Key Points
RUNX1a, but not RUNX1b, is overexpressed in CD34+ cells from patients with myelodysplastic/myeloproliferative neoplasms.
SRSF2P95H mutation induces RUNX1a overexpression and a monocytic phenotype in TF-1 cells.
Introduction
RUNX1 mutations have been shown to contribute to the development of myeloid neoplasms and are frequently identified in patients with myelodysplastic syndromes (MDS),1 myelodysplastic/myeloproliferative neoplasms (MDS/MPN), including chronic monocytic leukemia (CMML),2 and acute myeloid leukemia (AML).3 The biological function of these mutations has been elucidated in a mouse model4 and in human CD34+ cells.5 However, the relevance of the level of RUNX1 expression for the pathogenesis of hematological malignancies has not been explored.
The RUNX1 gene has several isoforms.6,7 Of 3 major isotypes, we focused on both functional full-length isoforms RUNX1b/c (lacking exon 7a and including exon 7b and 8), and the short isoform, RUNX1a (including exon 7a, which has a stop codon), which is found only in primates.8 RUNX1b and RUNX1c are functionally identical and RUNX1b is broadly expressed,9 whereas RUNX1a has a dominant-negative effect on RUNX1b.10 RUNX1a overexpression has been reported in AML.10,11 Enforced expression in murine hematopoietic cells enhances engraftment after transplantation.12 Consistent with this, RUNX1a expands hematopoietic stem cells,13,14 and dominant inhibition by RUNX1a results in monopoiesis.15 Therefore, expression levels of each RUNX1 isoform are likely to be important for normal and malignant hematopoiesis.
Accordingly, in the present study, we quantified expression levels of RUNX1 isoforms in CD34+ cells from patients with myelodysplastic disease. We found that RUNX1a was overexpressed in some MDS and MDS/MPN patients and that these levels of expression increased as the disease progressed. The aberrant splicing resulting in RUNX1a is believed to be caused by splicing factor mutations that are frequently detected in MDS and MDS/MPN.16 Therefore, we enforced the expression of the SRSF2 mutant in an MDS-derived cell line, and we show that this results in high expression of RUNX1a. Our results indicate that RUNX1a may have an important role in the progression of MDS and MDS/MPN and that splicing factor mutations may be one of the mechanisms contributing to RUNX1a overexpression.
Methods
Patients
Patients with hematological malignancies were examined as approved by the institutional review board at Juntendo University and Bunkyo Gakuin University. Patients gave written informed consent, in accordance with the Declaration of Helsinki. Gene mutation analysis was performed by Sanger sequencing as described previously5 using primer pairs listed in supplemental Table 1.
Quantitative reverse-transcription polymerase chain reaction
CD34+ cells were purified and total RNA was harvested as described previously.5 Gene expression levels were quantified with SYBR Select Master Mix and the following forward/reverse primer pairs: RUNX1a (5′-GAACCACTCCACTGCCTTTAAC-3′/5′-ATTCTGAGGGCTGTCATCTTTC-3′), RUNX1b (5′-GAACCACTCCACTGCCTTTAAC-3′/5′-GAAAGTTCTGCAGAGAGGGTTG-3′), and GAPDH (5′-GGAAGGTGAAGGTCGGAGTC-3′/5′-GGAAGATGGTGATGGGATTTC-3′).
Retrovirus transduction
Purified CD34+ cells were infected with retroviral RUNX1a vector or empty (control) vector. After GFP-based sorting, the cells were cultured on MS5 stromal cells as described previously.5 The MDS-derived cell line TF-1 was infected with retroviral SRSF2P95H vector, and transduced single cells were isolated using FACS Aria. Growing clones were then analyzed separately. Flow cytometry and immunoblot analysis were performed as reported previously.5 Details are provided in supplemental Methods.
Statistical analysis
For multiple pairwise comparisons, data were analyzed by 1-way analysis of variance followed by Dunnett’s multiple comparison test, or differences between individual groups were estimated using the Steel-Dwass test. P < .05 was considered statistically significant.
Results and discussion
Levels of expression of RUNX1a and RUNX1b in CD34+ cells from patients with hematological malignancies, including 10 lymphoma patients with normal bone marrow (controls), were analyzed. The average relative RUNX1a expression level in MDS patients (n = 55) and MDS/MPN patients (n = 27) was 9.6-fold and 11.1-fold that of controls, respectively, whereas RUNX1b expression was only approximately twofold that of the controls. In both disease groups, patients with excess blasts displayed a significantly higher level of RUNX1a expression (Figure 1A-B). During disease progression, the expression of RUNX1a increased, while it decreased during azacitidine treatment (shown for a single patient in Figure 1C-D). Interestingly, some patients had high expression of RUNX1a and repression of RUNX1b, even though both share a common promoter. RNA processing may explain why the expression levels of these isotypes do not change in parallel.
