Key Points
B-ALL development in the setting of lenalidomide treatment for MM is a distinct primary malignancy with high incidence of TP53 mutations.
Chronic lenalidomide therapy appears to be capable of expanding rare hematopoietic cells with acquired TP53 mutations.
Abstract
Patients with multiple myeloma (MM) who are treated with lenalidomide rarely develop a secondary B-cell acute lymphoblastic leukemia (B-ALL). The clonal and biological relationship between these sequential malignancies is not yet clear. We identified 17 patients with MM treated with lenalidomide, who subsequently developed B-ALL. Patient samples were evaluated through sequencing, cytogenetics/fluorescence in situ hybridization (FISH), immunohistochemical (IHC) staining, and immunoglobulin heavy chain (IgH) clonality assessment. Samples were assessed for shared mutations and recurrently mutated genes. Through whole exome sequencing and cytogenetics/FISH analysis of 7 paired samples (MM vs matched B-ALL), no mutational overlap between samples was observed. Unique dominant IgH clonotypes between the tumors were observed in 5 paired MM/B-ALL samples. Across all 17 B-ALL samples, 14 (83%) had a TP53 variant detected. Three MM samples with sufficient sequencing depth (>500×) revealed rare cells (average of 0.6% variant allele frequency, or 1.2% of cells) with the same TP53 variant identified in the subsequent B-ALL sample. A lack of mutational overlap between MM and B-ALL samples shows that B-ALL developed as a second malignancy arising from a founding population of cells that likely represented unrelated clonal hematopoiesis caused by a TP53 mutation. The recurrent variants in TP53 in the B-ALL samples suggest a common path for malignant transformation that may be similar to that of TP53-mutant, treatment-related acute myeloid leukemia. The presence of rare cells containing TP53 variants in bone marrow at the initiation of lenalidomide treatment suggests that cellular populations containing TP53 variants expand in the presence of lenalidomide to increase the likelihood of B-ALL development.
Introduction
Multiple myeloma (MM) is a malignant clonal expansion of plasma cells that exists on a spectrum with other plasma cell dyscrasias and monoclonal gammopathy of undetermined significance. It is the second most common hematologic malignancy and results in considerable morbidity and mortality owing to its tendency to affect older patients.1
Therapy for myeloma has rapidly evolved in the last two decades. Modern therapy involves a multimodal approach, often starting with a combination regimen involving immune modulators, proteasome inhibitors, biologics, and/or corticosteroids.2,3 Eligible patients are typically offered high-dose chemotherapy with autologous hematopoietic cell transplant (HCT) owing to its potential for prolonged remission.4 Long-term maintenance therapy with lenalidomide after HCT has been shown to improve survival, but several studies have noted an increased frequency of secondary malignancies.5-10 In one study, the hazard ratio for a second primary hematologic malignancy for patients with MM being treated with lenalidomide was 2.03 (P = .015).1 These malignancies have been described as clonally distinct from the primary tumor11 and in particular, B-ALL malignancies have demonstrated inferior outcomes relative to those developed in the absence of therapeutic pressure.9,12 As practice patterns have shifted, larger numbers of patients are receiving maintenance lenalidomide, and there has been a corresponding increase in secondary malignancies, especially hematologic malignancies.8
Secondary hematologic malignancies have been described in patients with MM since the 1970s but typically manifested as a treatment-related acute myeloid leukemia (t-AML) in patients who received alkylating agents.13,14 t-AML is a known high mortality long-term complication of cytotoxic chemotherapy, usually involving the selection and expansion of preexisting clones unrelated to the primary malignancy (ie, clonal hematopoiesis). These t-AMLs have characteristic cytogenetic abnormalities that are associated with their initiating cytotoxic agent (commonly alkylating agents or topo II inhibitors).15 Examples of common events are 11q23 (mixed lineage leukemia) translocations, del-7q, and TP53 mutations.15 In the era of lenalidomide, the development of B-cell acute lymphoblastic leukemia (B-ALL) has been observed in patients with MM on lenalidomide maintenance therapy.16,17 These lymphoid malignancies typically develop after years of lenalidomide maintenance. Analysis of these B-ALL samples often reveals normal cytogenetics.18 It is not clear how lenalidomide contributes to B-ALL pathogenesis.
