany); the global gene expressions of the 176 MDS patients in the training cohort, and the 30 MDS patients in the validation cohort, were profiled with Affymetrix GeneChip Human Transcriptome Array 2.0 (Affymetrix, Santa Clara, CA), following the manufacturer’s instructions.
We used the robust multichip average algorithm for microarray preprocessing and normalization. We then mapped the probes with the most updated version of the LNCipedia56 (V4.0) and GENCODE57 (release 25) databases (as in September 2016); 19 614 lncRNA probes were identified in the Affymetrix GeneChip Human Transcriptome Array 2.0 microarray.
Establishment of the lncRNA risk score
We first conducted probe-level Z transformation on the 19 614 lncRNAs across 176 MDS patients, which made zero mean and unit standard deviation (SD) of each lncRNA across the patients. We then used the univariate Cox proportional hazards regression model to analyze the association between overall survival (OS) and the expression levels of the individual lncRNAs. The lncRNAs with the most prominent significance on OS (univariate Cox P < 1 × 10−6) were selected for further multivariate Cox model to identify the lncRNAs whose expression levels could independently predict survival. The lncRNAs with significant association with OS in the multivariate test (multivariate Cox P < .05) were then selected to build the lncRNA risk-scoring system, in which the expression of component lncRNAs was subjected to a second multivariate Cox regression test to obtain the β values as their weights in the risk model equation. The lncRNA risk score was calculated as the summation of the normalized expression level of each component lncRNA multiplied by its weight:  where j denotes the patient accession number, lncRNAi represents the normalized expression level of the lncRNA probe i after Z transformation, and βi represents the weight of the particular lncRNA probe i. We also performed a 100 000-time random permutation test to ensure the strength of our risk model. For each iteration of the random permutation, the same number of lncRNAs was randomly selected from the 19 614 lncRNAs to construct a random scoring system. Then, the univariate Cox regression model was used for testing the prognostic significance of each randomly assigned lncRNA score. After 100 000 iterations, the empirical P value of our proposed risk model was calculated as the fraction of random scoring systems that achieved a better P value than our proposed risk model. The smaller the empirical P value, the greater probability that our proposed lncRNA risk score outperformed the other randomly assembled lncRNA combinations.
where j denotes the patient accession number, lncRNAi represents the normalized expression level of the lncRNA probe i after Z transformation, and βi represents the weight of the particular lncRNA probe i. We also performed a 100 000-time random permutation test to ensure the strength of our risk model. For each iteration of the random permutation, the same number of lncRNAs was randomly selected from the 19 614 lncRNAs to construct a random scoring system. Then, the univariate Cox regression model was used for testing the prognostic significance of each randomly assigned lncRNA score. After 100 000 iterations, the empirical P value of our proposed risk model was calculated as the fraction of random scoring systems that achieved a better P value than our proposed risk model. The smaller the empirical P value, the greater probability that our proposed lncRNA risk score outperformed the other randomly assembled lncRNA combinations.
Bioinformatics approaches for biological inference of the 4 lncRNAs
To obtain insights into the functionality of the 4 lncRNAs, we first investigated the differentially expressed mRNAs between patients with the highest (> average + 1 SD) and the lowest (< average − 1 SD) lncRNA risk scores, and then analyzed the correlated biological pathways with the Database for Annotation, Visualization and Integrated Discovery (DAVID; v6.8)58,59 and Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA).60
Statistical analysis
The Mann-Whitney U test was used to compare continuous variables and medians of distribution. The χ2 test or the Fisher exact test were used to compare the difference of discrete variables such as sex, WHO classification, cytogenetic changes, distribution of the Revised International Prognostic Scoring System (IPSS-R), and genetic alterations between patients with lower and higher lncRNA risk scores. OS was measured from the date of initial diagnosis to the date of last follow-up or death from any cause. Twofold cross-validation methodology was performed to validate the scoring system. In this procedure, we randomly divided our 176 MDS patients into 2 subgroups, half as the training set and the other half as the validation set. The training set was used to build a lncRNA risk-scoring system with the procedure stated in “Establishment of the lncRNA risk score.” This lncRNA risk-scoring system was then applied to the validation set to stratify patients into high-score and low-score subgroups. The survival prediction power of this particular lncRNA risk score was then evaluated by both the log-rank test and the univariate Cox proportional hazards model. After 100 000 iterations, the prediction rate of our proposed lncRNA risk score was calculated as the fraction of random scoring systems that achieved P < .05. Kaplan-Meier analysis was used to plot the survival curves, and the log-rank test was used to evaluate the statistical significance. The Cox proportional hazards model was used for multivariate regression analysis. P values <.05 were considered statistically significant. All statistical analyses were performed with BRB-ArrayTools (version 4.5.1; Biometric Research Branch, National Cancer Institute, Rockville, MD),61 IBM SPSS Statistics for Windows, version 20 (IBM Corp, Armonk, NY), and R software (https://cran.r-project.org/).
