TO THE EDITOR:
Molecular minimal residual disease (MRD) is an important disease biomarker for many hematological malignancies, including non-Hodgkin (NHL) lymphomas, predicting disease recurrence and response to therapy earlier than classical imaging-based detection methods.1-8 Although multiple approaches exist, MRD detection through Ig-HTS using the clonoSEQ assay from Adaptive Biotechnologies (formerly clonoSight from Sequenta, Inc before 20159) is increasingly integrated into clinical care for several lymphoid malignancies. Using tumor-involved tissue, Ig-HTS assays identify the dominant V(D)J rearranged heavy (IGH) or light chain (IGK/L) clonotype(s) that have undergone clonal expansion and are expected to uniquely identify a patient’s tumor given the theoretical diversity at each rearranged locus.10,11
The theoretical diversity at the human IgH and IgK/L loci represents the potential variety of antibody molecules that can be generated through V(D)J recombination. The precise number of V, D, and J gene segments in the human IgH locus can vary between individuals, but estimates include ∼38-46 functional V segments, 23 functional D segments, and 6 functional J segments.12 Considering only combinatorial diversity using these estimates, the theoretically expected VDJ diversity at the human IgH locus is ≈5796. In contrast, when considering the kappa and lambda immunoglobulin loci and the corresponding average numbers of functional segments (40 Vκ, 5 Jκ, 30 Vλ, and 4 Jλ), we can estimate substantially lower (∼20-fold to 40-fold less) theoretical diversity in light chains.13
Several important prior studies have evaluated the performance of Ig-HTS techniques for MRD detection.14-18 However, these studies have focused primarily on sensitivity and have not assessed performance when separately considering the heavy and light chains, which substantially differ not only in theoretical diversity (∼20-40×, as detailed above), but also in their observed junctional mutation and somatic hypermutation rates as paired clonotypes in B-cell malignancies.11,19 We therefore sought to empirically evaluate the clinical specificity of Ig-HTS for heavy-chain and light-chain MRD measurement in mature B-cell lymphomas.
All data were collected with informed consent from participants enrolled on studies approved by an institutional review board in accordance with all ethical regulations at their respective institutions, Stanford University (NCT00398177) or the National Cancer Institute (NCT00001563, NCT00001337, and NCT00006436). We generated a repository of lymphoma clonotypes from B-cell tracking or clonality reports of 284 patients with NHL, 129 of whom had undergone clonoSEQ profiling and 155 of whom had undergone clonoSight profiling. A control database was generated from all publicly available immuneACCESS20 (Adaptive Biotechnologies) heavy- and light-chain B-cell receptor (BCR) repertoires spanning malignant and healthy patient contexts (65 636 707 total clonotypes within the repertoires of 1769 subjects).
We assessed whether each lymphoma clonotype in the clinical repository was also present in any of the public repertoires in the control database. A match was considered positive if the sequence context of the clinical clonotype was an exact substring of any clonotype in the control database. Specificity was defined as the percent of all clonotypes (or patients) that had no match within the control database. V gene, J gene, and CDR3 assignments were made using the HighV-Quest (IMGT) platform.21
To assess if the degree of somatic hypermutation or CDR3 rearrangement mediated the degree of overlap between the clinical and control repertoires, the clinical lymphoma clonotypes were mapped against the human genome (build GRCh38/hg38) via BLAT22 (University of California, Santa Cruz), which allows for spliced alignments. Percent identity to germline was determined by dividing the highest BLAT match score23 by the length of the clonotype. Clonotypes with no germline match were assigned a percent identity score of zero.