In this case series, we evaluate 17 patients with MM treated with HCT and long-term lenalidomide maintenance, and who subsequently developed B-ALL. Whole exome sequencing (WES) and fluorescence in situ hybridization (FISH)/cytogenetic analysis identified that the MM and B-ALL cells in each individual were unique clonal malignancies, confirmed through immunoglobulin heavy chain (IgH) clonality assessment. In addition, we observed a high incidence of TP53 variants in B-ALL samples that were not initially detected in the MM samples. Specifically, 14 out of the 17 B-ALL tumors had either TP53 single-nucleotide variants (SNVs) detected by sequencing or deletions detected by cytogenetics/FISH. We also observed that in some MM samples, a small population of bone marrow cells contained the B-ALL TP53 variant at a low variant allele frequency (VAF), years before malignant transformation. This finding suggests that MM treatment with lenalidomide can select for hematopoietic cells that contain TP53 mutations, as previously shown for patients developing t-AML.19 Screening patients for the presence of rare cells with TP53 mutations could potentially inform the risk/benefit decisions of long-term lenalidomide use in patients with MM.
Methods
Sample collection
Bone marrow biopsy specimens and normal skin samples were obtained from subjects who provided written informed consent that indicated specific language, authorizing DNA sequencing and data sharing. This consent procedure was approved by the Washington University School of Medicine Institutional Review Board through the Human Research Protection Office. Samples were obtained from a single site (Siteman Cancer Center, Barnes Jewish Hospital, St Louis MO) where between 300 and 400 new patients with MM are seen per year. MM samples and matched normal samples were collected at time of MM diagnosis; B-ALL samples were collected at time of B-ALL diagnosis.
Cytogenetic and FISH analysis
Cytogenetic analysis was performed by the Cytogenetics and Molecular Pathology Laboratory or the Clinical Genomics Laboratory at the Washington University School of Medicine, Department of Pathology and Immunology. Analysis was performed in a laboratory regulated by CLIA. Data was extracted from cytogenetic reports including FISH and chromosome analysis.
IgH testing for clonality
DNA was extracted from formalin-fixed paraffin-embedded tissue and analyzed utilizing the LymphoTrack IGH FR2/3 Assay - S5/PGM reagents from Invivoscribe Technologies (San Diego, CA). Next-generation sequencing of libraries was performed on a ThermoFisher Scientific Ion Torrent S5 sequencer. The LymphoTrack Software - S5/PGM version 2.4.5 was used to analyze the IGH rearrangement sequences and the relative proportion of sequences corresponding to each IGH clonotype was determined as a percentage of total sequencing reads.
Whole exome sequencing analysis
Three samples were derived from each patient: primary MM tumor (decalcified and formalin fixed), B-ALL tumor (fresh frozen), and matched normal skin or buccal swab samples (fresh frozen). Each sample was subjected to exome library preparation using Integrated DNA Technologies Human Exome Library with a spike-in for a custom set of capture probes (manufactured by Integrated DNA Technologies) that were designed to tile across 264 genes found to be recurrently mutated in AML, including TP53.20 Exome sequencing was performed using the Illumina HiSeq platform with paired 2 × 100 bp reads. Sequencing success required that the mean coverage was greater than 30× and at least 70% of all bases within the target region had at least 20× coverage.
To ensure that sample trios were derived from the same individual and that each individual was unique, a single nucleotide polymorphism (SNP) profiling panel was employed.21 SNP panel coordinates were converted to the most recent reference genome (GRCh38) using LiftOver22 and the variant allele fraction (VAF) for each SNP was assessed using bam-readcount.23
Exome and custom capture reads were aligned to the human reference genome GRCh38 (NCBI build 38) using BWA-MEM. Somatic and germ line variant calling used common workflow language pipelines provided by the McDonnell Genome Institute (https://github.com/genome/analysis-workflows; last accessed on 2 May 2021). Somatic variants were called with Pindel,24 VarScan2,25 Mutect,26 and Strelka2,27 and germ line variants were called with GATK HaplotypeCaller.28
Filtering was employed on automated somatic variant calls obtained for all samples using heuristic cutoffs. Specifically, variant loci required at least 30 read counts in both the tumor and normal sample, a variant allele depth of at least 7 reads, and VAF of at least 5%. Variants also had to be observed by at least 2 different automated somatic variant callers. These filtering strategies mostly removed artifacts within MM samples that were associated with decalcified and formalin-fixed preparation. All variants were further filtered using a previously described standard operating procedure for manual review of aligned sequencing reads for somatic variants.29
Targeted sequencing
A 40-gene targeted sequencing panel was used to create a report for both genetic variants and VAF. The targeted panel sequencing was performed as previously described.30 Briefly, amplicon capture–based enrichment with unique molecular identifiers were employed. Variants were identified using a sequencing and annotation pipeline and reports were generated for physician review. Reports were reviewed to assess the presence of variants in regions of interest.