Results
Patient population
The median age of the 176 MDS patients was 67.8 years. The 164 patients who had karyotype data at diagnosis belonged to the following subgroups according to the IPSS-R: very-low risk, 3.7%; low risk, 32.9%; intermediate risk, 25.0%; high risk, 21.3%; and very-high risk, 17.1%. Most patients (68.8%) received supportive care only. Seventeen patients (9.7%) received hypomethylating agents (HMAs), 19 (10.8%) received low-dose cytarabine, 13 (7.4%) received AML-directed intensive chemotherapy either as a bridge to transplantation or at the time of leukemic transformation, and 19 (10.8%) underwent allogeneic hematopoietic stem cell transplantation. With a median follow-up duration of 37.3 months (range, 0.1-130.9 months), 83 mortalities were recorded, whereas 38 patients progressed to AML.
Computing the lncRNA risk score
To construct an lncRNA scoring system that is predictive of survival in MDS patients, we first identified 30 of the 19 614 lncRNA probes whose expression levels had significant association with OS (P < 1 × 10−6), as listed in supplemental Table 2. We then included expression levels of the 30 lncRNAs in a multivariate Cox model to select the lncRNAs whose expression levels had independent predictive power on survival. We discovered that high expressions of 4 lncRNA probes independently predicted poor OS (P < .05), namely TC07000551.hg.1, TC08000489.hg.1, TC02004770.hg.1, and TC03000701.hg.1, with multivariate Cox P values being <.001, <.001, .013, and .018, respectively (the details of the 4 lncRNAs are listed in supplemental Table 3). By incorporating the β values as statistical weights, we constructed the following equation: 4-lncRNA risk score = [TC07000551.hg.1] × 0.578 + [TC08000489.hg.1] × 0.526 + [TC02004770.hg.1] × 0.289 + [TC03000701.hg.1] ± 0.183. In a 100 000-time random permutation test, the 4-lncRNA risk score outperformed almost all of the other 100 000 random combinations of 4 lncRNAs in survival prediction, with an empirical P < 10−5, suggesting high performance and nonrandomness of our proposed scoring system.
Comparison of clinical characteristics and genetic alterations between patients with high vs low 4-lncRNA risk scores
The 176 MDS patients in the training cohort were divided into 2 groups by the median lncRNA risk score. The comparison of clinical and laboratory features between patients with higher and lower scores is listed in Table 1. The patients with higher lncRNA scores had higher BM blast percentages (P < .001) compared with low-score patients. There were no significant differences in distribution of age and sex, peripheral blood white blood cell count, absolute neutrophil count, hemoglobin, and platelet level between the 2 groups. High-score patients more frequently had refractory anemia with excess blasts-1 (RAEB-1) and RAEB-2 (27.3% vs 9.1%, P = .003 and 43.2% vs 6.8%, P < .001, respectively), but less commonly refractory cytopenia with unilineage dysplasia (RCUD), refractory anemia with ring sideroblasts (RARS), refractory cytopenia with multilineage dysplasia (RCMD), and RCMD and ring sideroblasts (RCMD-RS) compared with the low-score patients (14.8% vs 33.0%, P = .008; 2.3% vs 12.5%, P = .018; 12.5% vs 28.4%, P = .014; and 0% vs 10.2%, P = .003, respectively). High-score patients were more frequently assigned to IPSS-R high- and very-high-risk subgroups (36.0% vs 5.1%, P < .001 and 26.7% vs 6.4%, P = .001, respectively), but less frequently to the IPSS-R low-risk subgroup (17.4% vs 50.0%, P < .001).
Comparison of clinical and laboratory features between patients with high and low lncRNA scores
| Clinical characters . | Total, N = 176 . | Low lncRNA score, n = 88 . | High lncRNA score, n = 88 . | P . |
|---|---|---|---|---|
