Macrophages from tissues derive from adult HSPCs in macaques and clonally relate to circulating monocytes post-autologous transplantation.
In vivo labeling shows rapid turnover of macrophages in unmanipulated animals.
Visual Abstract
Macrophages orchestrate tissue immunity from the initiation and resolution of antimicrobial immune responses to the repair of damaged tissue. Murine studies demonstrate that tissue-resident macrophages are a heterogenous mixture of yolk sac–derived cells that populate the tissue before birth, and bone marrow–derived replacements recruited in adult tissues at steady-state and in increased numbers in response to tissue damage or infection. How this translates to species that are constantly under immunologic challenge, such as humans, is unknown. To understand the ontogeny and longevity of tissue-resident macrophages in nonhuman primates (NHPs), we use a model of autologous hematopoietic stem progenitor cell (HSPC) transplantation with HSPCs genetically modified to be marked with clonal barcodes, allowing for subsequent analysis of clonal ontogeny. We study the contribution of HSPCs to tissue macrophages, their clonotypic profiles relative to leukocyte subsets in the peripheral blood, and their transcriptomic and epigenetic landscapes. We find that HSPCs contribute to tissue-resident macrophage populations in all anatomic sites studied. Macrophage clonotypic profiles are dynamic and overlap significantly with the clonal hierarchy of contemporaneous peripheral blood monocytes. Epigenetic and transcriptomic landscapes of HSPC-derived macrophages are similar to tissue macrophages isolated from NHPs that did not undergo transplantation. We also use in vivo bromodeoxyuridine infusions to monitor tissue macrophage turnover in NHPs that did not undergo transplantation and find evidence for macrophage turnover at steady state. These data demonstrate that the life span of most tissue-resident macrophages is limited and can be replenished continuously from HSPCs.
Introduction
Conserved across most species of the Chordata phylum, macrophages are evolutionary the oldest leukocyte subsets and have the highest degree of plasticity.1 Macrophages play supportive roles in multiple aspects of physiology, including infections and tissue repair. Their function and phenotype depend upon their ontogeny; their anatomical location; and response to infection, tissue damage, adjacent tumor cells, or other environmental stressors, based on both intrinsic factors and degree of differentiation as well as extracellular cues.
Tissue macrophages are important targets for many RNA and DNA viruses (as previously reviewed2), and are hypothesized to be long-lived reservoirs for HIV and simian immunodeficiency virus.3 Tissue-resident macrophages infected with some viruses can resist immunological killing by natural killer cells and CD8 T cells.4,5 Design of strategies aimed at completely depleting reservoirs of these pathogenic viruses depends on the life span and ontogeny of infected macrophages and their capacity to be cleared by the immune system. Tissue macrophages, including central nervous system–resident microglial cells, are therapeutic targets of gene and drug therapies for various acquired and inherited disorders, with efficacy dependent on the ontogeny and life span of these cells.6,7
Murine macrophages first arise during embryonic development in the yolk sac, then populate the entire embryo after circulation is established. Yolk sac–derived macrophages can persist into adulthood in mice, as demonstrated by fate-mapping studies.8,9 Macrophages derived from the yolk sac or fetal liver that reside in organs such as the brain (as microglia cells), pancreas, spleen, liver (as Kuppfer cells), and kidney are thought to be very long lived, perhaps for the entire life span.9 Macrophages can also differentiate from hematopoietic stem progenitor cell (HSPC)-derived circulating monocytes and, particularly under stress, can replace or replenish tissue-resident macrophages.10-13 Recent murine studies suggest that, when present in tissues, macrophages can divide to maintain homeostasis.1,14,15 In specific pathogen–free (SPF) mice, the extent and timing of monocyte replacement varies dramatically based on tissue and experimental conditions, with some tissue-resident macrophages, such as brain microglial cells, having almost no turnover.16 By contrast, the dermis is thought to be primarily populated by monocyte-derived macrophages.17,18 Viral infection of some subsets of lymph node macrophages (those resident in lymphoid sinuses) leads to their rapid death as well as the death of uninfected macrophage neighbors.19 These macrophages are rapidly replaced by a combination of incoming monocytes and proliferation.20 Thus, there exists a continuum of scenarios for macrophage handling of pathogens that centers around their longevity, and understanding their turnover dynamics is of particular interest and importance.
Emerging data suggest that environmental variables can dramatically influence the immunological development and responses of the host. Inbred mouse strains maintained in captivity are immunologically disparate from wild mice and respond to pathogenic infections very differently.21,22 The interplay between endogenous host traits, the microbiome, other environmental factors, and infection represents an area of active research.23 It is unclear how macrophage turnover dynamics in laboratory mice correspond to longer-lived, outbred, pathogen-exposed species such as primates and humans.
To explore the ontogeny and longevity of tissue macrophages in outbred populations of nonhuman primates (NHPs) with high relevance to human biology and disease, we performed autologous HSPC transplantation in rhesus macaques (RMs) with genetically modified HSPCs genetically marked with integrating lentiviral vectors expressing green fluorescent protein (GFP) and containing trackable, highly diverse, genetic barcodes.24-26 This approach allowed us to explore the clonal composition, ontogeny, and stability of tissue-resident macrophages from multiple anatomic sites relative to the clonal composition of circulating blood cells.25,26 To assess the turnover of tissue macrophages in unmanipulated animals in the absence conditioning and transplantation, we infused 5′-bromo-2′-deoxyuridine (BrdU) into healthy adult RMs and tracked the emergence and loss of BrdU+ tissue-resident macrophages and blood monocytes longitudinally. Finally, we assessed the epigenetic landscapes and transcriptomics of transplanted tissue-resident macrophages compared with those of macrophages from healthy, RMs that did not undergo transplantation (unmanipulated).
Methods
Autologous barcoded HSPC transplantation
Tissue processing, cell purification, flow cytometry, and imaging
Four core biopsy specimen each of the spleen and liver were obtained via laparotomy, and 10 pinch biopsy specimen from the colon and jejunum were obtained via endoscopy. These samples were homogenized into single-cell suspensions.28 Bronchioalveolar lavage was performed by instilling 150 mL of saline into the lung followed by collection via suction. For further details of tissue processing, flow cytometry, and imaging, see supplemental Methods.
BrdU infusion and analysis
BrdU (Sigma) was prepared at a concentration of 10 mg/mL in Hanks balanced salt solution (Sigma), sterile filtered, and administered IV to RMs at 30mg/kg per day for 5 days. Blood was drawn before infusion through day 24. Biopsy samples of the spleen and lymph nodes were collected on days 10 and 24. Cells were analyzed for BrdU positivity by fluorescence-activated cell sorting.