TP53 immunohistochemistry (IHC)
Diagnostic bone marrow cores, 4-μm-thick paraffin-embedded tissue sections, were used for IHC, which was performed on Ventana BenchMark Ultra Slide Staining System using p53 antibody clone D07 (Ventana Medical Systems, Tucson, AZ). Quantitative scoring of p53 IHC slides was performed by a board-certified hematopathologist by manually counting any nuclear staining in 500 neoplastic cells.
Results
Data assembly, sequencing, and sample tracking
Samples from 17 patients were identified for the study. Tissue from the B-ALL and the MM samples were obtained for all patients. Matched normal tissue (5 skin and 3 buccal swab) for WES was obtained from 8 patients. Cytogenetics, FISH, IHC analysis, WES, IgH testing, and custom sequencing analysis was performed for patients based on availability of tissue (Table 1).
Clinical information
The average age at MM diagnosis across the whole cohort was 60 years (range, 45-74 years). For the 16 patients with information related to lenalidomide treatment duration, all had been on lenalidomide for at least 1 year, with an average treatment duration of 3.8 years (range, 1-13 years). There were 10 males and 7 females in the study. The majority of acute leukemias were B-ALL subtypes, with 1 case showing biphenotypic B/myeloid differentiation. All cases were Philadelphia chromosome negative (Ph−) (Table 1).
Clonality assessment between MM samples and matched B-ALL samples
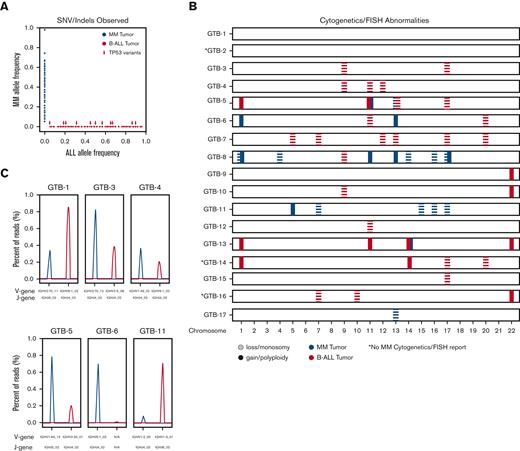
In total, 6 samples underwent IgH testing (GTB-1, GTB-3, GTB-4, GTB-5, GTB-6, and GTB-11). IgH clones were identified in all but 1 sample (GTB-6). When comparing the IgH clones from the original MM sample with the subsequent B-ALL sample, we noted that all 5 B-ALL samples revealed distinct dominant clones compared with those observed in the associated MM sample. In 2 of the 5 samples, the original MM clone was also observed at a lower frequency, along with a unique dominant B-ALL clone (Figure 1C).
Comparison of variants observed in MM and B-ALL for all samples based on data availability (seeTable 1). (A) Scatter plot indicating the VAFs for B-ALL (x-axis) and MM (y-axis) tumors (supplemental Table 1 for eligible samples and supplemental Table 2 for eligible variants). Arrows indicate variants in the TP53 gene. (B) Chromosome/FISH abnormalities between B-ALL (red) and MM (blue) tumors. Each row represents a single sample. Solid lines indicate gain or polyploidy in the associated chromosome (x-axis). Dashed lines indicate loss or monosomy in the associated chromosome (x-axis). (C) Top clone IgH analysis is plotted for B-ALL (red) and MM (blue) tumors. V-gene and J-gene alleles are provided on the x-axis. Peak height indicates clonality percentage based on sequencing reads.
Comparison of variants observed in MM and B-ALL for all samples based on data availability (seeTable 1). (A) Scatter plot indicating the VAFs for B-ALL (x-axis) and MM (y-axis) tumors (supplemental Table 1 for eligible samples and supplemental Table 2 for eligible variants). Arrows indicate variants in the TP53 gene. (B) Chromosome/FISH abnormalities between B-ALL (red) and MM (blue) tumors. Each row represents a single sample. Solid lines indicate gain or polyploidy in the associated chromosome (x-axis). Dashed lines indicate loss or monosomy in the associated chromosome (x-axis). (C) Top clone IgH analysis is plotted for B-ALL (red) and MM (blue) tumors. V-gene and J-gene alleles are provided on the x-axis. Peak height indicates clonality percentage based on sequencing reads.
