Key Points
BDCPs, including NBEA, LRBA, NBEAL1, and LYST, are required for efficient hematopoietic reconstitution after bone marrow transplantation.
NBEA regulates NOTCH receptor turnover in hematopoietic cells.
Visual Abstract
Hematopoietic stem cells (HSCs) can generate all blood cells. This ability is exploited in HSC transplantation (HSCT) to treat hematologic disease. A clear understanding of the molecular mechanisms that regulate HSCT is necessary to continue improving transplant protocols. We identified the Beige and Chediak-Higashi domain–containing protein (BDCP), Neurobeachin (NBEA), as a putative regulator of HSCT. Here, we demonstrated that NBEA and related BDCPs, including LPS Responsive Beige-Like Anchor Protein (LRBA), Neurobeachin Like 1 (NBEAL1) and Lysosomal Trafficking Regulator (LYST), are required during HSCT to efficiently reconstitute the hematopoietic system of lethally irradiated mice. Nbea knockdown in mouse HSCs induced apoptosis and a differentiation block after transplantation. Nbea deficiency in hematopoietic progenitor cells perturbed the expression of genes implicated in vesicle trafficking and led to changes in NOTCH receptor localization. This resulted in perturbation of the NOTCH transcriptional program, which is required for efficient HSC engraftment. In summary, our findings reveal a novel role for NBEA in the control of NOTCH receptor turnover in hematopoietic cells and supports a model in which BDCP-regulated vesicle trafficking is required for efficient HSCT.
Introduction
Hematopoietic stem cells (HSCs) are exploited routinely via bone marrow (BM) transplantation to treat hematologic disease and patients undergoing high-intensity chemotherapy. The ability of HSCs to reconstitute a healthy hematopoietic system depends on their vast self-renewal and differentiation potential, as well as their engagement with the BM niche.1-4 To identify novel molecular mechanisms that allow for efficient repopulation of the hematopoietic system by HSCs, we previously performed a transplantation-based screen for novel molecular regulators of HSC transplantation (HSCT).5 Thereby, we identified Neurobeachin (NBEA; encoded by the Nbea gene) as a putative positive regulator of HSCT.5
NBEA is a member of the Beige and Chediak-Higashi (BEACH) domain–containing protein (BDCP) family, which consists of 9 members: lipopolysaccharide-responsive beige-like anchor protein (Lrba), lysosomal trafficking regulator (Lyst), Nbea, Neurobeachin-like 1 (Nbeal1), Neurobeachin-like 2 (Nbeal2), neutral sphingomyelinase activation-associated factor (Nsmaf), WD and FYVE zinc finger domain–containing protein 3 (Wdfy3), Wdfy4, and WD repeat domain 81 (Wdr81) (reviewed by Cullinane et al6). BDCPs are generally large proteins of mostly unknown function and are defined by the presence of the BEACH7-9 domain, a ∼280 amino acid stretch whose specific role is largely unknown.6 The BEACH domain was first identified in LYST and is mutated in Chediak-Higashi syndrome, resulting in lysosome defects.8 BDCPs have been implicated as regulators of protein and vesicle trafficking by acting as scaffolding proteins that facilitate interactions between membranes, such as vesicle fusion and fission.6 They can also regulate the cellular trafficking of specific cargo via homologous or heterologous interactions with other BDCPs.6,10 Consistently, LYST, NBEA, and NBEAL2 affect the formation and secretion of platelet granules.6,11 BDCPs have also been implicated in additional cellular processes, such as apoptosis (Nsmaf), autophagy (Lyst, Wdfy3, and Lrba), synaptic transmission (Nbea), granule size (Lyst, Nbeal2, and Nbea), and lysosome size (Lyst and Nsmaf).6 Mutations in BDCPs have been linked to multiple human syndromes, including Chediak-Higashi syndrome (LYST),7-9 autism (NBEA),12-14 glioma (NBEAL1),15 gray platelet syndrome (NBEAL2),16-18 autoimmunity (LRBA and WDFY4),19,20 and syndrome of quadrupedal locomotion, mental retardation, and cerebro-cerebellar hypoplasia (WDR81)21 (reviewed in Cullinane et al6).
Consistent with a putative role in vesicle and protein trafficking, NBEA localizes mostly near the Golgi apparatus.22,Nbea-–/– mice develop normally in utero but die perinatally due to impaired respiration caused by poor synaptic transmission.22-24 Patients with autism with a disrupted NBEA gene also display platelets with abnormal morphology and dense granules.25 Interestingly, the NBEA locus on human chromosome 13 constitutes a fragile site in multiple myeloma cell lines that express high levels of NBEA,26,27 suggesting a potential role in hematopoietic cellular transformation. However, it is unclear whether perturbed NBEA expression is a passenger event in these patients.6,26,27
Here, we show that NBEA is required to maintain homeostasis in the hematopoietic system by regulating proper hematopoietic stem and progenitor cell (HSPC) differentiation. Moreover, our data reveal that multiple BDCP family members, including NBEA, LRBA, LYST, and NBEAL1 function as positive regulators of HSCT, because their acute knockdown compromises HSC in vivo hematopoietic repopulating activity. Furthermore, Nbea knockdown results in a loss of HSC survival and a block in differentiation after transplantation. Loss of Nbea perturbed NOTCH receptor turnover and transcriptional programs in hematopoietic cell lines. Thus, we propose a model in which BDCPs play a role in HSCT by regulating NOTCH signaling via their role in vesicle trafficking patterning.
Methods
Mice
Nbea–/– mice were kindly provided by Manfred W. Kilimann (Department of Otolaryngology, Göttingen University Medical Center; and Department of Molecular Neurobiology, Max-Planck Institute for Experimental Medicine, Göttingen, Germany). C57BL/6J and C57BL/6.SJL-PtprcaPep3b/BoyJ mice were acquired from The Jackson Laboratory (Bar Harbor, ME) and housed in a pathogen-free facility. All animal experiments were carried out according to procedures approved by the St. Jude Children’s Research Hospital Institutional Animal Care and Use Committee and complied with all relevant ethical regulations regarding animal research.
Genotyping
Genotyping of Nbea allele was done as previously described.24 Details are provided in supplemental Materials and Methods.
Transplants
For all transplants, 8- to 12-week-old CD45.2+/CD45.1+ C57BL/6J recipients were treated with 11 Gy of ionizing radiation in split doses of 5.5 Gy before transplant. All cells were transplanted by tail vein injection along with CD45.1+ C57BL/6.SJL BM cells into recipients. Please see details in text for specific cell doses for each experiment. For secondary transplantation, 5 × 106 whole bone marrow (WBM) cells were transplanted from primary recipients.
PB analysis
Peripheral blood (PB) was collected, stained, and analyzed as previously described.28 Details are provided in supplemental Materials and Methods.
Cell cycle analysis
After staining for surface antigens, cells were fixated with Cytofix/Cytoperm kit (BD Biosciences, San Diego, CA) and stained with Ki-67 and DAPI for cell cycle analysis.
Apoptosis analysis
To assess apoptosis, staining for surface proteins with fluorescently labeled antibodies was followed by labeling with AnnexinV-FITC (fluorescein isothiocyanate) (BD Biosciences) and DAPI (4',6-diamidino-2-phenylindole).
Cell culture
293T cells were cultured in Dulbecco minimal essential medium with 10% fetal calf serum. HSPCs were cultured in serum-free expansion medium (Stem Cell Technologies, Vancouver, BC, Canada) with 10 ng/mL recombinant murine (rm) stem cell factor, 20 ng/mL rm thrombopoietin, 20 ng/mL rm insulin-like growth factor 2 (Peprotech, Rocky Hill, NJ), and 10 ng/mL recombinant human fibroblast growth factor 1 (R&D Systems, Minneapolis, MN). OPM2 cells were cultured in RPMI1640 medium with 10% fetal calf serum.