| Sex (%) | .329 | |||
| Male | 121 (68.8) | 57 (64.8) | 64 (72.7) | |
| Female | 55 (31.3) | 31 (35.2) | 24 (27.3) | |
| Age, median (range), y | 67.8 (18.5-94.5) | 68.7 (18.5-94.5) | 68.7 (25.9-89.2) | .750 |
| Laboratory data, median (range) | ||||
| WBC, ×109/L | 3.285 (0.490-20.440) | 3.780 (1.490-10.880) | 3.895 (0.490-2.040) | .970 |
| ANC, ×109/L | 1.768 (0.103-12.728) | 1.971 (0.258-6.953) | 1.565 (0.103-12.728) | .395 |
| Hb, g/dL | 8.1 (3.5-14.6) | 8.0 (3.5-13.1) | 8.3 (4.0-14.6) | .409 |
| Platelet, ×109/L | 86 (3-721) | 104 (12-511) | 77 (3-721) | .091 |
| BM blast, % | 3.0 (0-18.8) | 1.5 (0-12.8) | 8.2 (0-18.8) | <.001* |
| 2008 WHO classification (%) | ||||
| RCUD | 42 (23.9) | 29 (33.0) | 13 (14.8) | .008* |
| RARS | 13 (7.4) | 11 (12.5) | 2 (2.3) | .018* |
| RCMD | 36 (20.5) | 25 (28.4) | 11 (12.5) | .014* |
| RCMD-RS | 9 (5.1) | 9 (10.2) | 0 (0) | .003* |
| RAEB1 | 32 (18.2) | 8 (9.1) | 24 (27.3) | .003* |
| RAEB2 | 44 (25) | 6 (6.8) | 38 (43.2) | <.001* |
| IPSS-R,†‡(%) | ||||
| Very low | 6 (3.7) | 5 (6.4) | 1 (1.2) | .103 |
| Low | 54 (32.9) | 39 (50.0) | 15 (17.4) | <.001* |
| Intermediate | 41 (25.0) | 25 (32.1) | 16 (18.6) | .070 |
| High | 35 (21.3) | 4 (5.1) | 31 (36.0) | <.001* |
| Very high | 28 (17.1) | 5 (6.4) | 23 (26.7) | .001* |
| Clinical characters . | Total, N = 176 . | Low lncRNA score, n = 88 . | High lncRNA score, n = 88 . | P . |
|---|---|---|---|---|
| Sex (%) | .329 | |||
| Male | 121 (68.8) | 57 (64.8) | 64 (72.7) | |
| Female | 55 (31.3) | 31 (35.2) | 24 (27.3) | |
| Age, median (range), y | 67.8 (18.5-94.5) | 68.7 (18.5-94.5) | 68.7 (25.9-89.2) | .750 |
| Laboratory data, median (range) | ||||
| WBC, ×109/L | 3.285 (0.490-20.440) | 3.780 (1.490-10.880) | 3.895 (0.490-2.040) | .970 |
| ANC, ×109/L | 1.768 (0.103-12.728) | 1.971 (0.258-6.953) | 1.565 (0.103-12.728) | .395 |
| Hb, g/dL | 8.1 (3.5-14.6) | 8.0 (3.5-13.1) | 8.3 (4.0-14.6) | .409 |
| Platelet, ×109/L | 86 (3-721) | 104 (12-511) | 77 (3-721) | .091 |
| BM blast, % | 3.0 (0-18.8) | 1.5 (0-12.8) | 8.2 (0-18.8) | <.001* |
| 2008 WHO classification (%) | ||||
| RCUD | 42 (23.9) | 29 (33.0) | 13 (14.8) | .008* |
| RARS | 13 (7.4) | 11 (12.5) | 2 (2.3) | .018* |
| RCMD | 36 (20.5) | 25 (28.4) | 11 (12.5) | .014* |
| RCMD-RS | 9 (5.1) | 9 (10.2) | 0 (0) | .003* |
| RAEB1 | 32 (18.2) | 8 (9.1) | 24 (27.3) | .003* |
| RAEB2 | 44 (25) | 6 (6.8) | 38 (43.2) | <.001* |
| IPSS-R,†‡(%) | ||||
| Very low | 6 (3.7) | 5 (6.4) | 1 (1.2) | .103 |
| Low | 54 (32.9) | 39 (50.0) | 15 (17.4) | <.001* |
| Intermediate | 41 (25.0) | 25 (32.1) | 16 (18.6) | .070 |
| High | 35 (21.3) | 4 (5.1) | 31 (36.0) | <.001* |
| Very high | 28 (17.1) | 5 (6.4) | 23 (26.7) | .001* |
ANC, absolute neutrophil count; Hb, hemoglobin; IPSS-R, Revised International Prognostic Scoring System; RAEB, refractory anemia with excess blasts; RARS, refractory anemia with ring sideroblasts; RCMD, refractory cytopenia with multilineage dysplasia; RCMD-RS, RCMD and ring sideroblasts; RCUD, refractory cytopenia with unilineage dysplasia; WBC, white blood cell.
Statistically significant if P < .05.
One hundred sixty-four patients, including 78 with low lncRNA scores and 86 with high lncRNA scores, had chromosome data at diagnosis.
IPSS-R: very low, ≤1.5; low, >1.5-3; intermediate, >3-4.5; high, >4.5-6; and very high, >6.
The incidence of cytogenetic abnormalities tended to be higher in patients with higher lncRNA risk scores than those with lower scores (46.5% vs 30.8%, P = .054). High-score patients had statistically significantly higher incidence of poor-risk cytogenetics (20.9% vs 9.0%, P = .049) and complex karyotypes (16.3% vs 5.1%, P = .026), but lower incidence of good-risk cytogenetics (58.1% vs 76.9%, P = .013) than low-score patients (supplemental Table 4).