ATAC-seq/RNA-seq
Assay for transposase-accessible chromatin with sequencing (ATAC-seq)/RNA sequencing (RNA-seq) processing, mapping, and analysis were performed as previously described29 and detailed in supplemental Methods.
Results
Marking frequencies of HSPC-derived leukocytes in tissues
We performed autologous transplantation of RMs with CD34+ HSPCs transduced with lentiviral vectors expressing copepod GFP and containing a high diversity library of genetic barcodes24,25 (Figure 1A). This allows for the use of GFP as a marker gene demonstrating derivation of cells in tissues from transplanted HSPCs as well as lineage tracing of HSPC output at a clonal level via quantitative barcode retrieval. Transplantation parameters are summarized in Table 1. As expected, numbers of circulating monocytes and neutrophils significantly declined in the first week after total body irradiation (TBI) and rapidly recovered (supplemental Figure 1). Analyses of peripheral blood (PB) and bone marrow longitudinal clonal dynamics in the 4 macaques that underwent transplantation included in this study have been, in part, published previously.25,26,30,31
We sampled leukocytes from multiple tissues including the liver, spleen, lymph node, jejunum and colon, lung (via bronchioalveolar lavage [BAL]), and PB at several time points after transplantation to assess the degree to which individual populations had been replaced from the transplanted, genetically marked HSPCs. Flow cytometry for GFP expression in mononuclear cell leukocyte populations including T cells, B cells, natural killer cells, and tissue myeloid cells (macrophages), was performed (Figure 1B-C). Consistent with previous reports from our group, and from others, lower fractions of T cells in both the PB and in most tissues as compared with other lineages were GFP+25,32 (Figure 1D), likely attributed to thymic involution and damage after TBI.
Notably, we were able to detect GFP+ tissue-resident macrophages in all anatomic sites (Figure 1C-D). The frequencies of GFP+ macrophages were higher than GFP+ T cells and equivalent to all other tissue leukocyte lineages (Figure 1D). Most importantly, the frequencies of GFP+ macrophages in tissues were no different than the frequency of GFP+ PB monocytes sampled concurrently (Figure 1D). Indeed, tissue-resident macrophages were as frequently GFP+ as circulating neutrophils (Figure 1D), which have an estimated half-life of <1 week in humans. Thus, tissue-resident macrophages sampled years after transplantation were equally derived from transplanted HSPCs compared with other leukocyte subsets resident in the liver, lymphoid tissues, and mucosal tissues, or circulating in the blood.
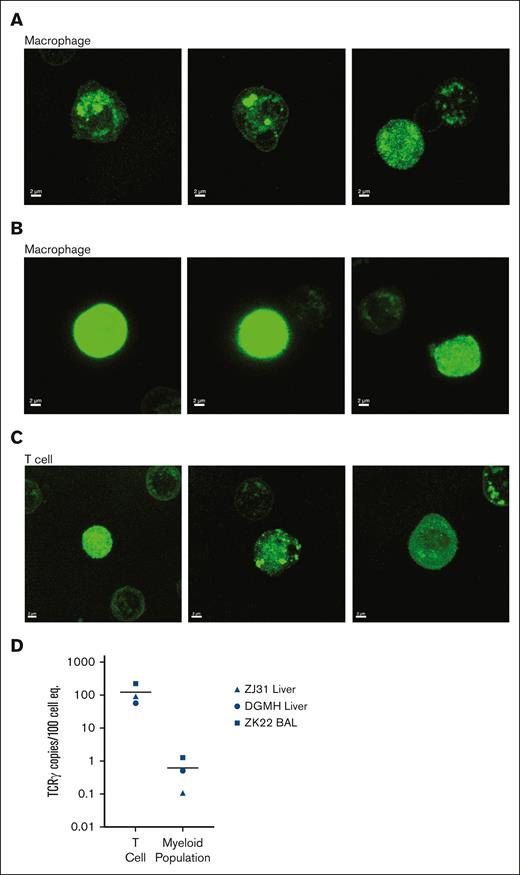
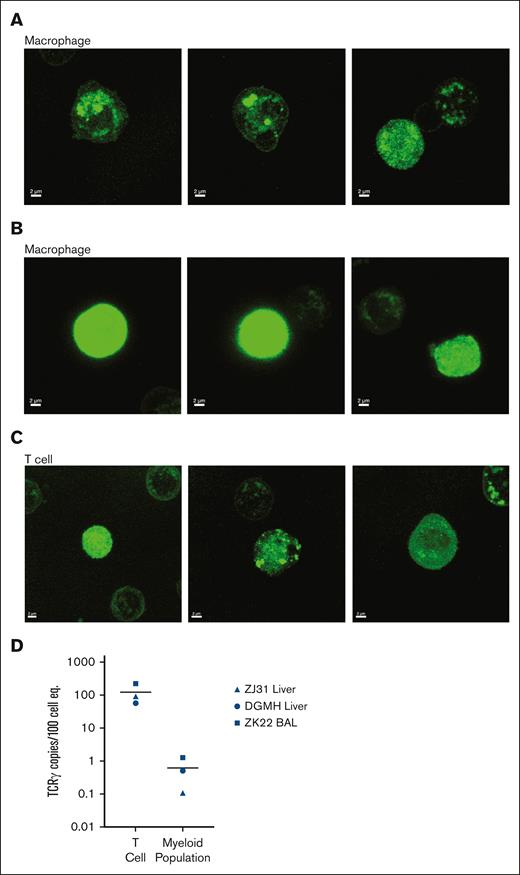
We have previously found that macrophage-mediated phagocytosis of other leukocyte subsets can confound analysis of tissue-resident macrophages.33,34 Phagocytosis would result in accumulation of GFP protein specifically within phagosomes and lysosomes, vs the more diffuse distribution across the cytoplasm in cells expressing endogenous GFP. We sorted T cells and macrophages from liver samples and analyzed GFP localization by confocal microscopy (Figure 2). Although some GFP was localized within intracellular organelles of tissue-resident macrophages (Figure 2A), there was also clear dispersed cytoplasmic GFP (Figure 2B) in these cells. Moreover, GFP localization in both organelles and cytoplasm was also seen in T cells (Figure 2C). Because myeloid cell phagocytosis of T cells would result in the accumulation of rearranged T-cell receptor (TCR) DNA in myeloid cells,33 we measured rearranged TCR DNA in T cells and macrophages from the liver and BAL of animals that underwent transplantation (n = 3). As expected, all T cells had detectable rearranged TCR DNA (Figure 2D). Only extremely low levels of rearranged TCR DNA were detected in tissue-resident myeloid cells, >2 logs lower than in T cells (Figure 2D). Taken together, these data indicate that phagocytosis of GFP+ leukocytes is unlikely to account for the majority of GFP positivity within tissue-resident macrophages.