Lentiviral vector preparation and lentiviral transduction
Vesicular stomatitis virus glycoprotein pseudotyped lentiviruses were prepared and collected as previously described.5,29 Nontissue culture–treated 96-well plates were spin loaded with lentiviral particles, and 15 000 cells were loaded in serum-free expansion medium, as previously described.5,29 Details are provided in supplemental Materials and Methods.
Isolation of HSPCs
BM cells were harvested from femurs, tibias, and pelvic bones of 6- to 10-week-old mice by crushing. c-Kit+ cells were enriched magnetically using anti–c-Kit microbeads (Miltenyi Biotech, San Diego, CA). Cells were then stained with fluorescently conjugated antibodies for lineage markers (B220, CD3, CD8, CD19, Gr-1, and TER119), SCA-1, and c-KIT and sorted on a FACSAria III (BD Biosciences). DAPI (Sigma-Aldrich) was used to exclude dead cells.
qRT-PCR analysis
Total RNA was isolated and reverse transcribed, and quantitative reverse transcription polymerase chain reaction (qRT-PCR) was carried out, as previously described.5,29 Details are provided in supplemental Materials and Methods. Differences in complementary DNA input were compensated by normalizing against Tbp or GAPDH expression levels, and changes in gene expression were calculated using the delta-delta Ct (DDCt) method. Primer sequences are provided in supplemental Table 1.
shRNAs
Short hairpin RNAs (shRNAs) were designed as previously described.29 Gene knockdown efficiency in HSPCs was quantified by qRT-PCR and normalized to transduction frequency. shRNA sequences are provided in supplemental Table 2.
Transcriptional profiling (RNA sequencing) and mRNA expression analysis
Isolated messenger RNA (mRNA) was sequenced at Vantage (Vanderbilt University Medical Center, Nashville, TN). mRNA expression was analyzed using R and Limma software packages. Gene set enrichment analysis was performed using the online tool WebGestalt30 and Ingenuity Pathway Analysis software (QIAGEN). Details are provided in supplemental Materials and Methods. The complete RNA sequencing data set was deposited in the Sequence Read Archive (BioProject ID: PRJNA1052655).
NOTCH flow cytometry analysis
OPM2 cell cultures were split into 2 samples for cell surface and total protein analysis. Total protein analysis samples were fixed and permeabilized following Cytofix/Cytoperm kit instructions (BD Biosciences). All samples were stained with anti-NOTCH1 (clone A6; Invitrogen) and anti-NOTCH2 (clone 605724; Invitrogen) primary antibodies. After washing the primary antibodies, all samples were incubated with anti-mouse Alexa488 and anti-rat Cy3 (cyanine3) secondary antibodies. Individual controls for primary and secondary antibodies were used for every experiment.
Immunofluorescence
OPM2 cells were fixed, permeabilized, blocked, and incubated with previously conjugated anti-NBEA (NBP1-90004; Novus Biologicals) and anti-RCAS1 (Receptor-binding cancer antigen expressed on SiSo cells) (clone D2B6N; Cell Signal). Cells were mounted and imaged on a Zeiss LSM780 confocal microscope (Zeiss, Oberkochen, Germany) using a ×63, 1.1NA oil immersion lens. Details are provided in supplemental Materials and Methods.
Statistics
Summary statistics, including mean and standard deviation, were reported for analyses. A two-sample t test, exact Wilcoxon rank-sum test, or Mann-Whitney test was used to test for differences between two groups, depending on the normality of the data, which was tested by the Shapiro-Wilk test. False discovery rate method, developed by Benjamini and Hochberg,31 was used to correct for multiple comparisons at a level of 0.05. Otherwise, P values <.05 were considered statistically significant. Analyses were conducted in R-3.3.1 and Prism. Sample size and experimental replicates are detailed in each figure legend.
CFU assays
For analysis of colony-forming unit (CFU) potential, cells were sorted and plated in M3434 methylcellulose (STEMCELL Technologies, Vancouver, Canada). Colonies were scored 10 days after plating.
Results
NBEA is a positive regulator of HSPC hematopoietic repopulating activity
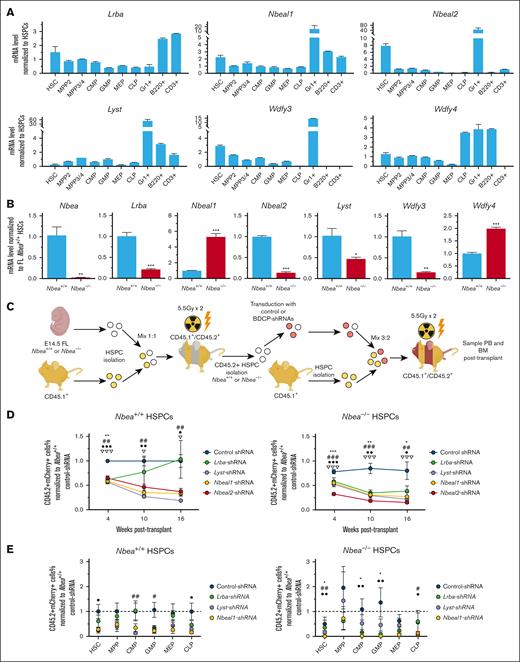
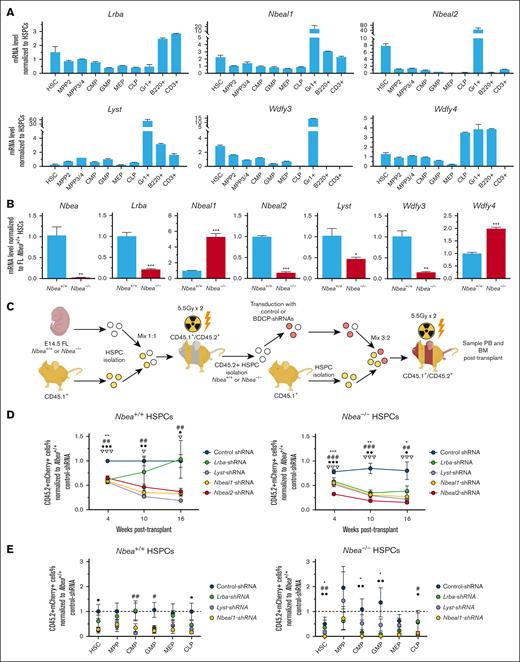
In a transplantation-based screen for regulators of HSCT, we identified 17 putative novel regulators of stable hematopoietic engraftment, including the BDCP and NBEA.5 In this previous study, shRNA-mediated knockdown of Nbea in HSPCs inhibited their ability to reconstitute hematopoiesis when transplanted into lethally irradiated mice.5 However, it is currently unknown how NBEA regulates HSCT and whether other related BDCP family members also play a role in this process. To explore these questions, we first examined Nbea expression in fetal liver (FL) and adult BM HSPCs. We observed that Nbea is most highly expressed by long-term HSCs (LT-HSCs; Lineage– SCA-1+c-KIT+CD150+CD48–) in adult BM, relative to committed progenitors and mature PB cells (Figure 1A). This pattern is preserved during fetal hematopoiesis, in which Nbea expression is also restricted to LT-HSCs in the E14.5 FL (supplemental Figure 1A). These data suggest a specific role for NBEA in HSCs.