RUNX1a overexpression in MDS, MDS/MPN, and AML patients with a history of MDS or MDS/MPN. (A-B) RUNX1 expression levels by quantitative reverse-transcription polymerase chain reaction in CD34+ cells from patients with MDS (A) or MDS/MPN (CMML and MDS/MPN-unclassifiable) (B). Relative RUNX1a and RUNX1b expression was calculated as a ratio relative to GAPDH expression; the ratio of RUNX1a to RUNX1b expression was also calculated. RNA from normal bone marrow CD34+ cells served as a control, their RNA level being defined as unity. *P < .05; **P < .01. Closed circles show RUNX1-mutated patients. BM, bone marrow. (C-D) Change in RUNX1 expression levels over time in a single patient with MDS (C) or MDS/MPN (D). Patient (Pt) 20, 42, 14, and 32 showed disease progression, whereas patient 302, who was treated with azacitidine (AZA), showed hematological improvement. The diagnosis was based on WHO2016.19 (E) Enforced RUNX1a expression in CD34+ cells from patients. Growth patterns of the GFP-sorted cells cultured on MS5 stromal cells are shown. EB, excess blasts; N.S., not significant; PB, peripheral blood.
RUNX1a overexpression in MDS, MDS/MPN, and AML patients with a history of MDS or MDS/MPN. (A-B) RUNX1 expression levels by quantitative reverse-transcription polymerase chain reaction in CD34+ cells from patients with MDS (A) or MDS/MPN (CMML and MDS/MPN-unclassifiable) (B). Relative RUNX1a and RUNX1b expression was calculated as a ratio relative to GAPDH expression; the ratio of RUNX1a to RUNX1b expression was also calculated. RNA from normal bone marrow CD34+ cells served as a control, their RNA level being defined as unity. *P < .05; **P < .01. Closed circles show RUNX1-mutated patients. BM, bone marrow. (C-D) Change in RUNX1 expression levels over time in a single patient with MDS (C) or MDS/MPN (D). Patient (Pt) 20, 42, 14, and 32 showed disease progression, whereas patient 302, who was treated with azacitidine (AZA), showed hematological improvement. The diagnosis was based on WHO2016.19 (E) Enforced RUNX1a expression in CD34+ cells from patients. Growth patterns of the GFP-sorted cells cultured on MS5 stromal cells are shown. EB, excess blasts; N.S., not significant; PB, peripheral blood.
To evaluate the effect of RUNX1a overexpression on long-term cell expansion, we transduced RUNX1a into CD34+ cells from MDS patients with low-level expression of RUNX1a. The RUNX1a-transduced cells proliferated more than the control cells (Figure 1E). Therefore, overexpression of RUNX1a may provide a growth advantage to CD34+ cells. Genomic mutations of RUNX1 were detected in 2 out of 39 (5.1%) MDS and 9 out of 25 (36%) MDS/MPN patients. Some patients had both a RUNX1 gene mutation and RUNX1a overexpression (Figure 1A-B).
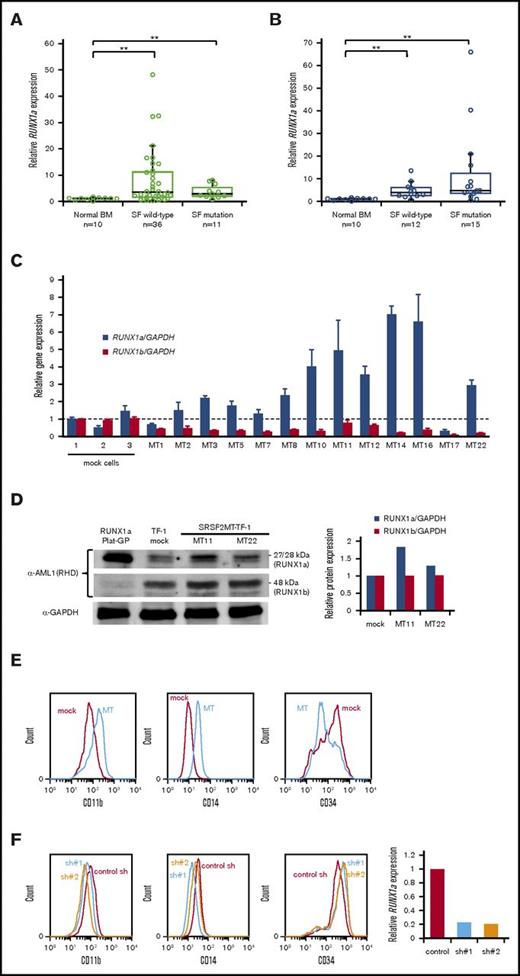
We next focused on the mechanism of RUNX1a overexpression. In MDS and MDS/MPN, splicing factor mutations are frequently found.16 We hypothesized that aberrant splicing of RUNX1 might explain our findings, and we therefore examined splicing factor mutations. SRSF2, U2AF1, and SF3B1 were detected in 11 out of 47 MDS patients (23%) and 15 out of 27 MDS/MPN patients (56%) (Figure 2A-B; supplemental Table 2). In MDS, patients with splicing factor mutations did not show high RUNX1a expression; however, some MDS/MPN patients with these mutations, most of which were SRSF2 mutations (supplemental Table 2), strongly overexpressed RUNX1a. Because SRSF2 contributes to myelodysplasia by mutation-specific effects on exon recognition,17 it was hypothesized that the SRSF2 mutation may affect the expression of RUNX1a. To investigate this, we analyzed SRSF2P95H-transduced TF-1 cells. Most of the clones exhibited higher expression of RUNX1a than mock-transduced cells, whereas RUNX1b expression was reduced in all of them (Figure 2C). Increased RUNX1a expression was confirmed by immunoblotting (Figure 2D). Furthermore, SRSF2P95H-transduced