In addition, 8 samples had whole exome sequencing (WES) for the MM sample, the B-ALL sample, and normal tissue (supplemental Table 1). After variant calling and filtering (see “Methods”), there were 973 somatic variants identified for all MM samples, with an average of 139 variants per sample (range, 6-295 variants). For B-ALL samples, there were 255 somatic variants identified, with an average of 32 variants per sample (range, 13-61 variants) (supplemental Table 2). To determine the relationship between the MM tumor and the B-ALL tumor for each patient, the union of variants was evaluated for sequence read support in matched MM/B-ALL samples (Figure 1). There were no somatic variants (including variants identified via cytogenetics/FISH) that were shared between the paired B-ALL and MM samples.
TP53-related aberrations on cytogenetics/FISH
Across the 17 B-ALL samples with karyotyping and/or FISH, 5 showed cytogenetic aberrations associated with TP53 mutations (Table 2). Specifically, GTB-3, GTB-5, and GTB-11 had monosomy of chromosome 17 on FISH. GTB-7 and GTB-14 had 17p deletions revealed by cytogenetics. These results indicate high incidence of TP53 structural variants in the B-ALL cohort.
Variants observed on whole exome/custom capture sequencing
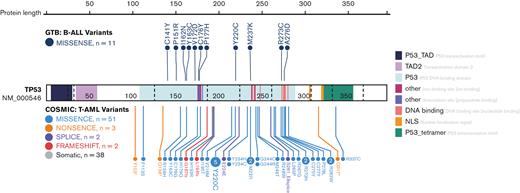
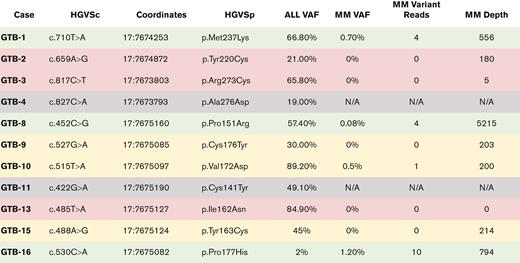
For the B-ALL samples, 8 samples had successful WES, and 9 had custom capture sequencing (Table 1). In the B-ALL samples, only TP53 was observed to be recurrently mutated (Table 2). For all B-ALL samples with sequencing data, 11 samples had a TP53 SNV (Table 2). In addition to these 11 samples, 1 sample (GTB-16) had a variant with low VAF and a predicted relevance of “uncertain significance.” All observed TP53 variants were within the DNA binding domain as per the Catalogue of Somatic Mutations in Cancer database31 under t-AML (Figure 2). When evaluating the cytogenetics or FISH analysis, 5 patients had 17p deletion or monosomy of chromosome 17. Therefore, 14 of the 17 patients (82%) harbored at least 1 somatic TP53 mutation in the B-ALL sample (Table 2). All variants observed, including 2 variants identified in the lenalidomide method of action pathway (IKAROS family regions) and 2 variants associated with B-ALL (eg, EZH2 and BCORL1), are shown in supplemental Table 3.
Assessment of TP53 variants observed on custom sequencing or WES for secondary B-ALL samples. Variant lolliplot shows the 11 TP53 missense variants identified in the 17 B-ALL samples with whole exome or capture sequencing. Catalogue of Somatic Mutations in Cancer TP53 variants associated with therapy related to acute myeloid leukemia are also shown below the gene.
Assessment of TP53 variants observed on custom sequencing or WES for secondary B-ALL samples. Variant lolliplot shows the 11 TP53 missense variants identified in the 17 B-ALL samples with whole exome or capture sequencing. Catalogue of Somatic Mutations in Cancer TP53 variants associated with therapy related to acute myeloid leukemia are also shown below the gene.