Nbea is a positive regulator of HSPC repopulating activity. (A) qRT-PCR of Nbea expression in hematopoietic cells from adult BM, including HSC (Lineage–SCA-1+c-KIT+[LSK]CD150+CD48–); MPPs (LSKCD150+CD48+ [MPP2] and LSKCD150–CD48+ [MPP3/4]); common myeloid progenitors (CMPs; Lineage–SCA-1–c-KIT+CD32/16LowCD34+); common lymphoid progenitors (CLPs; Lineage–c-KitmedSca-1medCD127+); megakaryocyte-erythroid progenitors (MEPs; Lineage–SCA-1–c-KIT+CD32/16–CD34–); granulocyte-myeloid progenitors (GMPs; Lineage–SCA-1–c-KIT+CD32/16–CD34–); granulocytes, (GR-1+), B cells (B220+), and T cells (CD3+). (B) CD45.2+ “test” BM-HSPCs were transduced with control or Nbea-shRNAs. mCherry+ cells were then sorted and transplanted along with mock-transduced CD45.1+ HSPCs at a 1:1 ratio into lethally irradiated CD45.1+/CD45.2+ recipients within 44 hours after isolation. Recipient PB was examined for >16 weeks after transplant by flow cytometry for CD45.2+ mCherry+ cells. (Bi) Schematic of competitive transplantation of Nbea-deficient BM-HSPCs. (Bii) Percentage of CD45.2+mCherry+ PB at 4 to 16 weeks after transplant. (Biii) Frequency of CD45.2+mCherry+ PB lineages at 4 to 16 weeks after transplant. (Biv) Contribution of CD45.2+mCherry+ progenitors to BM progenitors. Data in panel B are from 3 experiments; 5 mice per condition per transplant. Data are represented as mean ± standard error of the mean (SEM). ∗P < .05; ∗∗P < .005; ∗∗∗P < .001, relative to recipients of control-shRNA HSPCs.
Nbea is a positive regulator of HSPC repopulating activity. (A) qRT-PCR of Nbea expression in hematopoietic cells from adult BM, including HSC (Lineage–SCA-1+c-KIT+[LSK]CD150+CD48–); MPPs (LSKCD150+CD48+ [MPP2] and LSKCD150–CD48+ [MPP3/4]); common myeloid progenitors (CMPs; Lineage–SCA-1–c-KIT+CD32/16LowCD34+); common lymphoid progenitors (CLPs; Lineage–c-KitmedSca-1medCD127+); megakaryocyte-erythroid progenitors (MEPs; Lineage–SCA-1–c-KIT+CD32/16–CD34–); granulocyte-myeloid progenitors (GMPs; Lineage–SCA-1–c-KIT+CD32/16–CD34–); granulocytes, (GR-1+), B cells (B220+), and T cells (CD3+). (B) CD45.2+ “test” BM-HSPCs were transduced with control or Nbea-shRNAs. mCherry+ cells were then sorted and transplanted along with mock-transduced CD45.1+ HSPCs at a 1:1 ratio into lethally irradiated CD45.1+/CD45.2+ recipients within 44 hours after isolation. Recipient PB was examined for >16 weeks after transplant by flow cytometry for CD45.2+ mCherry+ cells. (Bi) Schematic of competitive transplantation of Nbea-deficient BM-HSPCs. (Bii) Percentage of CD45.2+mCherry+ PB at 4 to 16 weeks after transplant. (Biii) Frequency of CD45.2+mCherry+ PB lineages at 4 to 16 weeks after transplant. (Biv) Contribution of CD45.2+mCherry+ progenitors to BM progenitors. Data in panel B are from 3 experiments; 5 mice per condition per transplant. Data are represented as mean ± standard error of the mean (SEM). ∗P < .05; ∗∗P < .005; ∗∗∗P < .001, relative to recipients of control-shRNA HSPCs.
To confirm a role for NBEA in adult HSCs, we transduced adult BM HSPCs (Lineage–SCA-1+c-KIT+ cells) with shRNAs targeting Nbea (Figure 1B). For all shRNAs, we used microRNA30 (miR30)-embedded shRNAs driven by MSCV (murine stem cell virus) promoter in a lentiviral vector that also expresses mCherry from an independent PGK (phosphoglycerate kinase1) promoter (supplemental Figure 1B). Forty-four hours after transduction with Nbea-shRNA, mCherry+ adult CD45.2+ BM HSPCs (ie, “test” HSPCs) were transplanted along with CD45.1+ BM competitor HSPCs into lethally irradiated CD45.1+/CD45.2+ recipient mice (Figure 1B). Although 2 Nbea-shRNAs were tested, Nbea-shRNA “A” was used in all experiments, because it had no effect on Nbea–/– HSPCs while inhibiting the repopulating activity of Nbea+/+ HSPCs (supplemental Figure 1C-E). Particularly, to determine the presence of off-target effects driven by the Nbea-shRNAs, Nbea+/+ and Nbea–/– CD45.2+ HSPCs were recovered from CD45.1+/CD45.2+ recipients of E14.5 FL-HSPCs, transduced with control- or Nbea-shRNAs and transplanted into congenic CD45.1+/CD45.2+ recipients, demonstrating that Nbea-shRNA “A” did not affect the BM reconstitution ability of Nbea–/– HSPCs (supplemental Figure 1E). BM Nbea-deficient HSPCs display a dramatic loss of both short- and long-term hematopoietic repopulating activity, relative to HSPCs transduced with control shRNAs (Figure 1B; supplemental Figure 2A). Similar results were obtained when FL HSPCs were transduced with Nbea-shRNA (supplemental Figures 1D-E and 2A). Thus, these data indicate that Nbea is required by HSPCs at multiple developmental stages for efficient HSCT.
Nbea-deficient HSPCs display reduced survival and a block in differentiation after transplant
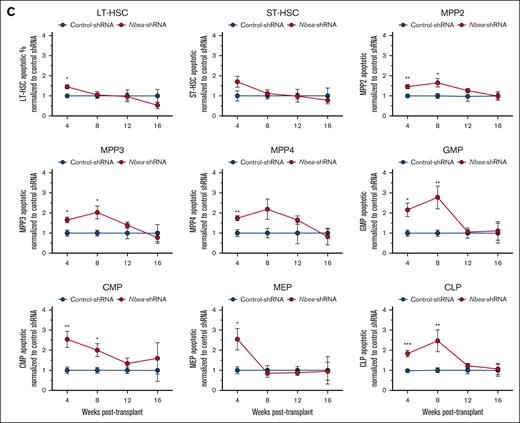
To explore why Nbea-deficient HSPCs fail to contribute robustly to the blood of transplanted recipients, CD45.2+ BM HSPCs transduced with either control- or Nbea-shRNA were transplanted into lethally irradiated CD45.1+CD45.2+ recipients 44 hours after transduction. Cohorts of recipients were then analyzed for CD45.2+ BM HSPCs at 10 days, 20 days, 4 weeks, 8 weeks, 12 weeks, and 16 weeks after transplant (Figure 2A; supplemental Figures 2B and 3). We did not observe significant differences in the acute contribution of Nbea-deficient HSPCs to reconstituted BM HSPCs at 10 and 20 days after transplant (supplemental Figure 3A). Importantly, we did not detect differences in the contribution of Nbea-deficient HSPCs to LT-HSCs and short-term HSCs (ST-HSCs), even up to 16 weeks after transplant (Figure 2A). However, we did observe that Nbea-deficient HSPCs progressively failed to contribute to committed downstream progenitors (multipotent progenitors [MPPs], common myeloid progenitors, granulocyte-myeloid progenitors, megakaryocyte-erythroid progenitors, and common lymphoid progenitors) relative to controls over time after transplant (Figure 2A). Cumulatively, these data indicate that Nbea-deficient HSPCs acutely engraft and generate BM HSPCs but fail to contribute to the blood efficiently after transplant (Figure 1Bii-iii), suggesting a potential defect in differentiation. In parallel, we found that Nbea-deficient LT-HSCs, ST-HSCs, MPP2, and MPP3 showed distinct CFU potential compared with controls (Figure 2B). Nbea-deficient progenitors produced fewer (Figure 2Bi) and smaller colonies (Figure 2Bii) than controls. These differences were more significant for LT-HSCs and ST-HSCs (Figure 2Bi-ii). Additionally, although not statistically significant, numbers of the most multipotent colony type (granulocyte, erythrocyte, monocyte, and megakaryocyte) from Nbea-deficient LT-HSCs and ST-HSCs trended down (Figure 2Biii).