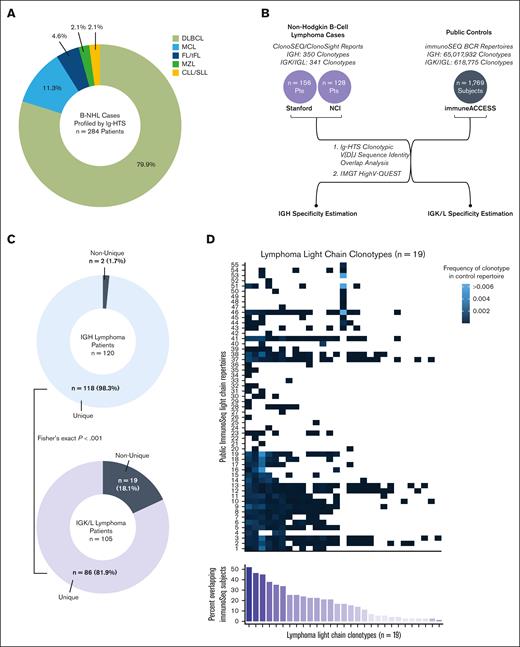
We considered B-NHL cases profiled by Ig-HTS within a cohort that included 227 patients with DLBCL, 32 mantle cell lymphoma, 13 follicular/transformed follicular lymphoma, 6 small lymphocytic lymphoma/chronic lymphocytic leukemia, and 6 marginal zone lymphoma (Figure 1A). The corresponding B-NHL clonotype database included 350 heavy chain clonotypes from 235 patients and 341 light chain clonotypes from 187 patients (Figure 1B). The IGK/L subset consisted of 66 IGL sequences from 49 patients and 275 IGK sequences from 180 patients. Of the IGH clonotypes, 153 (43.7%) had been determined by the clonoSight assay from 115 subjects, whereas the remaining 197 (56.3%) sequences had been determined by the clonoSEQ assay from 120 subjects. In the light chain database, clonoSight sequencing data accounted for 104 (30.5%) clonotypes from 82 patients whereas clonoSEQ accounted for 237 (69.5%) clonotypes from 105 patients. The control database consisted of 71 light-chain repertoires with 618 775 BCR sequences and 1,698 heavy chain repertoires with 65 017 932 BCR sequences (Figure 1B).
Ig-HTS clonotypic V(D)J sequence identity overlap analysis overview. (A) Histologic representation within the clinical cohort. (B) Schematic depication of analyses performed. (C) Donut charts depicting the proportion of patients with unique or non-unique heavy chain (blue) and light chain (purple) clonotypes detected by clonoSEQ. (D) Each column depicts a non-unique dominant lymphoma light chain clonotype, as determined by clonoSEQ. Each row represents an individual immunoSEQ BCR repertoire from the control immuneACCESS database. The frequency at which the clonoSEQ clonotype occurs within each control repertoire is depicted in blue. Clonotypes not found in a given control repertoire occupy white squares. The bottom bar chart sums up the percent of immuneACCESS subjects (out of 71 total) that overlap with each clonotype.
Ig-HTS clonotypic V(D)J sequence identity overlap analysis overview. (A) Histologic representation within the clinical cohort. (B) Schematic depication of analyses performed. (C) Donut charts depicting the proportion of patients with unique or non-unique heavy chain (blue) and light chain (purple) clonotypes detected by clonoSEQ. (D) Each column depicts a non-unique dominant lymphoma light chain clonotype, as determined by clonoSEQ. Each row represents an individual immunoSEQ BCR repertoire from the control immuneACCESS database. The frequency at which the clonoSEQ clonotype occurs within each control repertoire is depicted in blue. Clonotypes not found in a given control repertoire occupy white squares. The bottom bar chart sums up the percent of immuneACCESS subjects (out of 71 total) that overlap with each clonotype.
Two IGH lymphoma clonotypes from 2 separate patients (both sequenced with clonoSEQ) in the clinical database were also present in the BCR repertoire of 1 control patient. Accordingly, heavy chain MRD detection via clonoSEQ demonstrated a sequence-level specificity of 99.0% and a patient-level specificity of 98.3%. In comparison, 29 IGK/L clonotypes from 19 patients, all of whom had been sequenced with clonoSEQ, demonstrated overlap with 55 different control BCR repertoires (77% of all control repertoires). The specificity of light chain MRD by clonoSEQ was significantly lower than that of heavy chain: 87.8% at the sequence-level (Fisher exact P value < .001) and 81.9% at the patient-level (P < .001) (Figure 1C).