Confocal imaging of GFP localization and analysis of TCR rearrangement within tissue-resident cells excludes phagocytosis as major source of GFP in tissue macrophages. Confocal microscopic images of cellular GFP protein localization in sorted GFP+ tissue macrophages from the liver of animals that underwent transplantation and BAL (lung) samples demonstrate both (A) cellular organelle organization and (B) cytosolic GFP expression. (C) Confocal microscopic analysis of GFP expression patterns in T cells from the same samples are both localized to organelles and/or widespread throughout the cytoplasm. (D) Fraction of rearranged TCR DNA in purified T cells and macrophages from the liver and BAL of 2 animals that underwent transplantation (ZJ31 and ZK22) and 1 unmanipulated (DGMH) animal.
Confocal imaging of GFP localization and analysis of TCR rearrangement within tissue-resident cells excludes phagocytosis as major source of GFP in tissue macrophages. Confocal microscopic images of cellular GFP protein localization in sorted GFP+ tissue macrophages from the liver of animals that underwent transplantation and BAL (lung) samples demonstrate both (A) cellular organelle organization and (B) cytosolic GFP expression. (C) Confocal microscopic analysis of GFP expression patterns in T cells from the same samples are both localized to organelles and/or widespread throughout the cytoplasm. (D) Fraction of rearranged TCR DNA in purified T cells and macrophages from the liver and BAL of 2 animals that underwent transplantation (ZJ31 and ZK22) and 1 unmanipulated (DGMH) animal.
Clonal derivation of tissue-resident macrophages and T cells
We examined the clonal ontogenies of GFP+ tissue major histocompatibility complex class II+CD11b+ macrophages and CD3+ T cells as compared with PB CD14+ monocytes, PB CD14+CD163+ myeloid cells, and CD3+ T cells sampled at 2 to 4 years after transplantation in 4 lentivirally barcoded RMs (Table 1). Barcode retrieval and quantitation was performed to identify the fractional contributions of individual transplanted HSPC clones to each sample.26,35 This approach accurately quantitates individual HSPC barcode contributions even within highly polyclonal samples, with each barcode representing the output from a single transduced and transplanted HSPCs. We previously reported a pattern of contributions from lineage-restricted short-term engrafting HSPCs (ST-HSPCs) for the first several weeks to months after transplantation, transitioning to stable contributions derived from multipotent long-term repopulating HSPCs (LT-HSPCs) producing blood neutrophils, monocytes, and B cells for at least the next 7 to 12 years26 (C. Wu and C.E. Dunbar, unpublished data, 2023). Blood T-cell clonal patterns included LT-HSPC–derived contributions matching other lineages, along with waxing and waning oligoclonal expansions limited to T cells, representing peripheral memory homotypic responses.26
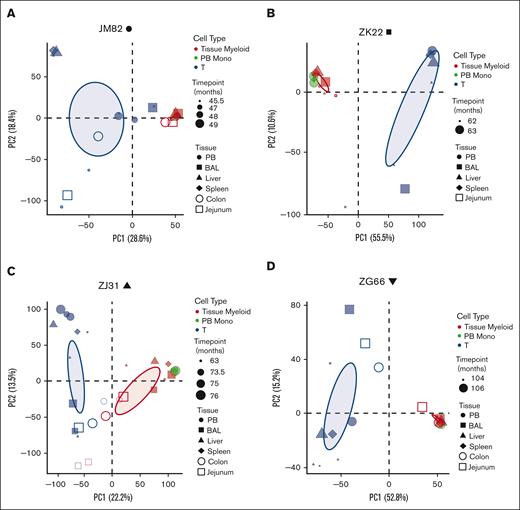
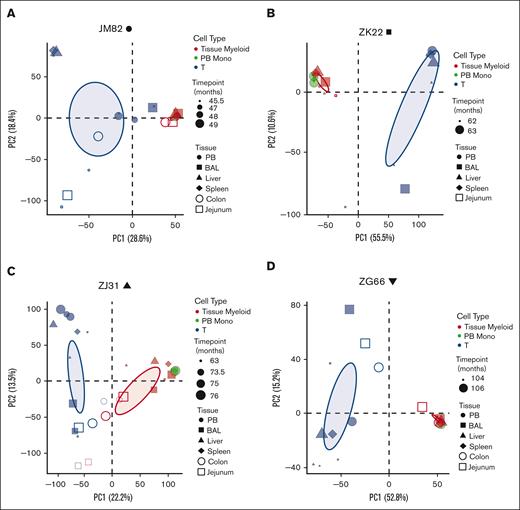
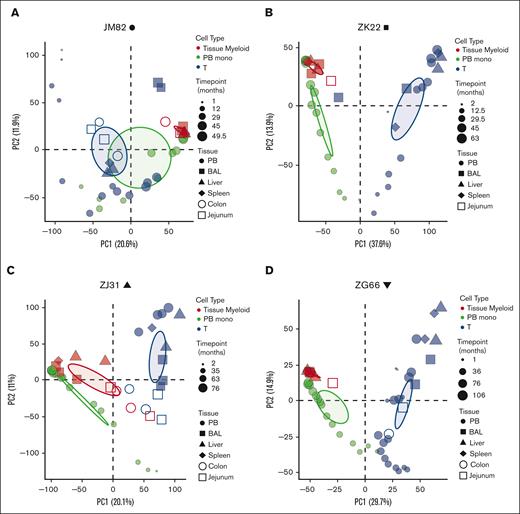
Principal coordinate analysis (PCA) was used to determine the clonal relatedness of tissue-resident macrophages, PB monocytes, and PB and tissue T cells, and the stability of contributions over time. The first principal coordinate separated T cells from myeloid cells. Clonal contributions to tissue-resident macrophages were very similar to contemporaneous blood monocytes (Figure 3A-D). There was little clonal change in macrophages over time, at least for the 2 to 10 months elapsing between serial sample collections in each animal, and similar clonal patterns between macrophages collected from different anatomic sites. Jejunum and colon samples from animal ZJ31 were less related; however, the low numbers of cells analyzed from these tissues likely introduced sampling artifacts. T cells from the blood clustered separately, as expected, as did T cells from various tissue sites, likely due to peripheral T-cell responses to local environmental stimuli.