Overall, 108 patients (61.4%) had at least 1 gene mutation, 42 (47.7%) in the low-score subgroup, compared with 66 (75.0%) in the high-score subgroup (P < .001). As listed in Table 2, the most common mutation in high-score patients was the ASXL1 mutation (31.8%), followed by RUNX1 (24.7%), SRSF2 (20.7%), DNMT3A (19.5%), and ZRSR2 mutations (16.3%). Compared with patients with lower lncRNA scores, those with higher scores more frequently harbored RUNX1, ASXL1, TP53, SRSF2, and ZRSR2 mutations (24.7% vs 4.6%, P < .001; 31.8% vs 10.2%, P = .001; 12.8% vs 1.1%, P = .002; 20.7% vs 6.8%, P = .009; and 16.3% vs 1.2%, P = .001, respectively), but had lower incidence of SF3B1 mutation (9.2% vs 23.9%, P = .014). We discovered distinctive patterns of concurrent gene mutations in MDS patients with either higher or lower lncRNA risk scores (Figure 1). Patients with higher lncRNA scores had more complex interrelationships among the various mutations (Figure 1A), compared with those with lower lncRNA scores (Figure 1B). In addition, the mutation co-occurrences involving SF3B1 and TET2 were more frequent in patients with lower lncRNA scores (P = .019 and P = .015, respectively), whereas those involving KRAS/NRAS, IDH1/2, SETBP1, TP53, MLL/PTD, and FLT3/ITD were only found in patients with higher lncRNA scores.
Comparison of genetic alterations between patients with low and high lncRNA scores
| Gene . | No. of specimens examined . | Mutated (%) . | P . | ||
|---|---|---|---|---|---|
| Total . | Low lncRNA score . | High lncRNA score . | |||
| SF3B1 | 175 | 29 (16.6) | 21 (23.9) | 8 (9.2) | .014* |
| FLT3/ITD | 174 | 1 (0.6) | 0 (0) | 1 (1.2) | .494 |
| NRAS/KRAS | 175 | 5 (2.9) | 1 (1.1) | 4 (4.6) | .211 |
| RUNX1 | 172 | 25 (14.5) | 4 (4.6) | 21 (24.7) | <.001* |
| MLL/PTD | 173 | 1 (0.6) | 0 (0) | 1 (1.2) | .497 |
| IDH1/2 | 175 | 4 (2.3) | 0 (0) | 4 (4.6) | .059 |
| ASXL1 | 173 | 36 (20.8) | 9 (10.2) | 27 (31.8) | .001* |
| TET2 | 175 | 22 (12.6) | 10 (11.4) | 12 (13.8) | .655 |
| DNMT3A | 175 | 25 (14.3) | 8 (9.1) | 17 (19.5) | .054 |
| TP53 | 174 | 12 (6.9) | 1 (1.1) | 11 (12.8) | .002* |
| SETBP1 | 175 | 4 (2.3) | 0 (0) | 4 (4.6) | .059 |
| EZH2 | 176 | 10 (5.7) | 4 (4.5) | 6 (6.8) | .538 |
| U2AF1 | 175 | 14 (8) | 7 (8) | 7 (8) | >.999 |
| SRSF2 | 175 | 24 (13.7) | 6 (6.8) | 18 (20.7) | .009* |
| ZRSR2 | 171 | 15 (8.8) | 1 (1.2) | 14 (16.3) | .001* |
| Gene . | No. of specimens examined . | Mutated (%) . | P . | ||
|---|---|---|---|---|---|
| Total . | Low lncRNA score . | High lncRNA score . | |||
| SF3B1 | 175 | 29 (16.6) | 21 (23.9) | 8 (9.2) | .014* |
| FLT3/ITD | 174 | 1 (0.6) | 0 (0) | 1 (1.2) | .494 |
| NRAS/KRAS | 175 | 5 (2.9) | 1 (1.1) | 4 (4.6) | .211 |
| RUNX1 | 172 | 25 (14.5) | 4 (4.6) | 21 (24.7) | <.001* |
| MLL/PTD | 173 | 1 (0.6) | 0 (0) | 1 (1.2) | .497 |
| IDH1/2 | 175 | 4 (2.3) | 0 (0) | 4 (4.6) | .059 |
| ASXL1 | 173 | 36 (20.8) | 9 (10.2) | 27 (31.8) | .001* |
| TET2 | 175 | 22 (12.6) | 10 (11.4) | 12 (13.8) | .655 |
| DNMT3A | 175 | 25 (14.3) | 8 (9.1) | 17 (19.5) | .054 |
| TP53 | 174 | 12 (6.9) | 1 (1.1) | 11 (12.8) | .002* |
| SETBP1 | 175 | 4 (2.3) | 0 (0) | 4 (4.6) | .059 |
| EZH2 | 176 | 10 (5.7) | 4 (4.5) | 6 (6.8) | .538 |
| U2AF1 | 175 | 14 (8) | 7 (8) | 7 (8) | >.999 |
| SRSF2 | 175 | 24 (13.7) | 6 (6.8) | 18 (20.7) | .009* |
| ZRSR2 | 171 | 15 (8.8) | 1 (1.2) | 14 (16.3) | .001* |
Statistically significant if P < .05.
Circos diagrams illustrating the different comutation patterns. Panels A and B depict the co-occurrence of mutations in patients with higher vs lower lncRNA scores, respectively. Ribbon widths are proportional to the frequencies of mutation co-occurrence.
Circos diagrams illustrating the different comutation patterns. Panels A and B depict the co-occurrence of mutations in patients with higher vs lower lncRNA scores, respectively. Ribbon widths are proportional to the frequencies of mutation co-occurrence.