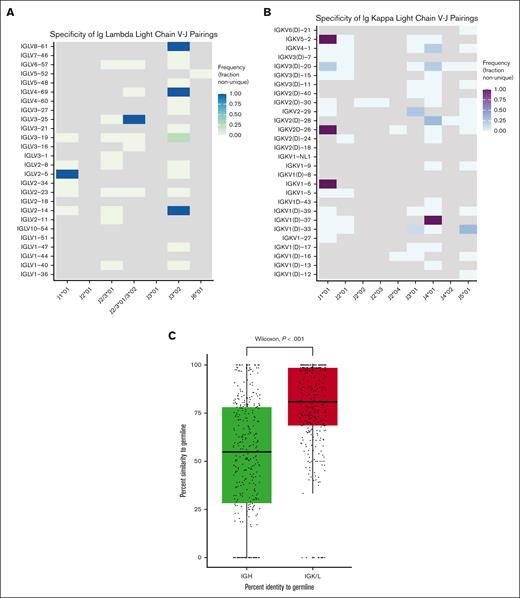
In 18.1% of patients, the IGK/L clonotypes that clonoSEQ had identified as the dominant lymphoma clone were also present in the unselected control BCR repertoire data from immunoSEQ, suggesting a large percentage of tracked clonotypes could be present within a patient’s healthy cells. Most non-unique clonotypes were shared across multiple control subjects (range, 1-37; median, 12) and present in the control database at a frequency ranging from 3.31 × 10-7 to 0.521 (median 9.44 × 10-5) (Figure 1D). Across both generations of Ig-HTS assays, precise V and J gene assignments could be determined for 181 (53.1%) of all light chain clonotypes. HighV-Quest was able to identify a CDR3 junction in 65.5% of non-unique clonotypes and 63.1% of unique clonotypes (Fisher exact P = .84). In terms of predicting clonotype uniqueness, V and J gene usage patterns were unrevealing (Figure 2A-B).
V-J pairing analyses and somatic hypermutation assessment. V-J gene pairing for all IGL (A) and IGK (B) lymphoma clonotypes, in which a precise V and J gene assignment could be made by HighV-Quest. Frequency is the number of times a given V-J gene pair is present in a non-unique clonotype over the total number of times that V-J gene pair is seen across all the lymphoma clonotypes in the clinical database. Gray squares represent V-J gene combinations not present within the data set. For a large proportion of clonotypes, HighV-Quest was unable to differentiate between IGLJ2∗01, IGLJ3∗01, and sometimes IGLJ3∗02. These are grouped together in panel A when applicable as depicted along the x-axis. Likewise, when HighV-Quest was unable to differentiate between a V gene and its ‘D’ paralog, these were grouped together in panel B. (C) Boxplot of the percent identity to germline scores as determined by BLAT between heavy and light chain clonotypes. Clonotypes with no genomic match by BLAT to their respective heavy or light chain region were assigned a percent identity score of zero.
V-J pairing analyses and somatic hypermutation assessment. V-J gene pairing for all IGL (A) and IGK (B) lymphoma clonotypes, in which a precise V and J gene assignment could be made by HighV-Quest. Frequency is the number of times a given V-J gene pair is present in a non-unique clonotype over the total number of times that V-J gene pair is seen across all the lymphoma clonotypes in the clinical database. Gray squares represent V-J gene combinations not present within the data set. For a large proportion of clonotypes, HighV-Quest was unable to differentiate between IGLJ2∗01, IGLJ3∗01, and sometimes IGLJ3∗02. These are grouped together in panel A when applicable as depicted along the x-axis. Likewise, when HighV-Quest was unable to differentiate between a V gene and its ‘D’ paralog, these were grouped together in panel B. (C) Boxplot of the percent identity to germline scores as determined by BLAT between heavy and light chain clonotypes. Clonotypes with no genomic match by BLAT to their respective heavy or light chain region were assigned a percent identity score of zero.
