TO THE EDITOR:
Cytokine release syndrome (CRS) is a potentially life-threatening toxicity that can occur due to high-level immune activation. CRS can be triggered non-specifically by T-cell engaging therapies administered to treat hematologic malignancies.1-3 Clinical manifestations vary, from fever to more severe symptoms requiring hospitalization.2 Although CRS severity is influenced by disease status and underlying histology, prediction of CRS risk is currently not possible.4 Identification of high-grade CRS risk factors is critical to inform decisions on patient management when administering T-cell engaging therapies.5
Glofitamab, a CD20 × CD3 T-cell engaging bispecific antibody with a novel 2:1 format, demonstrated high complete response rates with manageable safety in a phase 1/2 study (NP30179; NCT03075696) in patients with large B-cell lymphoma.6,7 CRS observed with glofitamab is typically Grade 1/2 by American Society for Transplantation and Cellular Therapy (ASTCT) criteria and is mostly confined to the first treatment cycle.4,8,9
Komanduri et al. outlined a model using 8 baseline factors (CRS-risk score [CRS-RS]) that accurately classified the risk of Grade ≥2 CRS after the first glofitamab dose.10 Patients classified as low-risk (CRS-RS <5, 60% of the cohort) had a 5% chance of experiencing Grade ≥2 CRS (negative predictive value [NPV] 0.95; standard error [SE] 0.03).10 In aggressive non-Hodgkin lymphomas (aNHL), the parameters requiring assessment of bone marrow infiltration, peripheral blood infiltration, and cardiac comorbidities could be excluded from CRS-RS thereby offering improved clinical applicability without compromising performance. We report a streamlined model using 5 baseline factors (CRS-RS.5p) derived from the original CRS-RS (Table 1). To determine whether CRS-RS.5p could accurately predict Grade ≥2 CRS events after the first glofitamab dose (step-up dosing [SUD], 2.5/10/30 mg), we compared CRS-RS.5p with CRS-RS in the NP30179 data set and then subsequently analyzed CRS-RS.5p performance in both NP30179 and the GO43921 non-interventional study (NIS).
Model parameters and risk classification
For CRS-RS and CRS-RS.5p, to predict the risk of developing Grade ≥2 CRS, a multivariate model including the first glofitamab dose and a weighted sum of baseline, clinical, laboratory, and imaging parameters based on disease and CRS pathophysiology was used (Table 1).10 Weights reflected the predictive strength of risk factors in a dose-adjusted logistic regression model and the relative stability of risk parameters in a multivariate model resulting from random forest analysis.
Patients were classified as low-risk (CRS-RS <5, CRS-RS.5p <4) or high-risk (CRS-RS ≥5, CRS-RS.5p ≥4) for CRS with glofitamab (2.5 mg); cut-off values were selected to achieve the required performance.
To control the rate of false negative predictions, missing data were input at the highest levels for each respective parameter. Following the identification of patients as low-risk for CRS, NPV was used to demonstrate the likelihood that a patient classified as low-risk would not develop Grade ≥2 CRS after the first glofitamab dose.
Assessment of model performance
The predictive model that includes drug dose and CRS-RS.5p risk score was trained on dose-finding (not part of this communication) and validated in SUD cohorts of NP30179 with glofitamab (2.5 mg). CRS-RS.5p model performance was further analyzed in the GO43921 NIS, which consisted of 7 clinical studies in patients with aNHL (excluding mantle-cell lymphoma) who received glofitamab SUD (2.5/10/30 mg) monotherapy or as a combination regimen (supplemental Table 1). At the time of the first analysis, 3 studies from the NIS had data available (NP39488, BP41072, and NP40126). Patients enrolled in NP39488 and BP41072 received obinutuzumab pretreatment before glofitamab (2.5 mg). Patients enrolled in NP40126 received either obinutuzumab or rituximab and chemotherapy prior to glofitamab (2.5 mg). The GO43921 NIS primary objective was to determine the predictive performance of CRS-RS.5p in identifying low-risk (no CRS or Grade 1 CRS) and high-risk (Grade ≥2 CRS) patients after the first glofitamab dose.