To further test the consequences of Nbea knockdown in transplanted HSPCs, we examined cell cycle status and apoptosis in CD45.2+mCherry+ BM of mice that received transplant. As expected, no differences were seen in the level of apoptotic cells or cell cycle status of Nbea-deficient HSPCs relative to controls acutely after transplant (supplemental Figure 3B-C). However, by 4 weeks after transplant, we observed an increase in the frequency of apoptotic cells in most Nbea-deficient reconstituted hematopoietic progenitors relative to controls (Figure 2C). Interestingly, more apoptotic cells were apparent and persisted longer after transplant in downstream and committed progenitor compartments than HSCs (Figure 2C). No differences in cell cycle status of Nbea-deficient HSPCs after transplant were observed, relative to controls (supplemental Figure 3D).
Overall, our data reveal that Nbea-deficient HSPCs stably engraft the BM but exhibit a differentiation block downstream of multipotent and committed progenitors. Nbea-deficient derived progenitors also display a survival loss over time, which exacerbates as they transition through the hematopoietic hierarchy.
NBEA is dispensable for native embryonic hematopoiesis
We next examined a role for Nbea in hematopoiesis by interrogating Nbea–/– mice.24,Nbea–/– mice die perinatally due to lack of synaptic transmissions,22-24 precluding an analysis of hematopoiesis in Nbea–/– adult mice. Hence, we examined E14.5 FL, because transplantable HSCs are concentrated in the FL at this developmental stage.32-35 Here, we observed no differences in the frequency of HSPCs in Nbea–/– and Nbea+/+ littermates (supplemental Figure 4A). Additionally, NBEA loss did not perturb FL HSPC in vitro differentiation (supplemental Figure 4B). To test the ability of FL Nbea–/– HSPCs to repopulate the hematopoietic system, 300 to 600 CD45.2+ E14.5 FL Nbea–/– or Nbea+/+ HSPCs were transplanted into lethally irradiated congenic CD45.1+CD45.2+ mice, along with 2 × 105 CD45.1+ WBM cells (supplemental Figure 4C-E). FL Nbea–/– HSPCs displayed multilineage repopulating activity equivalent to that of FL Nbea+/+ HSPCs isolated from littermate controls (supplemental Figure 4D). Although not statistically significant, secondary transplantation of WBM from primary recipients of E14.5 CD45.2+ FL Nbea–/– HSPCs resulted in a modest reduction of multilineage PB output relative to controls (supplemental Figure 4E). Because serial transplantation imposes additional regenerative stress on HSPCs and only HSPCs capable of preserving self-renewal potential after primary transplantation persist to repopulate secondary recipients,28,36-40 these suggest a potential role for Nbea in the maintenance of durable self-renewing HSCs. However, additional BDCP family members may functionally compensate for Nbea loss in this genetic model.
Nbea, Lrba, Lyst, and Nbeal1 BDCPs play overlapping roles in HSPC engraftment ability
Because genetic loss of Nbea only modestly perturbed their function, we next interrogated BM and FL HSPCs for the expression of additional BDCP family members (Figure 3A; supplemental Figure 4F). Here, we focused on the BDCPs structurally similar to NBEA, including LRBA, LYST, NBEAL1, NBEAL2, WDFY3, and WDFY4.6,Lrba was highly expressed by B and T cells in adult mice (Figure 3A), consistent with a role in these cell types.19,41 Lrba was also upregulated in mouse HSCs, compared with downstream HSPCs (Figure 3A). As expected, Lyst expression was upregulated in BM granulocytes42 (Figure 3A). Nbeal1, Nbeal2, Wdfy3, and Wdfy4 expression was also upregulated in granulocytes.
Lrba, Lyst, and Nbeal1 are positive regulators of BM-HSPC engraftment. (A) qRT-PCR of Nbea, Lrba, Lyst, Nbeal1, Nbeal2, Wdfy3, and Wdfy4 mRNA expression in the hematopoietic hierarchy of adult BM. (B) qRT-PCR of Nbea, Lrba, Lyst, Nbeal1, Nbeal2, Wdfy3, and Wdfy4 expression in Nbea+/+ and Nbea–/– E14.5 FL-HSCs. (C-E) Nbea+/+ and Nbea−/− CD45.2+ HSPCs recovered from CD45.1+/CD45.2+ recipients of E14.5- CD45.2+ FL-HSPCs 4 months after transplant were transduced with shRNAs targeting Lrba, Lyst, Nbeal1, or Nbeal2 and transplanted together with CD45.1+ HSPCs into CD45.1+/CD45.2+ (ratio 3:2). (C) Experimental schematic. (D) Percentage of mCherry+CD45.2+ PB after transplant of recipients of shRNA treated cells normalized to the average of control recipients. (E) Percentage of mCherry+CD45.2+ chimerism in the BM HSPCs at 20 weeks after transplant. Data are represented as mean ± SEM. ∗/#/•/∇P < .05; ∗∗/##/••/∇∇P < .005; ∗∗∗/###/•••/∇∇∇P < .001, relative to recipients of control-shRNA HSPCs. ∗ refers to Lrba-shRNA; # refers to Nbeal1-shRNA; • refers to Nbeal2-shRNA; and ∇ refers to Lyst-shRNA.
Lrba, Lyst, and Nbeal1 are positive regulators of BM-HSPC engraftment. (A) qRT-PCR of Nbea, Lrba, Lyst, Nbeal1, Nbeal2, Wdfy3, and Wdfy4 mRNA expression in the hematopoietic hierarchy of adult BM. (B) qRT-PCR of Nbea, Lrba, Lyst, Nbeal1, Nbeal2, Wdfy3, and Wdfy4 expression in Nbea+/+ and Nbea–/– E14.5 FL-HSCs. (C-E) Nbea+/+ and Nbea−/− CD45.2+ HSPCs recovered from CD45.1+/CD45.2+ recipients of E14.5- CD45.2+ FL-HSPCs 4 months after transplant were transduced with shRNAs targeting Lrba, Lyst, Nbeal1, or Nbeal2 and transplanted together with CD45.1+ HSPCs into CD45.1+/CD45.2+ (ratio 3:2). (C) Experimental schematic. (D) Percentage of mCherry+CD45.2+ PB after transplant of recipients of shRNA treated cells normalized to the average of control recipients. (E) Percentage of mCherry+CD45.2+ chimerism in the BM HSPCs at 20 weeks after transplant. Data are represented as mean ± SEM. ∗/#/•/∇P < .05; ∗∗/##/••/∇∇P < .005; ∗∗∗/###/•••/∇∇∇P < .001, relative to recipients of control-shRNA HSPCs. ∗ refers to Lrba-shRNA; # refers to Nbeal1-shRNA; • refers to Nbeal2-shRNA; and ∇ refers to Lyst-shRNA.
Significantly, Nbeal1, Nbeal2, and Wdfy3 were upregulated in BM and FL HSCs relative to HSPCs (Figure 3A; supplemental Figure 4F).