TP53 IHC staining is correlated with presence of pathogenic variants
TP53 missense mutations often produce dominant-negative proteins,32 stabilizing TP53 in cells and allowing detection of TP53 protein by IHC. Similar to myeloid neoplasms, mutant samples displayed significantly increased nuclear TP53 staining in B-ALL cases with missense TP53 mutations (mean 58.6%), compared with TP53 wild-type cases (mean 1.7%) (Table 2). Using a cut-off previously established in our laboratory, of the 11 samples with an observed SNV, 10 showed elevated TP53 IHC staining consistent with a missense TP53 mutation.33 One sample (GTB-16) with a missense mutation had a very low VAF (2%), and the protein was not detected by IHC. For the 3 samples with deletion of TP53 detected by cytogenetic or FISH analysis, TP53 abundance by IHC staining was not increased. For the 3 patients with no TP53 mutations, IHC staining was not increased. These results indicate that the presence of p53 IHC staining was predictive of the presence of TP53 variants in the B-ALL cohort.
MM bone marrow sequencing suggests that B-ALL arose from distinct clones with TP53 mutations, unrelated to the MM founding clone
MM samples underwent high-depth custom capture sequencing to determine whether the TP53 variants observed in B-ALL samples were present in rare cells in the bone marrow before lenalidomide treatment (Figure 3). Given low quality of DNA obtained from the aged formalin-fixed paraffin-embedded tissue samples, only 3 samples had sequencing depths >500× at loci of interest (GTB-1, GTB-8, and GTB-16) and an additional 3 samples had sequencing depths >200× at loci of interest (GTB-9, GTB-10, GTB-15). All 3 MM samples with sufficient sequencing depth (>500×) showed evidence of the B-ALL-specific TP53 mutation in the MM bone marrow. The sample with the highest B-ALL VAF (GTB-10) also showed evidence of B-ALL-specific variant-supporting reads in the MM bone marrow. Samples with low coverage (GTB-2, GTB-3, GTB-13) or no sequencing data (GTB-4, GTB-11) could not be accurately assessed.
Detectionof B-ALL-associated TP53 variants in MM samples usinghigh-depthsequencing. Samples in green had sufficient MM sequencing depth (>500×), samples in yellow had low MM sequencing depth (200×-500×), and samples in red had insufficient sequencing depth (<200×). Samples in gray did not have sufficient DNA material for sequencing.
Detectionof B-ALL-associated TP53 variants in MM samples usinghigh-depthsequencing. Samples in green had sufficient MM sequencing depth (>500×), samples in yellow had low MM sequencing depth (200×-500×), and samples in red had insufficient sequencing depth (<200×). Samples in gray did not have sufficient DNA material for sequencing.
Discussion
The use of lenalidomide is increasingly common for patients with MM, especially as a maintenance agent after autologous stem cell transplant.7 Rarely, patients develop B-ALL during lenalidomide maintenance treatment for MM, a devastating, high-mortality event for an elderly population.17,34 Two separate groups, the Intergroupe Francophone du Myelome and the Cancer and Leukemia Group (CALGB), conducted studies that demonstrated an increase in second primary malignancies (SPMs) after lenalidomide treatment.3,35,36 However, they looked at all SPMs, including cases of any hematologic malignancy (as well as solid tumors) and no clear mechanism of SPM development was established. In these studies, AML and myelodysplastic syndrome were the most common secondary hematologic malignancies, with B-ALL being reported in less than 1% of patients treated with lenalidomide.3,35,36 The data presented here support the hypothesis that B-ALL that develop after lenalidomide treatment is a SPM, and that B-ALL development is potentially associated with the expansion of preexisting cells within the bone marrow that contain TP53 mutations, a phenomenon that was described for secondary AML several years ago.19 Recently, a separate group published a case report of 2 patients with a history of MM and secondary B-ALL after exposure to lenalidomide; both patients had TP53 mutations in their B-ALL samples.37
The role of lenalidomide in the development of a secondary myeloid malignancy after therapy for MM cannot be ascertained for many patients because patients with MM often receive high-dose chemotherapy during their initial treatment courses. Many patients are treated with steroids, high-dose melphalan, and autologous stem cell transplant. Of the 17 patients in this study, 14 had a single stem cell transplant and 1 patient had 2 stem cell transplants. In addition, 12 patients were treated with dexamethasone, and 8 were treated with bortezomib. As such, it is possible that lenalidomide only partially contributed to the development of B-ALL in this cohort.