Correlation of the 4-lncRNA risk score with survival and leukemic transformation
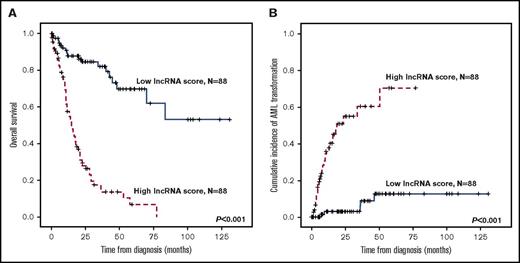
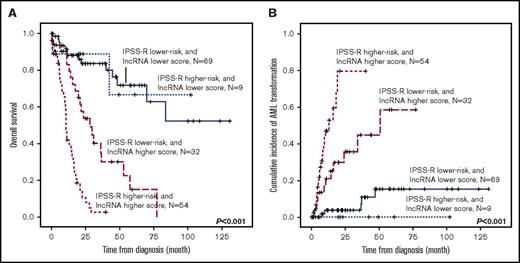
Patients with higher lncRNA scores had inferior OS than those with lower scores (median, 15.2 months vs not reached [NR], P < .001; Figure 2A). The cumulated incidence of transformation to AML at 5 years was 70.2% for higher-score patients and 12.5% for lower-score patients (P < .001; Figure 2B). The differences in outcomes remained significant in both IPSS-R lower-risk (very low, low, and intermediate risk) and IPSS-R higher-risk (high- and very-high-risk) subgroups (supplemental Figures 1 and 2). Likewise, when survival analysis was performed in the subgroup of patients with either a normal karyotype (N = 100), unfavorable cytogenetics (including complex karyotypes, monosomy 7, and del(7q); N = 24), WHO lower-risk subtypes (RCUD, RARS, RCMD, and RCMD-RS; N = 100), or WHO higher-risk subtypes (RAEB-1 and RAEB-2; N = 76), patients with higher lncRNA scores consistently fared worse, and had a significantly higher incidence of acute leukemic transformation at 5 years than those with lower scores (supplemental Figures 3-6). By incorporating the lncRNA scoring system into the traditional IPSS-R classification, we could further refine the clinical outcome prediction in MDS patients (Figure 3), as we note that no matter which IPSS-R subgroup the patients had been originally assigned to, their clinical outcomes are more in line with the lncRNA score risk groups.
Kaplan-Meier plots, stratified by lncRNA scores. (A) OS and (B) cumulative incidence of AML transformation of the 176 MDS patients in the training cohort. Patients with higher lncRNA scores had worse clinical outcomes than those with lower scores.
Kaplan-Meier plots, stratified by lncRNA scores. (A) OS and (B) cumulative incidence of AML transformation of the 176 MDS patients in the training cohort. Patients with higher lncRNA scores had worse clinical outcomes than those with lower scores.
Kaplan-Meier plots, stratified by the integration of IPSS-R and lncRNA scores. (A) OS and (B) cumulative incidence of AML transformation of the 164 patients in the training cohort who had cytogenetic data at diagnosis (thus IRSS-R could be calculated). The incorporation of the lncRNA scoring system into the traditional IPSS-R classification could refine risk stratification of MDS patients.
Kaplan-Meier plots, stratified by the integration of IPSS-R and lncRNA scores. (A) OS and (B) cumulative incidence of AML transformation of the 164 patients in the training cohort who had cytogenetic data at diagnosis (thus IRSS-R could be calculated). The incorporation of the lncRNA scoring system into the traditional IPSS-R classification could refine risk stratification of MDS patients.
Furthermore, as the treatment strategies and options of MDS patients have evolved in the past 2 decades, with the advent of HMA being the most prominent advance, to test whether our lncRNA scoring system would continue to hold up in the populations managed with HMA, we also specifically looked into this selected subgroup in our cohort. Within the HMA-treated patient subset (N = 17), those patients with higher lncRNA scores did have significantly shorter median OS (supplemental Figure 7A; 17.3 months vs NR, P = .028), along with a marginally higher incidence of projected AML transformation at 5 years than those with lower scores (supplemental Figure 7B; 52.9% vs 16.7%, P = .129). A similar trend could also be observed in the patient subgroup that, beyond supportive care, had received active treatment of their MDSs during disease course (supplemental Figure 8).