To assess possible genetic compensation by BDCP family members, we examined Lrba, Nbea, Nbeal1, Nbeal2, Wdfy3, and Wdfy4 expression in E14.5 FL Nbea–/– HSCs (Figure 3B). Lrba was downregulated in Nbea–/– HSCs. Interestingly, Nbeal1 is highly upregulated in Nbea–/– HSCs, whereas Nbeal2 is downregulated in Nbea–/– HSCs. These data highlight a complex transcriptional response of BDCP family members to genetic loss of Nbea mRNAs.43
To functionally evaluate any compensatory role for BDCP family members in Nbea–/– HSPCs, we individually knocked down each of the 4 BDCPs structurally most similar to Nbea in Nbea–/– cells (i.e., Lrba, Lyst, Nbeal1, and Nbeal2).6 To do this, 4 months after transplant, Nbea+/+ and Nbea–/– CD45.2+ HSPCs were isolated from primary recipients of E14.5 FL Nbea+/+ and Nbea–/– HSPCs and lentivirally transduced with shRNAs targeting Lrba, Lyst, Nbeal1, or Nbeal2 (Figure 3C-D; supplemental Figure 4G). As expected, Nbea–/– FL-HSPCs transduced with control shRNAs displayed a mild loss of serial repopulating activity (Figure 3C-D). Knockdown of Lrba, Lyst, Nbeal1, and Nbeal2 impaired the short- and long-term serial repopulating activity of Nbea+/+ FL-HSPCs (Figure 3C-D). The negative effect of Lrba knockdown was transient, because these cells repopulated PB similar to control by 16 weeks after transplant. This effect was exacerbated when these genes were knocked down in FL Nbea–/– HSPCs (Figure 3C-D), demonstrating the essential role of this family of proteins in HSPC transplantation and functional redundancy with Nbea.
NBEA effects a NOTCH transcriptional program in hematopoietic cells via regulation of NOTCH cell surface turnover
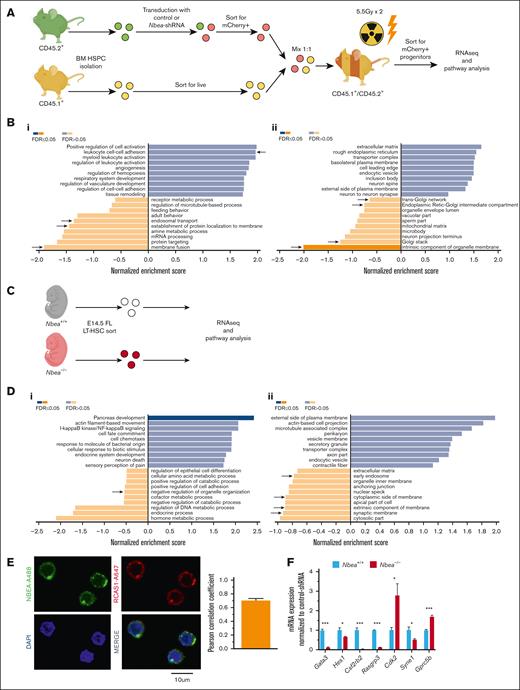
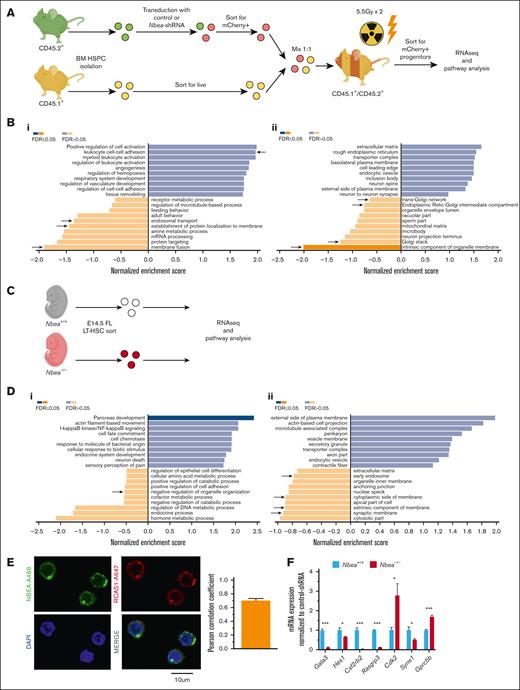
To unveil the role of Nbea in hematopoiesis, we isolated and amplified total mRNA from Nbea-shRNA–derived LT-HSCs (Figure 4A). Differential gene expression showed a general downregulation of transcriptional activity in Nbea-deficient cells compared with control (supplemental Figure 5A). Although very few pathway changes met the critical threshold of <0.05 false discovery rate, pathway analysis revealed alterations in gene sets related to vesicle traffic, membrane regulation, and Golgi activity, consistent with NBEA’s known roles in vesicle trafficking control6 (Figure 4B). We also examined global gene expression in FL LT-HSCs isolated from E14.5 Nbea+/+ and Nbea–/– littermates (Figure 4C; supplemental Figure 5B). Here, pathway analyses confirmed that Nbea deficiency perturbs components associated with vesicle traffic (Figure 4D). Indeed, colocalization with RCAS1 suggests that NBEA is largely confined to the Golgi apparatus in HSCs, consistent with reports in other cell types (Figure 4E).22 Accordingly, NBEA has been implicated as a regulator of vesicle trafficking in other cellular contexts, such as synaptic transmission.24,44 Moreover, reduced levels of Nbea increase vesicle secretion.25 Thus, our results suggest a possible role for NBEA in vesicle trafficking in HSCs.
Nbea deficiency affects vesicle trafficking components at the transcriptional level. (A-B) Control and Nbea-shRNA–treated progenitors were transplanted into lethally irradiated recipients. mCherry+ cells were recovered from transplanted mice 10 weeks after transplant and subjected to bulk RNA sequencing. (A) Experimental schematic. (B) Pathway analysis based on transcriptional differences among control and Nbea-shRNA–treated progenitors in (i) biological processes and (ii) cellular components. Differences related to vesicle trafficking highlighted with an arrow. (C-D) E14.5 FL LT-HSCs were isolated from Nbea+/+ and Nbea–/ – embryos and subjected to bulk RNA sequencing. (C) Experimental schematic. (D) Pathway analysis including (i) biological processes and (ii) cellular components showing differences among Nbea+/+ and Nbea–/– E14.5 FL LT-HSCs. (E) Representative confocal images of BM HSC and NBEA/RCAS1 colocalization analysis (Pearson correlation coefficient). NBEA-A488 shown in green, RCAS1-A647 (Golgi) in red, and nuclei (DAPI) in blue. (F) qPCR analysis of NOTCH-regulated genes in E14.5 FL-HSC in Nbea+/+ and Nbea–/–. Data in panel B are from 2 independent samples. Data in panel D are from 1 sample. Data in panel F are from 3 samples, represented as mean ± SEM. ∗P < .05; ∗∗P < .005, relative Nbea+/+.
Nbea deficiency affects vesicle trafficking components at the transcriptional level. (A-B) Control and Nbea-shRNA–treated progenitors were transplanted into lethally irradiated recipients. mCherry+ cells were recovered from transplanted mice 10 weeks after transplant and subjected to bulk RNA sequencing. (A) Experimental schematic. (B) Pathway analysis based on transcriptional differences among control and Nbea-shRNA–treated progenitors in (i) biological processes and (ii) cellular components. Differences related to vesicle trafficking highlighted with an arrow. (C-D) E14.5 FL LT-HSCs were isolated from Nbea+/+ and Nbea–/ – embryos and subjected to bulk RNA sequencing. (C) Experimental schematic. (D) Pathway analysis including (i) biological processes and (ii) cellular components showing differences among Nbea+/+ and Nbea–/– E14.5 FL LT-HSCs. (E) Representative confocal images of BM HSC and NBEA/RCAS1 colocalization analysis (Pearson correlation coefficient). NBEA-A488 shown in green, RCAS1-A647 (Golgi) in red, and nuclei (DAPI) in blue. (F) qPCR analysis of NOTCH-regulated genes in E14.5 FL-HSC in Nbea+/+ and Nbea–/–. Data in panel B are from 2 independent samples. Data in panel D are from 1 sample. Data in panel F are from 3 samples, represented as mean ± SEM. ∗P < .05; ∗∗P < .005, relative Nbea+/+.
sel-2, the Nbea/Lrba homolog in Caenorhabditis elegans, negatively regulates LIN-12/NOTCH activity by controlling its turnover at the cell surface in vulval precursor cells.45 Although the role of NOTCH signaling in HSCs in adult steady hematopoiesis remains controversial, NOTCH signaling constitutes an important modulator of HSC activity during stress hematopoiesis.46-49 We hypothesized that this regulatory mechanism may be evolutionarily preserved in HSCs. In this scenario, reduced NBEA levels might alter normal NOTCH turnover by perturbing vesicular trafficking, thus impairing NOTCH signaling and HSC engraftment. Consistently, in both of our Nbea-deficient experimental contexts, NOTCH1 is predicted to be perturbed according to the transcriptional differences with control cells (supplemental Figure 5C). Moreover, we confirmed the downregulation of multiple NOTCH1 targets in Nbea–/– HSCs in E14.5 FL Nbea–/– embryos (Figure 4F).