Clonal relationships between tissue-resident cell lineages and PB monocytes in HSPC-barcoded RMs. (A-D) PCA of the relationship of the level of all individual barcode reads retrieved, normalized to the total numbers of reads, from tissue-resident macrophages (red), tissue-resident T cells (blue), and PB monocytes (PB monos; green) in each animal. Symbol size corresponds to sampling time point relative to transplantation, and symbol shape to the tissue type. Ellipses correspond to 95% confidence intervals.
Clonal relationships between tissue-resident cell lineages and PB monocytes in HSPC-barcoded RMs. (A-D) PCA of the relationship of the level of all individual barcode reads retrieved, normalized to the total numbers of reads, from tissue-resident macrophages (red), tissue-resident T cells (blue), and PB monocytes (PB monos; green) in each animal. Symbol size corresponds to sampling time point relative to transplantation, and symbol shape to the tissue type. Ellipses correspond to 95% confidence intervals.
We also performed pairwise Pearson correlation analyses of clonal contributions to PB monocytes, PB macrophages, T cells, and tissue-resident macrophages from various anatomic sites (supplemental Figure 2A-D). These correlation analyses confirmed the PCA, documenting highly significant clonal correlations between PB monocytes and tissue-resident macrophages as well as between different anatomic sites and over several time points and less correlation with circulating or tissue-resident T cells. These data suggest that tissue-resident macrophages develop along the same lineage ontogeny as circulating monocytes via differentiation from the same set of stable LT-HSPC clones. The similarities in clonal composition between different anatomic sites argues against initial seeding with a small number of precursor cells and ongoing local proliferation/differentiation.36 The low correlation between tissue-resident macrophages and T cells also argues against phagocytosis of T cells as an explanation for GFP positivity of tissue-resident macrophages, supporting aforementioned confocal microscopy and TCR rearrangement results.
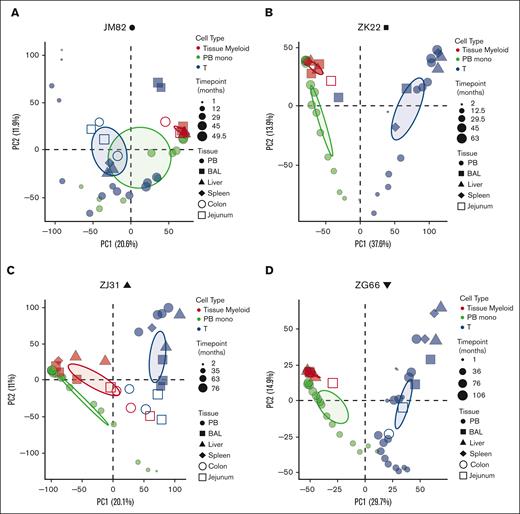
Next, we asked whether seeding of tissues with macrophage precursors (ie, monocytes) occurs only immediately after tissue damage resulting from pretransplantation ablative conditioning, or occurs on an ongoing basis. Although barcodes detected in tissue-resident macrophages and T cells were fairly consistent between the time points sampled (supplemental Figure 2; Figure 3), these time points were 46 months to 106 months after transplantation. Short-lived lineage-biased HSPCs contribute to blood lineages for the first several months after transplantation and are then replaced by contributions from stable multipotent LT-HSPCs.25,26 These dynamics allowed us to ask whether HSPC-derived tissue-resident macrophages were primarily progeny of short-term HSPCs cells seeding tissues in the first several months after TBI conditioning due to depletion of tissue-resident macrophages at that time and/or tissue damage signals, or derived via ongoing replacement from LT-HSPCs. We used PCA to compare the clonal composition of PB monocytes sampled 1 to 2 months after transplantation with that of tissue-resident macrophages sampled years after transplantation (supplemental Figure 3; Figure 4). The clones contributing to PB monocytes and T cells early after transplantation were significantly different from clones contributing to tissue-resident macrophages and tissue-resident T cells at later time points (Figure 4A-D). The clones contributing to tissue-resident macrophages most closely resembled the LT-HSPC clones contributing to contemporaneously sampled PB monocytes and other circulating lineages (supplemental Figure 4). Thus, although TBI conditioning might have facilitated initial tissue-resident macrophage turnover and replacement, our model suggests ongoing replacement of these cells from LT-HSPCs.
Continual development of tissue-resident macrophages and T cells from HSPCs. (A-D) PCA of the relationship of the level of all individual barcode reads retrieved, normalized to the total numbers of reads, from tissue-resident macrophages (red), PB monos (green), and T cells (blue) from the barcoded animals at multiple time points after transplantation. Symbol size represents biopsy time points relative to transplantation, symbol shapes to different tissues, and ellipses correspond to 95% confidence intervals.
Continual development of tissue-resident macrophages and T cells from HSPCs. (A-D) PCA of the relationship of the level of all individual barcode reads retrieved, normalized to the total numbers of reads, from tissue-resident macrophages (red), PB monos (green), and T cells (blue) from the barcoded animals at multiple time points after transplantation. Symbol size represents biopsy time points relative to transplantation, symbol shapes to different tissues, and ellipses correspond to 95% confidence intervals.
Phenotypic, epigenetic, and transcriptomic characteristics of HSPC-derived tissue macrophages
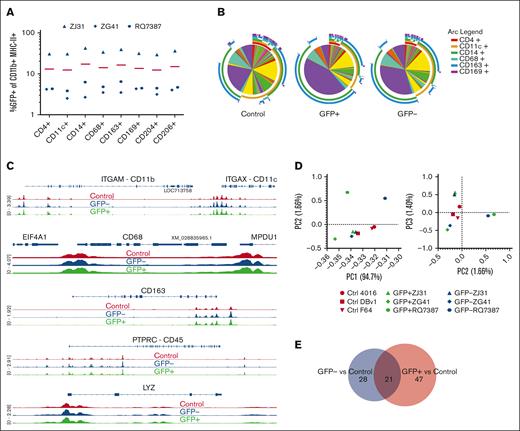
Although the clonal composition of GFP+ tissue macrophages closely matched contemporaneously sampled PB myeloid cells, we wanted to know whether the cell surface, epigenetic, and gene expression characteristics of these cells resembled those of steady state macrophages from control unmanipulated animals, or GFP− cells from the animals that underwent transplantation. We found no difference in the percentages of GFP+ cells in any subset of liver macrophages using CD4, CD11c, CD14, CD68, CD163, CD169, CD204, or CD206 (Figure 5A), and the expression of each marker was similar on GFP+ and GFP− liver macrophages (supplemental Figure 5). We used Simplified Presentation of Incredibly Complex Evaluations to analyze the expression of all 8 markers in combination in GFP+ or GFP− liver macrophages from RMs that underwent transplantation as well as unmanipulated control RMs.37 No significant differences were found between any of the pairwise combinations by permutation testing (Figure 5B).