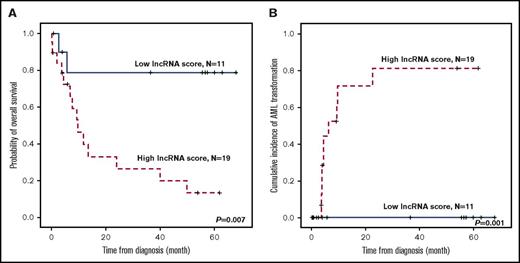
To validate our 4-lncRNA scoring system, we first applied the twofold cross-validation method and found that the P values derived from log-rank and univariate Cox regression models were both <.05. To further confirm the prognostic significance, we profiled the lncRNA expression of an independent validation cohort of 30 MDS patients with the same Affymetrix GeneChip HTA 2.0 platform. The clinical characteristics of patients in the validation cohort were comparable to those in the training cohort, as shown in supplemental Table 5, and the detailed description of the steps in the validation procedure is presented in “Methodology used to externally validate the 4-lncRNA score” in the supplemental Data. Similar to the training cohort, in the validation cohort, patients with higher lncRNA scores had a significantly shorter OS (P = .007), and a higher cumulated incidence of transformation to AML at 5 years (P = .001), compared with lower-score patients (Figure 4). In multivariate analysis of OS and leukemia-free survival in the 164 MDS patients who had diagnostic cytogenetic data (for calculating IPSS-R), we included age, karyotype, IPSS-R, and gene mutations that had P values <.10 in univariate Cox regression analysis (supplemental Table 6) as covariables. Higher lncRNA risk score was identified as an independent unfavorable prognostic factor for both OS (relative risk, 4.783; 95% confidence interval [CI], 2.491-9.182; P < .001) and leukemia-free survival (relative risk, 7.670; 95% CI, 2.702-21.772; P < .001) after adjusting for other clinical and molecular parameters (Table 3).
Kaplan-Meier plots of an independent validation cohort stratified by lncRNA scores. (A) OS and (B) cumulative incidence of AML transformation of the 30 MDS patients in the validation cohort.. Patients with higher lncRNA scores had shorter OS and higher AML transformation rates.
Kaplan-Meier plots of an independent validation cohort stratified by lncRNA scores. (A) OS and (B) cumulative incidence of AML transformation of the 30 MDS patients in the validation cohort.. Patients with higher lncRNA scores had shorter OS and higher AML transformation rates.
Multivariate analysis for OS and LFS in 164 MDS patients who had cytogenetic data at diagnosis
| Variable . | OS . | LFS . | ||||||
|---|---|---|---|---|---|---|---|---|
| RR . | 95% CI . | P . | RR . | 95% CI . | P . | |||
| Lower . | Upper . | Lower . | Upper . | |||||
| Age* | 1.022 | 1.006 | 1.039 | .006† | 0.984 | 0.964 | 1.004 | .122 |
| Karyotype‡ | 0.590 | 0.251 | 1.387 | .226 | 0.454 | 0.118 | 1.740 | .249 |
| IPSS-R§ | 1.621 | 1.220 | 2.153 | .001† | 1.820 | 1.206 | 2.746 | .004† |
| ASXL1 | 1.039 | 0.491 | 2.201 | .920 | 4.065 | 1.576 | 10.483 | .004† |
| TP53 | 3.735 | 1.327 | 10.514 | .013† | 4.795 | 0.790 | 29.081 | .088 |
| EZH2 | 1.847 | 0.643 | 5.300 | .254 | 0.960 | 0.205 | 4.485 | .958 |
| SRSF2 | 0.985 | 0.448 | 2.168 | .971 | 0.678 | 0.214 | 2.151 | .510 |
| ZRSR2 | 1.068 | 0.502 | 2.272 | .863 | 0.442 | 0.121 | 1.614 | .216 |
| lncRNA score|| | 4.783 | 2.491 | 9.182 | <.001† | 7.670 | 2.702 | 21.772 | <.001† |
| Variable . | OS . | LFS . | ||||||
|---|---|---|---|---|---|---|---|---|
| RR . | 95% CI . | P . | RR . | 95% CI . | P . | |||
| Lower . | Upper . | Lower . | Upper . | |||||
| Age* | 1.022 | 1.006 | 1.039 | .006† | 0.984 | 0.964 | 1.004 | .122 |
| Karyotype‡ | 0.590 | 0.251 | 1.387 | .226 | 0.454 | 0.118 | 1.740 | .249 |
| IPSS-R§ | 1.621 | 1.220 | 2.153 | .001† | 1.820 | 1.206 | 2.746 | .004† |
| ASXL1 | 1.039 | 0.491 | 2.201 | .920 | 4.065 | 1.576 | 10.483 | .004† |
| TP53 | 3.735 | 1.327 | 10.514 | .013† | 4.795 | 0.790 | 29.081 | .088 |
| EZH2 | 1.847 | 0.643 | 5.300 | .254 | 0.960 | 0.205 | 4.485 | .958 |
| SRSF2 | 0.985 | 0.448 | 2.168 | .971 | 0.678 | 0.214 | 2.151 | .510 |
| ZRSR2 | 1.068 | 0.502 | 2.272 | .863 | 0.442 | 0.121 | 1.614 | .216 |
| lncRNA score|| | 4.783 | 2.491 | 9.182 | <.001† | 7.670 | 2.702 | 21.772 | <.001† |
Only variables with P ≤ .10 in univariate analysis were incorporated into the multivariate Cox proportional hazard regression analysis.
CI, confidence interval; LFS, leukemia-free survival; RR, relative risk.
Age as a continuous variable.
Statistically significant if P < .05.
Unfavorable cytogenetics vs others. Patients without chromosome data at diagnosis were not included in the analysis.