We next used a surrogate hematopoietic human cell line to further explore this mechanism in vitro, because it is difficult to expand and maintain HSCs in vitro.50 We selected OPM2 cells based on high NBEA mRNA levels and amenability to lentiviral infection (supplemental Figure 5D-E). We verified that, similar to HSCs, NBEA is mostly located in the Golgi apparatus in OPM2 cells, as revealed by RCAS1 staining22 (supplemental Figure 5F). To further study NBEA in this context, shRNAs capable of efficient human NBEA knockdown (1 and 2) were selected (supplemental Figure 5E). OPM2 cells transduced with NBEA-shRNAs (1 and 2) were sorted based on mCherry expression and expanded to establish OPM2 cell lines expressing different levels of NBEA mRNA.
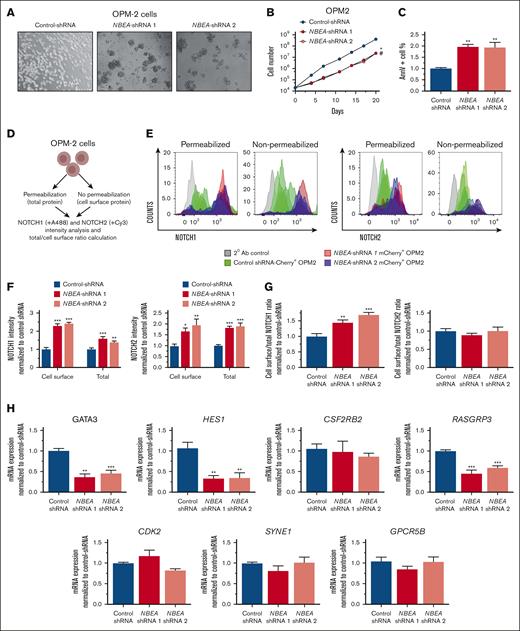
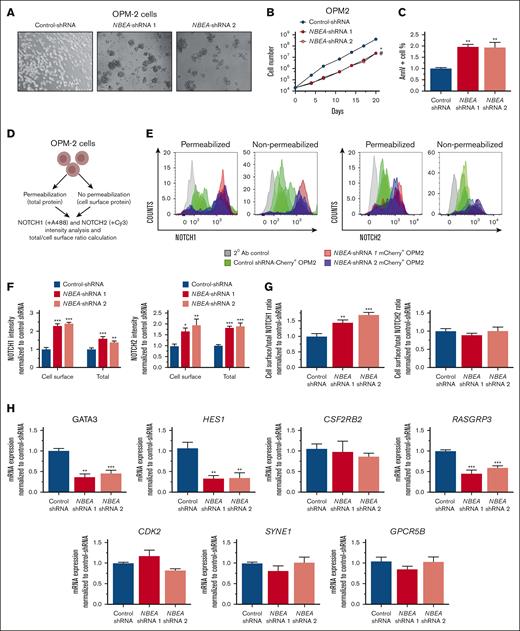
Interestingly, OPM2 cells transduced with multiple independent NBEA-shRNAs consistently grew as cell aggregates, in contrast to OPM2 cells transduced with control shRNAs (Figure 5A). Additionally, NBEA-deficient OPM2 cells showed slower growth and elevated apoptosis than controls (Figure 5B-C). Cell cycle was not perturbed in the absence of NBEA (Supplemental Figure 5G), recapitulating some of the main effects of Nbea deficiency in primary cells.
NBEA deficiency perturbs NOTCH proteins expression and cell localization. (A) Representative images of OPM2 cell lines expressing different NBEA-shRNAs (1 and 2) and control-shRNA. Transduced OPM2 cells for each shRNA were sorted based on mCherry+ expression. (B) Cell growth curves for OPM2 transduced cells. (C) Annexin V analysis in control- and NBEA-shRNA OPM2 cells. Percentage of annexin V+ cells is shown. (D-G) NOTCH1 and NOTCH2 cell surface and total expression in OPM2 transduced cells. (D) Experimental schematic. (E) Flow cytometry histograms showing NOTCH1 and NOTCH2 protein levels in the cell surface and total expression for each OPM2 cell line. (F) Quantification of NOTCH1 and NOTCH2 protein levels normalized to control-shRNA. (G) Ratio of cell surface and total levels of NOTCH1 and NOTCH2. (H) qRT-PCR analysis of NOTCH-regulated genes in OPM2 cell lines. Data are represented as mean ± SEM. ∗P < .05; ∗∗P < .005; ∗∗∗P < .001, relative to control-shRNA. ∗ refers to NBEA-shRNA 1; # refers to NBEA-shRNA 2. Ab, antibody.
NBEA deficiency perturbs NOTCH proteins expression and cell localization. (A) Representative images of OPM2 cell lines expressing different NBEA-shRNAs (1 and 2) and control-shRNA. Transduced OPM2 cells for each shRNA were sorted based on mCherry+ expression. (B) Cell growth curves for OPM2 transduced cells. (C) Annexin V analysis in control- and NBEA-shRNA OPM2 cells. Percentage of annexin V+ cells is shown. (D-G) NOTCH1 and NOTCH2 cell surface and total expression in OPM2 transduced cells. (D) Experimental schematic. (E) Flow cytometry histograms showing NOTCH1 and NOTCH2 protein levels in the cell surface and total expression for each OPM2 cell line. (F) Quantification of NOTCH1 and NOTCH2 protein levels normalized to control-shRNA. (G) Ratio of cell surface and total levels of NOTCH1 and NOTCH2. (H) qRT-PCR analysis of NOTCH-regulated genes in OPM2 cell lines. Data are represented as mean ± SEM. ∗P < .05; ∗∗P < .005; ∗∗∗P < .001, relative to control-shRNA. ∗ refers to NBEA-shRNA 1; # refers to NBEA-shRNA 2. Ab, antibody.
To assess the role of NBEA in controlling the subcellular localization and turnover of NOTCH receptors, we analyzed, by flow cytometry, the abundance of cell surface vs total NOTCH1 and NOTCH2 in control and NBEA-deficient OPM2 cells. We observed an accumulation of both cell surface and total NOTCH1 and NOTCH2 in NBEA-deficient OPM2 cells, relative to controls (Figure 5D-F). To identify changes in the subcellular distribution of these proteins, we calculated the ratio between the protein located at the cell surface vs total protein. Notably, this ratio was different only for NOTCH1, indicating a greater proportion of this protein in the cell membrane of NBEA-deficient OPM2 cells (Figure 5G). These data implicate NBEA in the regulation of NOTCH1 and NOTCH2 protein content and localization.
To confirm that altered subcellular NOTCH1 location has functional consequences on NOTCH transcriptional programs, we examined the expression levels of NOTCH target genes, including HES1, GATA3, RASGRP3, and CSF2RB2, in NBEA-deficient OPM2 cells. Here, we confirmed the downregulation of HES1, GATA3, and RASGRP3, relative to controls (Figure 5H). Remarkably, these NOTCH target genes were also downregulated in Nbea–/– E14.5 FL-HSCs (Figure 4F). We also verified the expression of NOTCH ligands in OPM2 cells, which can trigger the activation of the NOTCH pathway in neighboring cells (supplemental Figure 5H). Importantly, NOTCH activity has been implicated in modulating the repopulation activity of adult HSCs.46,47,51 This would suggest that NBEA might regulate the ability of HSCs to efficiently engraft at least in part through the fine modulation of NOTCH transcriptional programs.