We observed that 14 of the 17 patients in this study had at least 1 TP53 mutation in the B-ALL sample that was not initially observed in the MM sample. The incidence of TP53 variants in de novo adult B-ALL varies between 2% and 15%.38-40 The high frequency of TP53 mutations in the B-ALLs arising in patients with MM treated with lenalidomide is reminiscent of a well-described mechanism of t-AML.19 This study showed that some of the TP53 mutations detected in patients with secondary AML were detectable in hematopoietic cells that were not associated with the original MM tumor, many years before the development of leukemia, and that these small clones were potentially selected for by chemotherapy. The emergence of de novo TP53 mutations in patients with 5q− myelodysplastic syndrome being treated with lenalidomide has also been reported.41,42 Although we did not observe significant trends associated with the 3 samples that did not have a TP53 variant (GTB-6, GTB-12, and GTB-17) we noted that all individuals were female and relatively young (age < 55 years) at MM diagnosis.
Despite the median age of the patients in this study (60 years), none of the B-ALL cases had a detectable BCR-ABL fusion.43 In a study that defined the incidence of Ph+ B-ALL, patients aged between 65 and 74 years had an average Ph+ B-ALL incidence of 43.3%. We did not identify other recurrent cytogenetic abnormalities or distinct immunophenotypic profiles in these secondary B-ALL cases. This could imply that lenalidomide-induced ALL in patients with MM is a molecularly distinct Ph− ALL subtype.
Limitations of this study included highly degraded DNA from many MM samples, which precluded quality exome sequencing; many of the MM samples had low coverage (<200×) at the TP53 locus. At this depth, we can expect an 86% chance of detecting variants with a VAF of >2.5%, but TP53 variants in rare cells in the MM sample could easily be missed with this level of sensitivity. Furthermore, it is possible that some variants were missed or mislabeled using this analysis owing to potential tumor contamination of normal tissue.
Future studies with larger cohorts of lenalidomide-treated MM will be required to fully understand the incidence of this complication of therapy. Regardless, these data, and similar observations by other groups, strongly suggest that chronic lenalidomide therapy appears to be capable of expanding rare hematopoietic cells with acquired TP53 mutations. Because clonal hematopoiesis is not rare in elderly patients and can clearly be caused by TP53 mutations, this complication will certainly occur in many similarly treated patients with MM in the years to come. Increased awareness of this syndrome may allow for improved approaches for monitoring such patients, and for earlier intervention if a secondary B-ALL does occur.
Acknowledgments
The authors would like to thank the patients and their families for their participation in this study. The authors would also like to thank Timothy Ley for his helpful discussions regarding these data and his review of the manuscript.
K.F.N. was supported by the Washington University School of Medicine Surgical Oncology Basic Science and Translational Research Training Program grant T32CA009621, from the National Cancer Institute (NCI). M.G. was supported by the National Human Genome Research Institute under award number R00HG007940. O.L.G. was supported by the National Cancer Institute under award number K22CA188163. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authorship
Contribution: E.K.B., K.F.N., M.C., K.M.C., M.B.R., and L.D.W. drafted the manuscript; E.K.B., Z.L.S.M., K.F.N., M.C., K.M.C., K.C.C., N.C.S., M.B.R., E.J.D., M.G., O.L.G., and L.D.W. performed analysis and interpretation of data; E.K.B., Z.L.S.M., K.F.N., M.C., K.M.C., K.C.C., N.C.S., M.B.R., T.W., B.A., F.K., B.A.P., J.F., Y.-S.L., A.H., J.A.K., D.R.K., K.V., M.A.F., G.L.U., R.V., M.G., O.L.G., and L.D.W. provided critical revision of the manuscript for important intellectual content; E.K.B., Z.L.S.M., K.F.N., M.C., N.C.S., M.B.R., E.J.D., and J.S.W. contributed to data acquisition; M.B.R., T.W., B.A., F.K., B.A.P., E.J.D., J.L.F., Y.-S.L., A.H., J.A.K., D.R.K., M.A.F., J.S.W., G.L.U., K.V., and R.V. provided clinical correlations for data; O.L.G. and L.D.W., provided study supervision; M.G., O.L.G., and L.D.W., advised on study development.
Conflicts-of-interest disclosure: These authors disclose the following: E.K.B. is an owner, employee, and member of Geneoscopy Inc, and is an inventor of the intellectual property owned by Geneoscopy Inc. The remaining authors disclose no competing financial interests.
Correspondence: Obi L. Griffith, Washington University School of Medicine; e-mail: obigriffith@wustl.edu; and Lukas D. Wartman, Washington University School of Medicine; e-mail: lwartman@wustl.edu.
References
Author notes
Sequencing data can be found in the dbGaP (accession number phs003069.v1.p1).
The full-text version of this article contains a data supplement.