GFP+ and GFP− liver macrophages are substantially similar to each other and liver macrophages from unmanipulated control animals. (A) The expression of GFP in CD11b+ major histone compatitibilty class II–positive liver macrophages expressing each of the listed surface proteins was examined by flow cytometry. There were no significant differences by 2-way ANOVA. (B) Surface protein phenotypes were displayed as Simplified Presentation of Incredibly Complex Evaluations pie charts, in which each slice is a particular combination of markers as defined by the arc legend. Plots were not significantly different by permutation test. (C) Examples of chromatin accessibility at gene loci by ATAC-seq. Traces are averages of 3 animals for each group. (D) PCA analyzing variation in reads across consensus peaks in ATAC-seq samples. (E) The number of differentially accessible regions associated with a gene between either GFP+ cells and controls or GFP− cells and controls.
GFP+ and GFP− liver macrophages are substantially similar to each other and liver macrophages from unmanipulated control animals. (A) The expression of GFP in CD11b+ major histone compatitibilty class II–positive liver macrophages expressing each of the listed surface proteins was examined by flow cytometry. There were no significant differences by 2-way ANOVA. (B) Surface protein phenotypes were displayed as Simplified Presentation of Incredibly Complex Evaluations pie charts, in which each slice is a particular combination of markers as defined by the arc legend. Plots were not significantly different by permutation test. (C) Examples of chromatin accessibility at gene loci by ATAC-seq. Traces are averages of 3 animals for each group. (D) PCA analyzing variation in reads across consensus peaks in ATAC-seq samples. (E) The number of differentially accessible regions associated with a gene between either GFP+ cells and controls or GFP− cells and controls.
ATAC-seq29 analysis of flow cytometrically sorted GFP+ and GFP− liver macrophages from animals that underwent transplantation and liver macrophages from unmanipulated controls revealed minimal differences in chromatin accessibility across the genome. Individual loci of genes expressed in macrophages from both groups had accessibility peaks at corresponding locations (Figure 5C). Additionally, CD45 and LYZ, peaks that could differ depending on the ontogeny of the cell,38 appear similar between these macrophage populations (Figure 5C). PCA of read variations across consensus ATAC-seq peaks showed no clear separation of GFP+, GFP−, and control macrophages (Figure 5D). Significantly different accessibility between GFP+ and control macrophages was found for only 68 regions. GFP− and control cells showed 59 differing regions, and 21 of these regions were shared (Figure 5E). Only 13 regions associated with any gene significantly differed between GFP+ and GFP− macrophages. To determine whether there were any biological pathways associated with these differentially accessible genes, we performed overrepresentation analysis (supplemental Table 2); however, no pathways reached a false discovery rate of <0.05 (likely because of the few genes identified).
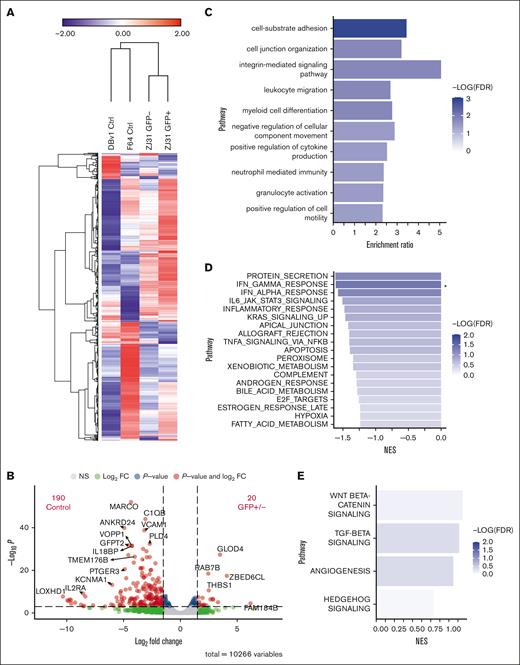
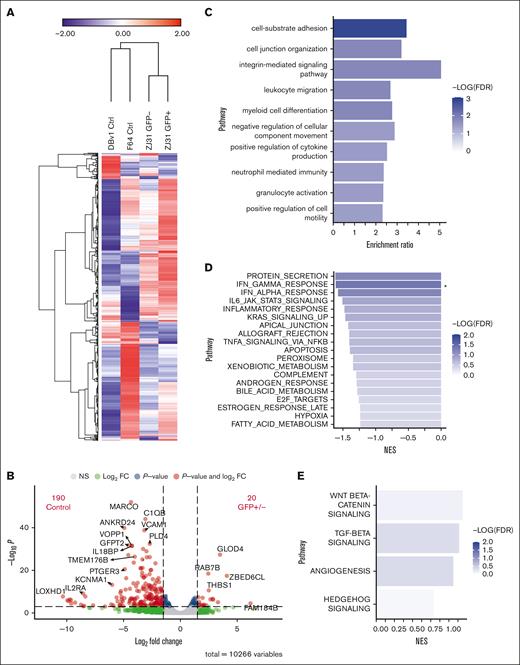
We also performed RNA-seq on GFP+ and GFP− liver macrophages from an animal that underwent transplantation and on 2 control animals’ sorted cells. The standardized expression of the top 1500 genes revealed strong similarities in gene expression patterns between the GFP+ and GFP− macrophages (Figure 6A). Differential expression analysis identified more genes with increased expression in control macrophages compared with those in both cell types from the animal that underwent transplantation than between GFP+ and GFP− cells from that animal (Figure 6B). Genes found in pathways for cell adhesion, leukocyte migration, and granulocyte activation were identified in the small subset of 276 genes that were more highly expressed in macrophages from samples from the animal that underwent transplantation than in the unmanipulated control samples, with a P < 0.1 using overrepresentation analysis (Figure 6C). These differences could reflect changes in the liver environment resulting from transplantation more generally, rather than derivation or not from adult HSPCs.