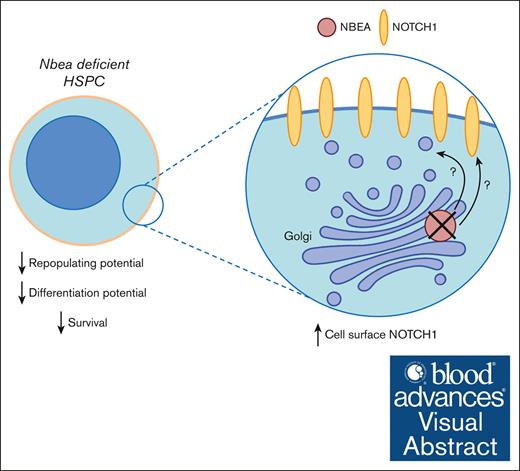
In summary, here, we reported, for the first time, the functional redundancy of multiple BDCP family members in HSCT, because their acute loss perturbs HSC in vivo repopulating activity (eg, Nbea, Nbeal1, Lrba, and Lyst). Moreover, we implicated NBEA in the regulation of NOTCH protein levels and cell surface localization, with commiserate effects on NOTCH-regulated transcription. These effects could contribute to the loss of repopulating activity seen after NBEA loss.46 Our work further shows that complex mRNA compensatory mechanisms modulate the expression of BDCPs to preserve the integrity of the hematopoietic system, supporting an important role for BDCPs in hematopoiesis.
Discussion
We previously identified Nbea as a putative positive regulator of HSCT.5 Here, we report that Nbea expression is highly restricted to HSCs within the hematopoietic hierarchy, suggesting a specific function in HSCs (Figure 1A; supplemental Figure 1A). We found that Nbea is required for the efficient engraftment of both FL and BM HSPCs (Figure 1B; supplemental Figure 1D-E). Nbea knockdown increased apoptosis in acutely engrafted HSCs, which resulted in reduced PB chimerism (Figures 1ii-iv and 2). The milder transplantation defects observed in Nbea–/– HSCs compared with acute knockdown of Nbea (Figures 1B; supplemental Figure 1D-E) likely results from compensation by other BDCP family members, given the dramatic changes in gene expression of multiple BDCPs in Nbea–/– E14.5 FL-HSCs (Figure 3B). This is possibly related to a phenomenon known as transcriptional adaptation.43,52,53 Transcriptional adaptation is a new form of genetic adaptation that has been described in zebrafish, mouse, and C elegans and that highlights, for instance, the poor correlation among the phenotypes induced by acute gene induction in zebrafish via morpholinos and those detected in mutant zebrafish.43,52,53 Additionally, knockdown of Lrba, Lyst, and Nbeal1 in HSPCs each induced a transplantation defect, confirming a positive role for these genes in HSCT (Figure 3C-E). Moreover, knockdown of Lrba, Lyst, or Nbeal1 in Nbea–/– HSPCs exacerbated these repopulating defects (Figure 3C-E). Importantly, BDCPs are predicted to form homodimers and heterodimers.6 Thus, it is possible that different pairs of BDCPs might have specific patterns of transported cargoes, whereas other BDCP dimers might share common cargoes and compensate for other BDCPs, as previously suggested.6
We found NBEA mainly localized to the Golgi apparatus in HSCs and leukemia cell lines, as seen in other cell contexts (Figure 4E; supplemental Figure 5F).6,22 Consistently, genes implicated in vesicular transport were upregulated in Nbea–/– FL-HSCs (Figure 4B,D), which is also consistent with the increased vesicle trafficking previously reported in Nbea–/– neurons24,44,54 and with a critical role for NBEA in endocytic recycling of N-methyl-D-aspartate (NMDA) receptors.10
The perturbation of vesicle trafficking can affect many processes including those related to vesicle biogenesis, cargo incorporation, and downstream signaling because of changes in receptor levels present in the cellular membrane.55,56 Our data showed NOTCH1 accumulated in the cytoplasmic membrane of Nbea-deficient OPM2 cells (Figure 5D-F). Accordingly, the NOTCH transcriptional program was perturbed in Nbea–/– HSCs and in Nbea-deficient cells (Figures 4F and 5H). Thus, although several cargoes and downstream pathways could be affected in these cells, our data support that, among those, NBEA regulates NOTCH cell surface turnover, modulating NOTCH transcriptional activity in HSPCs. Because NOTCH activity governs the repopulating potential of HSPCs,46,51 this model is consistent with an NBEA requirement for optimal HSCT. Supporting this, sel-2 (Nbea/Lrba homolog in C elegans) regulates the activity of LIN-12/NOTCH via the regulation of endosomal traffic that controls efficient delivery of cell surface proteins to the lysosome including NOTCH.45 After the interaction of NOTCH ligands with NOTCH receptors, the extracellular domain (ECD) of NOTCH is normally endocytosed and may be degraded or recycled.48 Although NOTCH protein levels in the membrane of Nbea-deficient cells are higher (Figure 5D-G), it is likely that some of these receptors are nonrecycled NOTCH-ECDs and thus noncompetent to transduce a signal. These nonrecycled NOTCH-ECDs can likely compete with functional NOTCH receptors, resulting in lowered NOTCH signaling (Figures 4F and 5H).
Additionally, although our results support a role of NOTCH signaling in stress hematopoiesis as described by others,46-49,51 the role of NOTCH signaling in HSPCs in adult steady-state hematopoiesis remains contentious.46-49 Supporting a role for NOTCH signaling in adult native hematopoiesis, in myeloid progenitor cell lines, it was shown that NOTCH activity was required for differentiation.57,58 Furthermore, induced increased levels of JAGGED1 in the BM niche led to HSC expansion.59 In contrast, multiple mouse models including Rbpj-deficient mice, transgenic mice expressing a dominant negative mutant form of Mastermind (which blocks NOTCH signaling), and Notch1 and Notch2 inducible knockout mice in the hematopoietic system support a dispensable role of NOTCH activity in HSCs.46,60-62 Accordingly, we observed that Nbea is dispensable for native hematopoiesis in the FL (supplemental Figure 4A).
Overall, our data showed a major and conserved role of the BDCP family of proteins in regulating proper HSCT and of NBEA in maintaining optimal steady-state hematopoiesis. We propose a model in which BDCPs sustain proper NOTCH receptor activity by regulating NOTCH turnover in the cytoplasmic membrane. Our results indicate that clinical intervention aimed to modulate NOTCH activity through the manipulation of vesicle trafficking activity operated by BDCPs could lead to a better HSC engraftment during BM transplantation.
Acknowledgments
The authors thank Wilson Clements, the McKinney-Freeman laboratory, and Department of Hematology at St. Jude Children’s Research Hospital (SJCRH) for critical discussions and reading of the manuscript; Richard Ashmun, Sandy Schwemberger, and Jonathan Laxton for fluorescence-activated cell sorting support; Cara Davis-Goodrum, K. Millican, Amber Reap, and Chandra Savage for help with transplants; Victoria Frohlich, Sharon King, Jennifer Peters, and Aaron Pitre for help with confocal imaging; Melanie Lloyd and John Morris of the Hartwell Center at SJCRH for assistance with gene expression arrays; and Manfred W. Kilimann (Department of Otolaryngology Göttingen University Medical Center & Department of Molecular Neurobiology Max-Planck-Institute for Experimental Medicine) for kindly providing Nbea–/– mice.