cells showed phenotypic changes, with higher CD11b and CD14 expression than mock cells (Figure 2E). The phenotypic changes were partially reverted by RUNX1a knockdown (Figure 2F), suggesting that SRSF2P95H may induce monocytic differentiation via RUNX1a overexpression, at least in part. RUNX1 mutations in intron 6 and exon 7a were also analyzed. A 5′ splice site change just after exon 6 (c.724+1_2dup) was detected in a CMML patient with RUNX1a overexpression. This may represent another mechanism responsible for RUNX1a overexpression. No mutations of exon 7a or changes in the 3′ splice site just before exon 7a were detected. The possibility that other mechanisms are involved also needs to be considered.18
RUNX1a overexpression induced by splicing factor mutations. (A-B) RUNX1a expression levels in CD34+ cells from MDS (A) or MDS/MPN (B) patients with or without splicing factor (SF) mutations. **P < .01. (C) RUNX1 expression levels of 13 SRSF2 mutant (MT)–transduced clones were compared with those of 3 mock-transduced cells. Average transcript expression and standard deviation from triplicate experiments is shown. (D) RUNX1 protein expression in TF-1 cells was verified by immunoblot, and the relative protein level was quantified. RUNX1a-transfected Plat-GP cells were used as a positive control for RUNX1a protein. (E) Representative flow cytometry analysis of mock-transduced cells and SRSF2 mutant (MT)–transduced cells. (F) Phenotypic changes by RUNX1a knockdown in SRSF2 mutant–transduced cells. RUNX1a knockdown was verified by relative RUNX1a expression to GAPDH expression. BM, bone marrow.
RUNX1a overexpression induced by splicing factor mutations. (A-B) RUNX1a expression levels in CD34+ cells from MDS (A) or MDS/MPN (B) patients with or without splicing factor (SF) mutations. **P < .01. (C) RUNX1 expression levels of 13 SRSF2 mutant (MT)–transduced clones were compared with those of 3 mock-transduced cells. Average transcript expression and standard deviation from triplicate experiments is shown. (D) RUNX1 protein expression in TF-1 cells was verified by immunoblot, and the relative protein level was quantified. RUNX1a-transfected Plat-GP cells were used as a positive control for RUNX1a protein. (E) Representative flow cytometry analysis of mock-transduced cells and SRSF2 mutant (MT)–transduced cells. (F) Phenotypic changes by RUNX1a knockdown in SRSF2 mutant–transduced cells. RUNX1a knockdown was verified by relative RUNX1a expression to GAPDH expression. BM, bone marrow.
Pathogenic functions of RUNX1a have been reported in mouse models. However, this is not clear in patients with myeloid neoplasms. Here, we show for the first time that in addition to RUNX1 mutations, overexpression of RUNX1a may have an important role in disease progression in MDS and MDS/MPN patients. Splicing factor mutations are suggested to contribute to the mechanism of RUNX1 dysregulation.
The full-text version of this article contains a data supplement.
Acknowledgments
This work was carried out in part at the Laboratory of Molecular and Biochemical Research, Research Support Center, Juntendo University Graduate of Medicine. The staff provided excellent technical support in carrying out these experiments.
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (grants 24591398, 25461422, 15K09460, and 16K09831) (H.H. and Y.H.).
Authorship
Contribution: H.S. collected data and wrote the paper; Y.H. assembled and analyzed the data and wrote the paper; Y.O. and Y.K. collected data; N.S., N.D., K.O., and N.K. provided patient samples and clinical information; T.K. contributed helpful discussion; and H.H. designed and performed research and revised the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hironori Harada, Laboratory of Oncology, School of Life Sciences, Tokyo University of Pharmacy and Life Sciences, 1432-1 Horinouchi, Hachioji, Tokyo 192-0392, Japan; e-mail: hharada@toyaku.ac.jp.