Similar gene expression profiles between GFP+ and GFP− liver macrophages. (A) Heat map displaying expression of the top 1500 expressed genes by RNA-seq. Fragments per kilobase of transcript per million mapped reads expression data are mean-centered and standardized. Hierarchical clustering was performed using 1-minus Pearson correlation, average linkage. (B) Volcano plot showing the number of differentially expressed genes with at least a 1.5-fold change and P < .001 between combined GFP+ and GFP− cells compared with control cells. (C) Overrepresentation analysis using the 276 genes with higher relative expression in both GFP+ and GFP− cells compared with controls with a P < .1 finds genes enriched in multiple pathways including cell adhesion and granulocyte activation. (D) Gene set enrichment analysis results for genes more highly expressed in controls than GFP+/− cells. ∗FDR < 0.05. (E) Gene set enrichment analysis results for genes more highly expressed in GFP+/− compared with controls. FDR, false discovery rate; NES, normalized enrichment score.
Similar gene expression profiles between GFP+ and GFP− liver macrophages. (A) Heat map displaying expression of the top 1500 expressed genes by RNA-seq. Fragments per kilobase of transcript per million mapped reads expression data are mean-centered and standardized. Hierarchical clustering was performed using 1-minus Pearson correlation, average linkage. (B) Volcano plot showing the number of differentially expressed genes with at least a 1.5-fold change and P < .001 between combined GFP+ and GFP− cells compared with control cells. (C) Overrepresentation analysis using the 276 genes with higher relative expression in both GFP+ and GFP− cells compared with controls with a P < .1 finds genes enriched in multiple pathways including cell adhesion and granulocyte activation. (D) Gene set enrichment analysis results for genes more highly expressed in controls than GFP+/− cells. ∗FDR < 0.05. (E) Gene set enrichment analysis results for genes more highly expressed in GFP+/− compared with controls. FDR, false discovery rate; NES, normalized enrichment score.
We compared GFP+ and GFP− macrophages together with the 2 control macrophage samples to assess differential gene expression. Gene set enrichment analysis for genes more highly expressed in controls revealed protein secretion, interferon-gamma, and interferon-alpha responses as the top pathways (Figure 6D). In contrast, the Wnt/β-catenin and transforming growth factor β pathways were most highly enriched in GFP+/− cells (Figure 6E). In general, inflammation-associated genes were not more highly expressed in liver macrophages from animals that underwent transplantation than in unmanipulated animals. Thus, the epigenetic and transcriptomic analyses suggest that many tissue-resident macrophages in adult NHPs are bone marrow derived, even long after transplantation, given the similarities between GFP+ cells from animals that underwent transplantation, which must be adult HSPC–derived, to both GFP− and control nontransplanted macrophages.
Turnover of tissue-resident macrophages at steady state
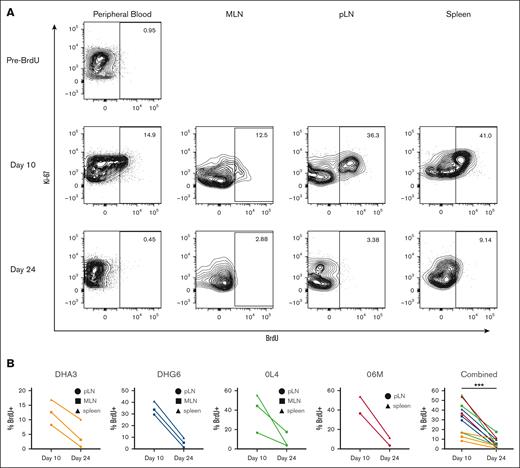
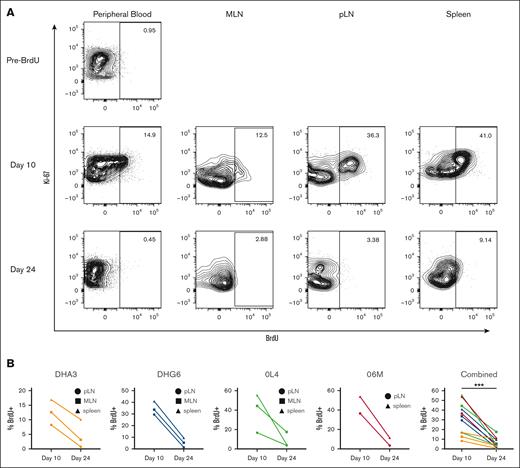
Our data suggest robust ongoing replacement of tissue-resident macrophages from genetically marked HSPCs after TBI conditioning and autologous transplantation, given the similar clonal patterns between tissue-resident macrophages and contemporaneous blood cells, reflecting multipotent LT-HSPCs rather than ST-HSPCs to be responsible for initial hematopoietic reconstitution. However, the impact of the conditioning regimen could confer conditions contributing to increased tissue myeloid cell turnover and replacement.10-12 To investigate the degree of tissue-resident macrophage turnover at steady state, independent of TBI, we infused 4 healthy, young adult, unmanipulated RMs with 30 mg/kg IV BrdU per day for 5 days. On day 10 after the start of infusion, we sampled the spleen and both peripheral and mesenteric lymph nodes. PB myeloid cells and tissue-resident macrophages were analyzed for genome incorporation of BrdU (Figure 7). To determine whether BrdU was lost from these cells either by cell death or ongoing proliferation, we sampled the same anatomic sites 24 days after initiation of BrdU (19 days after BrdU discontinuation; Figure 7). BrdU incorporation was detectable in an appreciable percentage of PB myeloid cells (41.4% ± 15.7% at day 7; 11.2% ± 4.3% at day 10) and, importantly, at high levels in tissue-resident macrophages at day 10 (31.7% ± 16.4%) but decreased to significantly lower levels by day 24 (6.4% ± 5.1%; Figure 7B). Although this type of analysis could not differentiate between ongoing replacement from HSPC-derived circulating cells vs local proliferation of tissue macrophages, the findings demonstrate rapid turnover of lymphoid tissue-resident macrophages at steady state and support the concept of continuous replacement of tissue-resident macrophages, even in the absence of conditioning.
Tissue macrophages turnover at steady state. Healthy, unmanipulated RMs were infused with BrdU intravenously for 5 days. Five days from the conclusion of administration (day 10) and 14 days later (day 24), blood and tissue cells were analyzed by flow cytometry to assess BrdU incorporation. (A) Representative plots showing percent BrdU+ cells of at least 1000 cells defined as single, live, CD45+, CD11b+, HLA-DR+, CD20− mononuclear cells from the same animal at each time point. (B) BrdU levels from all 4 healthy unmanipulated animals in tissue macrophages. ∗∗∗P < .001, using Wilcoxon matched-pairs test.