IPSS-R risk groups: very good, good, intermediate, poor, very poor.
High vs low lncRNA risk scores.
Correlation of the lncRNA signature with gene expression and potential functionality
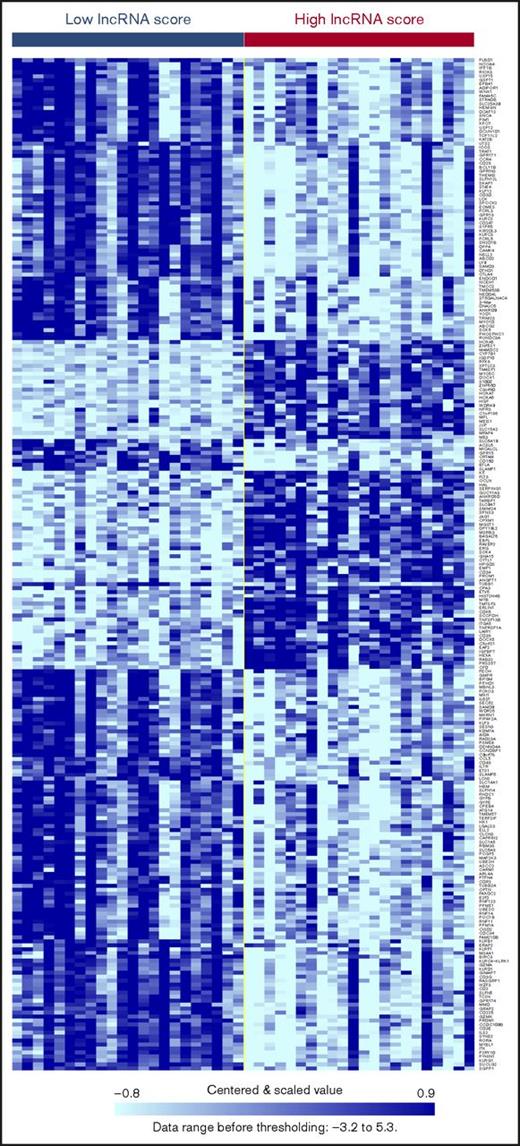
We first profiled the microarray data of 19 867 mRNA probes (corresponding to 18 638 unique coding genes) in the 176 patients and compared the expression levels between patients with the highest (> average + 1 SD; n = 22) and the lowest (< average − 1 SD; n = 22) lncRNA risk scores. Using the threshold of a fold-change of 2 and the Student t test P < .001, we identified 255 genes that were differentially expressed between these 2 groups of patients (listed in supplemental Table 7; corresponding heatmap in Figure 5). Of note, several genes in the homeobox (HOX) family, such as HOXA5, HOXA6, HOXA7, and MEIS1, as well as stem cell markers such as CD34 and KIT, were significantly upregulated in the patients with high lncRNA scores. We used DAVID for functional annotation of the 255 genes and identified a number of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways that were enriched in patients with higher lncRNA scores (supplemental Table 8), with “hematopoietic cell lineage” and “T-cell receptor signaling” pathways showing the top statistical significances.
Heatmap demonstrating differential gene expression. Depicted are the 255 genes that are differentially expressed between patients with the highest (> average +1 SD, n = 22) and the lowest (< average − 1 SD, n = 22) lncRNA risk scores.
Heatmap demonstrating differential gene expression. Depicted are the 255 genes that are differentially expressed between patients with the highest (> average +1 SD, n = 22) and the lowest (< average − 1 SD, n = 22) lncRNA risk scores.
In IPA analysis, we also noted several implicated biological pathways related to the 255 differentially expressed genes (supplemental Figure 9). The pathways with top 5 significance levels were “T-cell receptor (TCR) signaling” (P < .001), “inducible T-cell costimulator (iCOS)-iCOS ligand (iCOSL) signaling in T helper cells” (P < .001), “natural killer (NK) cell signaling” (P < .001), “cytotoxic T-lymphocyte–associated antigen 4 (CTLA4) signaling in cytotoxic T lymphocytes” (P < .001), and “CD28 signaling in T helper cells” (P < .001).
Discussion
Establishing a powerful prognostic system to guide the appropriate treatment of MDSs is important because of the highly complex clinical presentations and the corresponding treatment strategies of this disease.
Gene expression profiling of mRNA levels had been found to outperform the traditional parameters, such as morphology and cytogenetics, in prognostication of MDSs whether CD34+ selected cells62-64 or unsorted bulk marrow cells65 were used for analysis. As lncRNAs have emerged as important participants in the pathogenesis and disease progression of AML,66-74 it is likely that profiling of their expression would have significant impact on MDS prognostication. Recently, it has been shown that lncRNA expression signatures were closely associated with specific mutations and a small subset of lncRNAs could predict clinical outcome in cytogenetically normal elderly AML patients,75 but the clinical significance of lncRNA profiling in MDSs has not yet been reported. To our knowledge, this is the first report to provide a lncRNA platform for prognostication in MDS patients. A scoring system was constructed based on the expression levels and weights of only 4 lncRNAs through well-organized statistical modeling to ensure its significant and independent prognostic implication.