Data were collected prospectively from the included studies since December 2021, with retrospective data from NP30179 (not available at model development stage) analyzed to allow for broader and more precise conclusions.
CRS-RS.5p performance in the NP30179 validation cohort
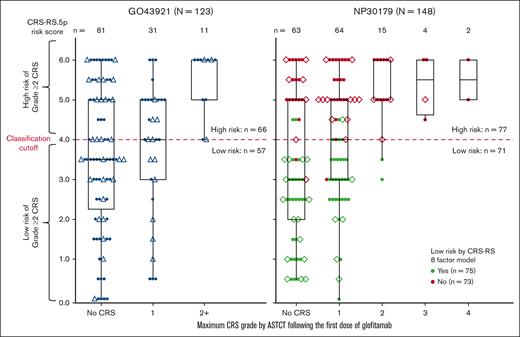
CRS-RS.5p performance was compared with the CRS-RS model in the NP30179 data set. Figure 1 shows the distribution of CRS-RS.5p baseline score values across ASTCT–graded CRS events after the first glofitamab dose. Cross-classification results for NP30179 by the original score and CRS-RS.5p are shown in supplemental Table 2. Based on data acquired since the introduction of CRS-RS in the NP30179 study (n = 44), both CRS-RS and CRS-RS.5p identified the same patients who experienced Grade ≥2 CRS as high-risk (Grade 2, n = 4; Grade 3, n = 1). Both models achieved NPVs of 0.97 (n = 148; SE, 0.02).
CRS-RS.5p and CRS-RS risk scores vs CRS grade by ASTCT in GO43921 + NP30179 data sets after the first dose of glofitamab in the second-line or later treatment of NHL. Triangles represent patients in the prospective part of GO43921, diamonds represent patients with NHL recruited to NP30179 between October 2021 and April 2022. Every patient from the complete analysis cohort is represented by a colored dot. In the right-hand side panel, green dots indicate patients with a low-risk classification as assessed by the CRS-RS model in the NP30179 study; red dots indicate patients that were not deemed low-risk by the CRS-RS model in the NP30179 study. In the left-hand side panel, black dots represent patients in the GO43921 study. For every patient, the ASTCT CRS grade category corresponds to the maximum ASTCT grade of all CRS events recorded in the first week after the first dose of glofitamab. NHL, non-Hodgkin lymphoma.
CRS-RS.5p and CRS-RS risk scores vs CRS grade by ASTCT in GO43921 + NP30179 data sets after the first dose of glofitamab in the second-line or later treatment of NHL. Triangles represent patients in the prospective part of GO43921, diamonds represent patients with NHL recruited to NP30179 between October 2021 and April 2022. Every patient from the complete analysis cohort is represented by a colored dot. In the right-hand side panel, green dots indicate patients with a low-risk classification as assessed by the CRS-RS model in the NP30179 study; red dots indicate patients that were not deemed low-risk by the CRS-RS model in the NP30179 study. In the left-hand side panel, black dots represent patients in the GO43921 study. For every patient, the ASTCT CRS grade category corresponds to the maximum ASTCT grade of all CRS events recorded in the first week after the first dose of glofitamab. NHL, non-Hodgkin lymphoma.
Performance of CRS-RS.5p in the GO43921 and NP30179 studies
Reliable prediction of Grade ≥2 CRS was determined using CRS-RS.5p in both GO43921 (n = 123) and NP30179 (n = 148; Figure 1; supplemental Table 3); as of 6 June 2022, 11 Grade ≥2 CRS events in the second-line or later setting for patients with aNHL (n = 123, data collected prospectively for 41 patients) were correctly predicted using CRS-RS.5p. In combined data from both studies, CRS-RS.5p accurately predicted Grade ≥2 CRS in 30 (CRS-RS.5p≥4) out of 32 (Grade ≥2 CRS) cases with the true positive prediction rate of 0.94 (95% confidence interval, 0.79-0.99). The majority (126/128) of patients predicted to be low-risk from both studies did not develop Grade ≥2 CRS 7 days after the first glofitamab dose (NPV = 0.98; 95% confidence interval, 0.94-1.00). Across all parameters, 11% and 3% of CRS-RS.5p values were missing for GO43921 and NP30179, respectively. In GO43921 most cases were missing baseline lactate dehydrogenase (LDH) levels. In NP30170, a single risk factor was missing in each case (n = 3, Ann Arbor stage; n = 1, sum of the product of perpendicular diameters [SPD]; n = 1, LDH).