Tissue macrophages turnover at steady state. Healthy, unmanipulated RMs were infused with BrdU intravenously for 5 days. Five days from the conclusion of administration (day 10) and 14 days later (day 24), blood and tissue cells were analyzed by flow cytometry to assess BrdU incorporation. (A) Representative plots showing percent BrdU+ cells of at least 1000 cells defined as single, live, CD45+, CD11b+, HLA-DR+, CD20− mononuclear cells from the same animal at each time point. (B) BrdU levels from all 4 healthy unmanipulated animals in tissue macrophages. ∗∗∗P < .001, using Wilcoxon matched-pairs test.
Discussion
Macrophages regulate tissue immunity, orchestrating the initiation and resolution of antimicrobial immune response as well as the maintenance of tissue integrity. Macrophage ontogeny and life span have been the subject of intense interest given their broad functions and phenotypes in health and disease,39 including regulation of tissue repair,40 susceptibility to infection by DNA and RNA viruses,2 control of local tissue immune responses with an ability to retain epigenetic “memory” of prior infection,41 transformation to an immunosuppressive role by tumors,42 and neoplastic transformation.43 Such understanding is necessary to harness these processes for clinical benefit.
Studies in mice have revealed that embryonic macrophages are first directly generated in the yolk sac without monocytic precursors. Upon establishment of blood circulation, yolk sac macrophages seed the whole embryo,44 as revealed by murine fate mapping. A paradigm emerged suggesting that embryonically derived macrophages persist into adulthood and continue to constitute the bulk of the adult tissue macrophage compartment.45 Murine models with specific impairment of definitive but not primitive hematopoiesis revelated that yolk sac–derived cells can indeed contribute to tissue-resident macrophage populations in adult mice.14 Parabiosis and some depletion studies suggested that once seeded with embryonic macrophages, tissue-resident populations are maintained by local proliferation, not via contributions from circulating monocytes.46 However, some models instead suggest that murine tissue–resident macrophages in adult mice require ongoing replenishment from HSPCs,47 and the topic remains controversial.
Although murine studies have driven our fundamental understanding of immunological processes, mammalian development, and mechanisms underlying multiple diseases, accumulating data suggest that highly inbred and caged mice housed under SPF conditions can fail to capture important genetic and environmental factors that influence physiology,48 including tissue-resident macrophage biology. Tissue-resident macrophages in SPF mice have been shown to die and then rapidly repopulate under stress conditions. Iron or aspirin toxicity results in toxic death and subsequent repopulation of Kupffer cell liver-resident macrophages.11,12 Similarly, CSF1R signaling is critically important for the maintenance of microglial cells in the brain, and blockade of CSF1R lead to loss of >99% of microglial cells, with rapid replenishment from nestin+ cells.10
Administration of chemotherapeutic agents aimed at killing rapidly dividing neoplastic cells but also leading to the death of multiple HSPC-derived cells that proliferate in vivo can result in loss of microglial cells in the brain, which are subsequently replenished from peripheral myeloid cells.13 Taken together, it is clear that in mice, macrophages can be short lived and rapidly replenished from monocyte precursors in the PB in certain settings.
Given the drawbacks of extrapolating murine results to human biology and disease, it is of great importance to consider these questions for humans and outbred NHP models. To date, there have been few studies of the ontogeny of tissue macrophages in humans or primates. Macrophages have been confirmed to develop embryonically in the human fetal liver as assessed by single-cell transcriptomics;49 however, their ontogenic relationships were not clear, and it is unknown how long these cells survive. At least some liver macrophages can be long lived in humans, as shown by a small pool of donor Kupffer cells persisting in allogeneic liver transplants for over a decade,50 although recipient cells were also shown to contribute to the total macrophage population.
In the context of allogeneic HSPC transplantation, a few early studies used low-resolution methodologies in sex-mismatched donor–recipient pairs to find donor-type perivascular myeloid cells in the brain.51 However, graft-versus-host immune ablation of host-type HSPCs and of mature hematopoietic cells makes interpretation of allogeneic tissue-resident macrophage chimerism difficult to extrapolate to a nonallogeneic transplantation setting. However, the efficacy of allogeneic transplantation and of autologous HSPC gene therapy in the treatment of some central nervous system metabolic storage disorders such as adrenoleukodystrophy suggest that replacement of microglial cells and other tissue-resident macrophages by HSPC-derived cells is possible.52,53 An early retroviral gene marking study in RMs demonstrated GFP-expressing perivascular macrophages in the brain.54 Finally, a study analyzing macrophages in 1 pigtail macaque that underwent allogeneic hematopoietic stem cell transplantation found marking of multiple macrophage subsets in many tissues55; however, in the aforementioned study a drug resistance gene was introduced into transplanted HSPCs followed by repeated chemotherapy cycles to greatly enrich for transplanted cells. Moreover, preliminary analysis of animals that underwent transplantation with GFP-transduced HSPCs suggested that GFP+ macrophages were found within the brain.54 We were unable to longitudinally sample brain-resident macrophages, and further work is certainly merited.
Our study provides detailed and unequivocal evidence that significant percentages (up to 50%) of macrophages resident in the lung (BAL), liver, gastrointestinal tract, and lymphoid tissues can be derived from HSPC precursors in adult NHPs. Clonal tracking strongly supported continuous replacement of macrophages by circulating LT-HSPC–derived cells. Even in the absence of transplantation, labeling studies with BrdU demonstrated rapid steady-state turnover of tissue-resident macrophages. Importantly, given that the transduction efficiency of HSPCs is <100%,25,36 our analysis underestimates the total number of tissue-resident macrophages derived from adult HSPCs. Given the marked similarities of the epigenetic, transcriptomic, and phenotypic characteristics of HSPC-derived tissue macrophages to those in unmanipulated healthy NHPs, our data suggest that many tissue macrophages are adult HSPC derived in health and are not, universally, embryonically derived by adulthood.
These data are consistent with mathematical modeling of viral production decay rates after macrophage-tropic HIV/simian immunodeficiency virus infections of immunocompromised mice that underwent transplantation with a human immune system or of NHPs.56,57 Virus-infected macrophages were shown to have a half-life as short as 1 day. In summary, these results stand to inform strategies to eliminate HIV reservoirs in infected humans.