This work was supported by the American Society of Hematology (S.M.-F.), the Hartwell Foundation (S.M.-F.), the National Institutes of Diabetes and Digestive and Kidney Diseases (grants K01DK080846 and R01DK104028 [S.M.-F.]), the American Lebanese Syrian Associated Charities (S.M.-F.), and the National Cancer Institute (grant P30 CA021765-35, SJCRH Cell and Tissue Imaging Center). M.G. is funded by the American Society of Hematology (Global Research Award [ASH GRA 2021]), Barts Charity (the Rising Stars Programme [grant MGU0459]), Greg Wolf Fund, Kay Kendall Leukaemia Fund (grant KKL1444), Leukaemia UK (John Goldman Fellowship [number 2020/JGF/001]), and Medical Research Council (MRC; MRC Career Development Award [number MR/V009222/1]). SMF is a Scholar of the Leukemia and Lymphoma Society.
The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Authorship
Contribution: M.G. and A.M.-H. designed the study, performed and analyzed transplants, collected and analyzed data, and wrote the manuscript; A.V.H. performed and analyzed in vitro experiments; T.H. and A.C. performed and analyzed mice that received transplantation; A.C. performed and analyzed confocal microscopy images; C.C. contributed to in vivo studies; D.F. analyzed expression data in the Cancer Cell Line Encyclopedia; M.W.K. provided Nbea-/- mice. S.M.-F. designed the study, analyzed data, and wrote the manuscript; and all authors discussed the results and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Miguel Ganuza, Barts Centre Cancer Research UK, Charterhouse Square, London, EC1M 6BQ, United Kingdom; email: m.ganuza@qmul.ac.uk; and Shannon McKinney-Freeman, St. Jude Children's Research Hospital. 262 Danny Thomas Place. Memphis, TN. 38105; email: shannon.mckinney-freeman@stjude.org.
References
Author notes
M.G. and A.M.-H. contributed equally to this study.
The complete RNA sequencing data set was deposited in the Sequence Read Archive database (BioProject ID: PRJNA1052655).
Original data are available on request from the corresponding authors, Miguel Ganuza (m.ganuza@qmul.ac.uk) and Shannon McKinney-Freeman (shannon.mckinney-freeman@stjude.org).
The full-text version of this article contains a data supplement.


![Nbea is a positive regulator of HSPC repopulating activity. (A) qRT-PCR of Nbea expression in hematopoietic cells from adult BM, including HSC (Lineage–SCA-1+c-KIT+[LSK]CD150+CD48–); MPPs (LSKCD150+CD48+ [MPP2] and LSKCD150–CD48+ [MPP3/4]); common myeloid progenitors (CMPs; Lineage–SCA-1–c-KIT+CD32/16LowCD34+); common lymphoid progenitors (CLPs; Lineage–c-KitmedSca-1medCD127+); megakaryocyte-erythroid progenitors (MEPs; Lineage–SCA-1–c-KIT+CD32/16–CD34–); granulocyte-myeloid progenitors (GMPs; Lineage–SCA-1–c-KIT+CD32/16–CD34–); granulocytes, (GR-1+), B cells (B220+), and T cells (CD3+). (B) CD45.2+ “test” BM-HSPCs were transduced with control or Nbea-shRNAs. mCherry+ cells were then sorted and transplanted along with mock-transduced CD45.1+ HSPCs at a 1:1 ratio into lethally irradiated CD45.1+/CD45.2+ recipients within 44 hours after isolation. Recipient PB was examined for >16 weeks after transplant by flow cytometry for CD45.2+ mCherry+ cells. (Bi) Schematic of competitive transplantation of Nbea-deficient BM-HSPCs. (Bii) Percentage of CD45.2+mCherry+ PB at 4 to 16 weeks after transplant. (Biii) Frequency of CD45.2+mCherry+ PB lineages at 4 to 16 weeks after transplant. (Biv) Contribution of CD45.2+mCherry+ progenitors to BM progenitors. Data in panel B are from 3 experiments; 5 mice per condition per transplant. Data are represented as mean ± standard error of the mean (SEM). ∗P < .05; ∗∗P < .005; ∗∗∗P < .001, relative to recipients of control-shRNA HSPCs.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/15/10.1182_bloodadvances.2023012426/2/m_blooda_adv-2023-012426-gr1.jpeg?Expires=1765005458&Signature=WwcYQ~j1dbiIw~e69ZUcB2ZjmwGlFRrdfKNyjwO0MluLasQy8dTWBFXcbhwEecY1xdXm3tj55JXq7XN2TZ2o5bnR5tdYzZo9LrwDyxK18PfZFwCKdIta~n3BMe7FS68-Q0YfZemrdIxHD02sqkE5vrzCQPbXIOegJ2SLHO69AA4re4ET0WCcxIIm6kjeeWDTT2GIMD~zLFVftDb4xOApyzDmypGhB~M-jfaL-YeVwQvAqGDT1X~INIk~GEFBKvfJfZdUxaQYK3iAzBEH8za2KNXH34ZfXI7rM-TiYUUfan1I1QyMJcaf-6fJfYn828vnBnqgTvY6RAzICprzzSrB3w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Nbea is a positive regulator of HSPC repopulating activity. (A) qRT-PCR of Nbea expression in hematopoietic cells from adult BM, including HSC (Lineage–SCA-1+c-KIT+[LSK]CD150+CD48–); MPPs (LSKCD150+CD48+ [MPP2] and LSKCD150–CD48+ [MPP3/4]); common myeloid progenitors (CMPs; Lineage–SCA-1–c-KIT+CD32/16LowCD34+); common lymphoid progenitors (CLPs; Lineage–c-KitmedSca-1medCD127+); megakaryocyte-erythroid progenitors (MEPs; Lineage–SCA-1–c-KIT+CD32/16–CD34–); granulocyte-myeloid progenitors (GMPs; Lineage–SCA-1–c-KIT+CD32/16–CD34–); granulocytes, (GR-1+), B cells (B220+), and T cells (CD3+). (B) CD45.2+ “test” BM-HSPCs were transduced with control or Nbea-shRNAs. mCherry+ cells were then sorted and transplanted along with mock-transduced CD45.1+ HSPCs at a 1:1 ratio into lethally irradiated CD45.1+/CD45.2+ recipients within 44 hours after isolation. Recipient PB was examined for >16 weeks after transplant by flow cytometry for CD45.2+ mCherry+ cells. (Bi) Schematic of competitive transplantation of Nbea-deficient BM-HSPCs. (Bii) Percentage of CD45.2+mCherry+ PB at 4 to 16 weeks after transplant. (Biii) Frequency of CD45.2+mCherry+ PB lineages at 4 to 16 weeks after transplant. (Biv) Contribution of CD45.2+mCherry+ progenitors to BM progenitors. Data in panel B are from 3 experiments; 5 mice per condition per transplant. Data are represented as mean ± standard error of the mean (SEM). ∗P < .05; ∗∗P < .005; ∗∗∗P < .001, relative to recipients of control-shRNA HSPCs.](https://ash.silverchair-cdn.com/ash/content_public/journal/bloodadvances/8/15/10.1182_bloodadvances.2023012426/2/m_blooda_adv-2023-012426-gr1.jpeg?Expires=1765591473&Signature=X4muPio0NeISP6DoSZJSLJ79nJN1OU3F906mBSF4rKHLOVtNe2aIqpKG2~GUt95dZ1bO4IOHa3xsmeH5tXzHrsuZfZl7QupFGL7M3GlliEAv1D7a1z3GrU0uQJsu8ntwSb0wN0OC7jO3-DuFeJ9UNtiRv7~efTDI9C51Sq-BncJIqaqF9D4aw6zdY5cljPBek7rpM0gd~kULBIXumEq3KGW7S7fYZSRt3JcRoTolwFmSQd04TpQoGJ3OrCo5fyUTCavmdLwc76nLHq~3y-bGItWSk7NhJj35veHNxPZ41N-05tZ~KvAkaGpzYNEmTZtdp6-Fupb2xyckT8RetabcYw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)