We next sought to assess if the lack of specificity with the light chain MRD assay could be related to biologic differences in the degree of somatic hypermutation or rearrangement from baseline during V(D)J recombination. Light chain clonotypes demonstrated significantly greater identity to the human germline reference sequence as determined by BLAT (mean percent identity 77.9%) compared with heavy chain clonotypes (mean percent identity 51.4%) (Wilcoxon P < .001) (Figure 2C). Although this result fits expectations, to our knowledge, this is the first study to demonstrate that biologic differences in diversity drive observed differences in the clinical specificity of Ig-HTS assays and that the magnitude of this effect is significant despite the intrinsic algorithmic adjustments to the limit of detection threshold that these assays include to attempt to account for differences in diversity.16,18 The impact and magnitude of this association has not been previously described and serves as a critical shortfall for diagnostic products that are in routine clinical use when considering the unmet needs of the patients and providers relying on the accuracy of the associated results. Further optimization and a clinical benchmark of observed clinical specificity of these US Food and Drug Administration–approved assays are needed. Our results suggest that a positive light chain MRD result has a high false-positive likelihood, suggesting impaired clinical utility to this result. Caution should be exercised when using Ig-HTS to monitor MRD when detection is driven by light chain.
Contribution: N.D.S., D.M.K., and A.A.A. designed the analyses, analyzed the data, and wrote the manuscript; J.S.M. and K.R.K. processed and interpreted the data; B.J.S., S.A., M.J.F., D.B.M., S.C., M.D., M.S.K., and M.R. contributed and critically interpreted the data; all authors revised the manuscript.
Conflict-of-interest disclosure: B.J.S. reports consultancy for Foresight Diagnostics. D.M.K. reports consultancy for Roche, Adaptive Biotechnologies, and Genentech and equity ownership interest in Foresight Diagnostics. S.K.A. reports speaker honoraria from Takeda. M.J.F. reports consultancy and research funding from Adaptive Biotechnologies, research funding from Kite/Gilead, stock options from Allogene Therapeutics, and equity in Roche/Genentech. M.S.K. reports research funding from CRISPR Therapeutics and Nutcracker Therapeutics, and advisory committee membership for Myeloid Therapeutics and Daiichi Sankyo. M.D. reports research funding from AstraZeneca, Genentech, Varian Medical Systems, and Illumina; ownership interest in CiberMed and Foresight Diagnostics; and consultancy from AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb (BMS), Genentech, Gritstone Oncology, Illumina, Novartis, and Roche. D.B.M. holds a patent with Pharmacyclics supporting ibrutinib for chronic graft-versus-host disease and receives consulting or research fees or serves as an advisor for Pharmacyclics, Kite Pharma, Adaptive Biotechnologies, Novartis, BMS, Janssen Pharmaceuticals, Roche, Genentech, Precision Bioscience, Allogene, Miltenyi Biotec, Fate Therapeutics, 2Seventy, and Adicet. A.A.A. reports consultancy for ADCT, Celgene, Chugai, Genentech, Gilead, Janssen, Pharmacyclics, and Roche; scientific advisory board membership in the Lymphoma Research Foundation; professional affiliations with the American Society of Hematology, American Society of Clinical Oncology, American Society of Clinical Investigation, and Leukemia & Lymphoma Society; research funding from the National Cancer Institute, National Heart, Lung, and Blood Institute, National Institutes of Health, Celgene, BMS, and Pfizer; patent filings, including patent issued, licensed, and with royalties paid from FortySeven, a patent pending and licensed to Foresight, a patent pending relating to MARIA, a patent issued and licensed to CiberMed, a patent issued, a patent pending to CiberMed, a patent issued to idiotype vaccines, and a patent issued, licensed, and with royalties paid from Roche; and equity ownership interests in CiberMed Inc, Foresight Diagnostics, FortySeven Inc, and CARGO Therapeutics. B.J.S. reports patent filings related to cancer biomarkers. D.M.K., M.D., and A.A.A. also report issued patents licensed to Foresight Diagnostics regarding cancer biomarkers and circulating tumor DNA. The remaining authors declare no competing interests.
Steven Coutre died on 9 November 2021.
Correspondence: Ash A. Alizadeh, Divisions of Oncology & Hematology, Department of Medicine, 265 Campus Dr, SIM1 Lorry Lokey Building, Suite G2120B, Stanford, CA 94305; email: arasha@stanford.edu.
References
Author notes
The heavy and light chain repertoires analyzed in this study are publicly available and obtained from immuneACCESS at https://clients.adaptivebiotech.com/immuneaccess. Please contact corresponding author Ash A. Alizadeh for other forms of data sharing: arasha@stanford.edu.