It remains possible that some tissue-resident GFP− macrophages in our NHPs that underwent transplantation are extremely long lived and represent residual embryonic macrophage–derived cells. Our analysis was by no means completely comprehensive for all anatomic sites in which macrophages reside. Moreover, given the high cost and long periods of time required for these types of experiments, we were unable to determine whether animal sex influenced the frequency of HSPC development into tissue-resident macrophages or whether sex influenced the epigenetic or transcriptomic landscape of the tissue-resident macrophages. With 2 female animals in our cohort (1 that received transplantation and 1 unmanipulated control animal; Table 1), we found significant variation in transcriptomic landscapes of the macrophages we sampled. Understanding the mechanisms responsible for these differences, particularly in outbred animals with significantly different immunological experiences will be important to understand. Irrespective, our results demonstrate that HSPCs continuously develop into tissue-resident macrophages in adult NHPs and that these cells turnover at steady state. These findings will aid the development of therapeutic interventions aimed at attenuating the function and life span of tissue-resident macrophages in health and disease.
Acknowledgments
The authors thank the staff of the National Heart, Lung, and Blood Institute, National Institute of Allergy and Infectious Diseases, and National Institutes of Health Divisions of Veterinary Resources nonhuman primate facilities for skilled animal care and tissue sampling, and the National Heart, Lung, and Blood Institute DNA Sequencing and Genomics and Flow Cytometry core facilities.
Funding for this study was provided by the Divisions of Intramural Research of the National Institute of Allergy and Infectious Diseases and the National Heart, Lung, and Blood Institute.
The content of this publication does not necessarily reflect the views or policies of the US government, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US government.
Authorship
Contribution: A.R.R., C.W., S.G.H., L.P., H.D.H., and J.M.B. performed experiments for the manuscript; A.R.R., C.W., L.P., T.E.M., and H.D.H. analyzed the data and created the figures; J.M.B. and C.E.D. conceived and supervised the study; and all authors contributed to the writing and editing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cynthia E. Dunbar, National Institutes of Health Translational Stem Cell Biology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bldg 10 CRC, Room 5E-3332 9000, Bethesda, MD 20892; email: dunbarc@nhlbi.nih.gov; and Jason M. Brenchley, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bldg 04, Room 201, Bethesda, MD 20892; email: jbrenchl@mail.nih.gov.
References
Author notes
∗A.R.R. and C.W. contributed equally to this study.
†C.E.D. and J.M.B. contributed equally to this study.
Sequencing data have been deposited in the Gene Expression Omnibus database (accession number GSE246351).
Data are available on request from the corresponding authors, Jason M. Brenchley (jbrenchl@mail.nih.gov) and Cynthia E. Dunbar (dunbarc@nhlbi.nih.gov).
The full-text version of this article contains a data supplement.

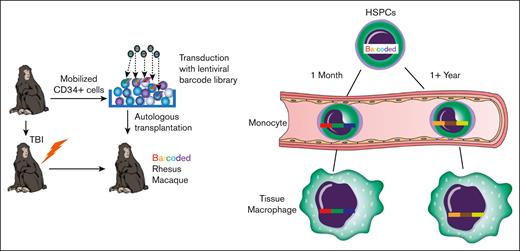

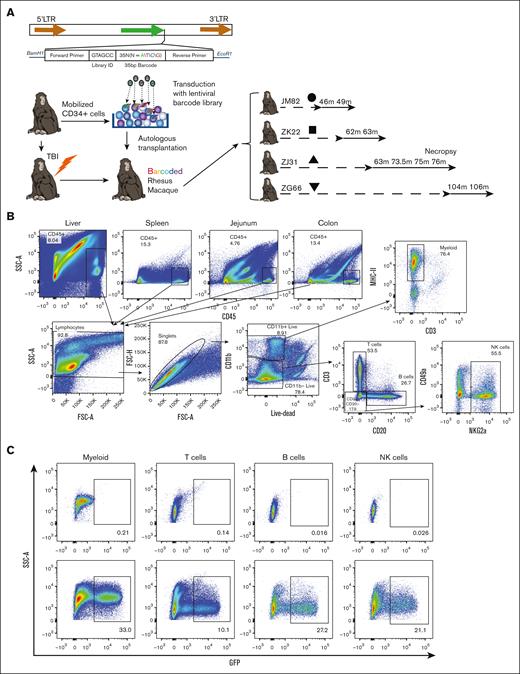
![RM autologous barcoded HSPC transplantation and detection of GFP+ cells in tissues. (A) RM HSPCs were transduced with a barcoded lentiviral library and transplanted into the autologous macaque after conditioning with 1000 rads TBI. The blood and tissue were sampled periodically to assess the clonal composition of HSPC-derived cells. (B) Representative flow cytometric gating strategy to measure GFP expression among leukocyte subsets. (C) The expression of GFP was determined by flow cytometry in tissue mononuclear cell suspensions after gating for CD45+ cells and lymphocytes and then lineage-defining markers. Representative liver samples from an unmanipulated control animal (top) and a sample from a barcoded animal ZJ31 collected 76 months after transplantation (bottom). (D) The percent GFP+ hematopoietic cells in the liver, gastrointestinal tract, or lymphoid tissues for the listed cell lineages is shown. Each individual animal is designated with a different symbol, including those that match animals reported in panel A, and a line connects samples obtained at the same time point. Tissue-resident myeloid cell percentage GFP positivity was not significantly different compared with B or natural killer (NK) cells in the same tissue and was significantly higher than in T cells in the tissue (using 2-way analysis of variance [ANOVA] with Dunnett multiple comparisons test, lines compare tissue-resident myeloid cells with other leukocyte subsets); ∗P < .05; ∗∗P < .01. GI, gastrointestinal; LN, lymph node.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/3/10.1182_bloodadvances.2023011499/2/m_blooda_adv-2023-011499-gr1d.jpeg?Expires=1768013633&Signature=4JbdDZDYTIiNXridYNEFGmOsL~0ibDeCpmvTZeYDz8AbBoSB1hQljvPs3PpZPf3hn6ULpzt4FGMmrTXB-7A4DOg9bswd5gZhzO~FaJU7Y~tUmUQUwB4ZaiJ0eCVV29h-Jxs6HVS9jPrAd3f-BrglM7eLkMtaaTadgI00HvH~XBcy0oqzfqvEb4oHCTs2dT~I6uyON2BjoCdMlDqPv6cjfmSObe~-0KGjNf8sJaAO~XZRKLKM9Lf7oZNawn3I-JxNgZF76QxGm2HMBLQlQFuC5RiGegxLmw9BMBX1qAvgCitl4UOqNyqdLCxze7NKs6xDhRvkWlwUCWOh-UR5Vi~lsQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)