Both studies showed similar association between CRS-RS.5p and maximal observed CRS grade following glofitamab (logistic regression for Grade ≥2 CRS; χ2 test P = 0.004 at R2 = 0.21 for GO43921; P < 0.001 at R2 = 0.16 for NP30179).
Discussion
Predictive models have the potential to accurately identify patients at risk of adverse outcomes, and early detection of high-risk patients can better enable the adoption of preventive interventions.12,13 Before the development of this model, identifying the patients most likely to develop Grade ≥2 CRS was not possible, although glofitamab SUD demonstrated a reduction in high-grade CRS when compared with fixed-dosing.4,14 This streamlined 5-parameter model, using cut-off CRS-RS.5p<4 accurately identified glofitamab-treated patients at low-risk of Grade ≥2 CRS. CRS-RS and CRS-RS.5p showed similar high performance in predicting patients with aNHL most at risk of Grade ≥2 CRS following the first glofitamab dose. CRS-RS.5p has reduced complexity than CRS-RS and patient risk can be determined through an easy-to-use scoring system using readily available clinical data. The main limitation of CRS-RS.5p is the imaging assessment required to calculate the SPD in a clinical setting; developing a surrogate marker for high tumor burden such as the sum of longest diameters of a maximum of 3 target lesions could improve its clinical applicability.15
In clinical trials, CRS-RS.5p enabled accurate prediction of Grade ≥2 CRS with 94% true positive prediction chance. Together with a 98% chance for a predicted low-risk patient to remain Grade ≥2 CRS-free after the first glofitamab dose, CRS-RS.5p can potentially support clinical management for patients at high risk of Grade ≥2 CRS following glofitamab therapy. CRS-RS.5p is an easy-to-use tool that could identify patients who require more intensive monitoring or hospitalization.
Acknowledgments: Third-party medical writing assistance, under the direction of all authors, was provided by Dikeledi Matlebjane of Ashfield MedComms, an Inizio company, and was funded by F. Hoffmann-La Roche Ltd.
This study was sponsored by F. Hoffmann-La Roche Ltd.
Contributions: A.B., J.R., M.D., and M.T. conceived and designed the study; A.B., M.D., and G.G. collected and assembled the data; A.B., J.R., M.D., M.T., G.G., and K.K. analyzed and interpreted the data; and all authors reviewed and revised the manuscript, approved the final version, and agreed to submit the manuscript for publication.
Conflict-of-interest disclosure: G.G. reports consulting/advisory fees from Takeda, Kite-Gilead, F. Hoffmann-La Roche Ltd, Clinigen Group, BeiGene, Incyte, Genmab, and Ideogen; travel expenses from Janssen, Sandoz, and BeiGene. A.B., M.D., and M.T. are employees of Roche Products Ltd and own equity in F. Hoffmann-La Roche Ltd. J.R. is an employee of Roche Products Ltd and owns equity in F. Hoffmann-La Roche Ltd and F-Star Therapeutics. K.K. has acted as a consultant for Genentech, Inc./F. Hoffmann-La Roche Ltd, Iovance, Incyte, Syncopation Life Sciences, and CRISPR Therapeutics, and attended scientific advisory boards for Aegle Therapeutics and Avacta Therapeutics.
Correspondence: Giuseppe Gritti, UC Ematologia Ospedale Papa Giovanni XXIII, Piazza OMS, 1, 24127 Bergamo, Italy; email: g.gritti@asst-pg23.it; and Krishna Komanduri, Division of Hematology and Oncology, The University of California San Francisco, 400 Parnassus Ave Fourth Floor, San Francisco, CA 94143; email: krishna.komanduri@ucsf.edu.
References
Author notes
Qualified researchers may request access to individual patient-level data through the clinical study data request platform (https://vivli.org/). Further details on Roche’s criteria for study eligibility are available at https://vivli.org/members/ourmembers/. Further details on Roche’s Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents are available at https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm.
The full-text version of this article contains a data supplement.