Although the marrow cells in MDSs are heterogeneous with regard to lineages and maturation stages, studies have demonstrated the clonal nature of these cells regardless of the blast counts.76 Although the amount of immature cells may influence the expression levels of lncRNAs, in our study, we clearly demonstrated that the prognostic significance of the lncRNA score held true not only in an unselected cohort of MDS patients, but also in various selected patient subsets, such as the lower- or higher-risk subgroups based on IPSS-R and WHO classifications. Furthermore, we found that this lncRNA score could maintain its performance on outcome prediction in the HMA-treated MDS patient subset. This observation is clinically relevant as HMAs are the mainstay treatment of the high-risk MDS patients nowadays. In multivariate analysis, we demonstrated that the lncRNA score remained an independent prognostic factor with regard to both OS and leukemia-free survival, after adjusting for other clinical parameters (which included IPSS-R, and thus taking the influence of marrow blast percentage into consideration) and gene mutations. Therefore, the poor prognostic implication of higher lncRNA score is indeed independent of the maturation stages of BM cells, or a more advanced disease status at diagnosis.
The finding that several HOX family genes were upregulated in patients with high lncRNA scores was intriguing. The HOX genes have been known to be a conserved family of homeodomain-containing transcription factors acting as critical effectors in cell-fate determination and differentiation, and consequently implicated in hematopoiesis and leukemogenesis.77 Overexpression of the HOX genes is associated with unfavorable prognosis in AML,78 and their deregulation is also linked to higher risk of AML transformation in MDSs.65 HOXA5 expression can inhibit erythropoiesis but promote myeloid lineage commitment.79 On the other hand, overexpression of HOXA6, HOXA7, HOXA9, and MEIS1 have been reported to correlate with AML with MLL rearrangements.80 The association of high lncRNA scores and upregulation of HOX genes could possibly contribute to the more advanced disease stages and worse clinical outcomes of MDS patients with high lncRNA scores. On the other hand, DAVID and IPA analysis showed that the 255 differentially expressed genes between highest and lowest lncRNA scores were highly associated with hematopoietic cell lineage–related pathways, and those related to T-cell and NK-cell functions. These findings might reflect recent discoveries that dysregulated innate and adaptive immunity lead to the pathogenesis of MDSs. For instance, clonal T-cell expansion and skewed TCR repertoire in CD4 and CD8 T cells have been implicated in the pathogenesis of MDSs.81,82 There is also evidence that the compromised regulatory T-cell and NK-cell functions can contribute to ineffective immune surveillance, culminating in the pathogenesis of MDSs, and govern disease risk and progression.83-87
In summary, we provide evidence that higher expression levels of the 4 lncRNAs in our scoring system are associated with distinct clinical and genetic features and could predict inferior outcomes of MDS patients. To our knowledge, this is the first study to develop a risk prediction model incorporating lncRNA expressions and their weights for MDS patients. We believe our lncRNA risk score, in combination with IPSS-R, may further improve risk stratification of MDS patients and guide treatment options accordingly. In this era of high-throughput technologies, the incorporation of multidisciplinary parameters including molecular biomarkers would undoubtedly move us one step further toward the holy grail of precision medicine. We acknowledge that further basic research would be necessary to delve into the biological functions and networking of these lncRNAs, and prospective clinical studies are anticipated to validate the prognostic power of our lncRNA risk score.
The raw and normalized microarray data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE97064).
The full-text version of this article contains a data supplement.
Acknowledgments
The work was supported by a National Taiwan University Hospital (NTUH)−National Taiwan University joint research grant (UN103-051), the Ministry of Science and Technology of Taiwan (MOST102-2325-B-002-028, 103-2314-B-002-130-MY3, 103-2314-B-002-131MY3, and 104-2923-B-002-001), Far Eastern Hospital and NTUH joint grant 105-FTN24, NTUH and National Taiwan University College of Medicine joint grant UN106-024, and the Ministry of Health and Welfare of Taiwan (MOHW102-TD-C-111-001 and MOHW103-TD-B-111-04).
Authorship
Contribution: C.-Y.Y. was responsible for data collection and management, statistical analysis and interpretation, literature research, and manuscript writing; C.-H.C. was responsible for data management and statistical analysis; H.-H.H. assisted in statistical analysis; H.-A.H. and C.-C.L. were responsible for data collection and management; M.-H.T. and C.-J.K. performed the gene mutation studies; and T.-P.L., W.-C.C., and H.-F.T. planned, designed, and coordinated the study over the entire period and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hwei-Fang Tien, Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan; e-mail: hftien@ntu.edu.tw; Wen-Chien Chou, Department of Laboratory Medicine, National Taiwan University Hospital, Taipei, Taiwan; e-mail: wchou@ntu.edu.tw; and Tzu-Pin Lu, Institute of Epidemiology and Preventive Medicine, National Taiwan University, Taipei, Taiwan; e-mail: tplu@ntu.edu.tw.
References
Author notes
C.-Y.Y. and C.-H.C. contributed equally to this work.